Abstract
Plasminogen activator inhibitor-1 (PAI-1) is the primary physiological inhibitor of both tissue-type and urokinase-type plasminogen activators. The balance between plasminogen activators and PAI-1 plays an important role in several physiological and pathophysiological processes such as atherosclerosis or thrombosis. Because these conditions are associated with hypoxia, it was the aim of the present study to investigate the influence of low O2tension on the expression of PAI-1 mRNA and protein using primary cultured rat hepatocytes as a model system. We found that PAI-1 mRNA and protein were induced by mild hypoxia (8% O2). The hypoxia-dependent PAI-1 mRNA induction was transcriptionally regulated because it was inhibited by actinomycin D (ActD). Luciferase (LUC) reporter gene constructs driven by about 800 bp of the 5′-flanking region of the rat PAI-1 gene were transiently transfected into primary rat hepatocytes; mild hypoxia caused a 3-fold induction, which was mediated by the PAI-1 promoter region -175/-158 containing 2 putative hypoxia response elements (HRE) binding the hypoxia-inducible factor (HIF-1). Mutation of the HRE-1 (-175/-168) or HRE-2 (-165/-158) also abolished the induction by mild hypoxia. Cotransfection of a HIF-1 vector and the PAI-1–LUC constructs, as well as gel shift assays, showed that the HRE-2 of the PAI-1 promoter was most critical for induction by hypoxia and HIF-1 binding. Thus, PAI-1 induction by mild hypoxia via a HIF-1 binding HRE in the rat PAI-1 promoter appears to be the mechanism causing the increase in PAI-1 in many clinical conditions associated with O2deficiency.
PLASMINOGEN ACTIVATOR inhibitors (PAIs) play a crucial role in physiological and pathophysiological processes, which involve the tissue-type plasminogen-activator (tPA) and the urokinase (uPA) catalyzed conversion of plasminogen to plasmin.1,2 Two types of PAIs named PAI-13 and PAI-24 could be identified on the basis of biochemical, immunologic, and molecular biological properties. Whereas the physiological function of PAI-2 has not been clearly elucidated,5 PAI-1 is the major plasminogen activator inhibitor.6 PAI-1 can be produced and secreted by platelets, vascular endothelial cells,7 vascular smooth muscle cells,8 and several nonvascular cell types,9,10 among them hepatocytes.11Furthermore, PAI-1 is found also in the extracellular matrix.12 Thus, PAI-1 appears to be a key inhibitor of fibrinolysis3,13 and of proteolytic processes, which are associated with neovascularization, tissue remodelling, and regeneration, such as inflammation14 and fibrosis.15
The importance of the balance between plasminogen activators and PAI-1 has been emphasized by several clinical studies: decreased PAI-1 levels are associated with bleeding diathesis,16-18 whereas increased PAI-1 levels have been found in a number of clinical conditions, such as sepsis,19 atherosclerotic lesions leading to myocardial infarction,20 and deep vein thrombosis.21 In a rabbit model of thrombosis, increased levels of PAI-1 promoted thrombus extension,22 and inhibition of PAI-1 activity by a monoclonal antibody against PAI-1 partially prevented thrombus formation.23 Moreover, in rats, hemorrhage increased the expression of PAI-1 in various tissues, such as lung, kidney, and liver.24 Thus, PAI-1 may promote thrombus extension and the development of thromboembolic diseases.
Because prethrombotic events, hemorrhage, and thrombus formation are associated with hypoxia, it was the goal of this study to investigate the influence of reduced O2 tensions on the expression of PAI-1. Primary cultured rat hepatocytes were used as a model system. We found that PAI-1 mRNA and protein were induced by mild hypoxia (8% O2). The enhancement of PAI-1 gene expression by mild hypoxia was mainly regulated on the level of transcription via an O2 responsive sequence (-175/-158) containing the potential hypoxia responsive elements -1 and -2 (HRE-1, -175/-168 and HRE-2, -165/-158) binding the hypoxia-inducible factor-1 (HIF-1). Transfection of PAI-1 promoter luciferase (LUC) gene constructs and gel shift assays showed that the HRE-2 had a key role in PAI-1 gene activation by hypoxia and HIF-1 binding. Thus, HIF-1 appears to be most critical for the increase in PAI-1 under hypoxia.
MATERIALS AND METHODS
All biochemicals and enzymes were of analytical grade and were purchased from commercial suppliers.
Animals.
Male Wistar rats (200 to 260 g) were kept on a 12-hour day/night rhythm (light from 7 am to 7 pm) with free access to water and food. Rats were anesthetized with pentobarbital (60 mg/kg body weight) before preparation of hepatocytes between 8 am and 9am.
Cell culture experiments.
Liver cells were isolated by collagenase perfusion. Cells (1 × 106 per dish) were maintained under standard conditions in a normoxic atmosphere of 16% O2, 79% N2, and 5% CO2 (by vol) in medium M199 containing 0.5 nmol/L insulin added as a growth factor for culture maintenance, 100 nmol/L dexamethasone required as a permissive hormone, and 4% fetal calf serum for the initial 4 hours of culture. Cells were then cultured in serum-free medium from 4 to 24 hours at normal arterial 16% O2. At 24 hours, medium was changed and culture was continued at normoxia 16% O2 or at mild hypoxia 8% O2 (87% N2, 5% CO2 [by vol]). Sixteen percent O2 and 8% O2 at the medium surface correspond to 13% O2 = arterial pO2and 6% O2 = venous pO2 at the cell surface.25 In actinomycin and cycloheximide experiments, hepatocytes were pretreated for 30 minutes with ActD (1 μg/mL) or cycloheximide (10 μg/mL) before the cells were cultured under normoxia or mild hypoxia for 3 hours.
RNA preparation and Northern analysis.
Isolation of total RNA and Northern analysis were performed as described.26 Digoxigenin (DIG)-labelled antisense RNAs served as hybridization probes; they were generated by in vitro transcription from pBS-PAI-1 using T3 RNA polymerase and RNA labeling mixture containing 3.5 mmol/L 11-DIG-uridine triphosphate (UTP), 6.5 mmol/L UTP, 10 mmol/L guanosine triphosphate (GTP), 10 mmol/L cytidine triphosphate (CTP), 10 mmol/L adenosine triphosphate (ATP). Hybridizations were performed with 50 ng/mL transcript at 68°C for 6 hours according to the manufacturer's application notes of the DIG-nucleic acid detection kit (Roche, Mannheim, Mannheim, Germany). Detection of hybrids was performed as described previously.26 Blots were quantified with a videodensitometer (Biotech Fischer, Reiskirchen, Germany).
Plasmid constructs.
The rat PAI-1 promoter 5′-flanking region27 from -766 to +31 was amplified by polymerase chain reaction (PCR) from rat genomic DNA by using the oligonuclotide 5′-gacagagctctctgtggtaacccctgtgctc-3′ (-766/-736) as forward and 5′-tcggagatctgcagcagcctgatccagctgtg-3′ (-1/+31) as reverse primer, respectively. The resulting PCR product was digested with SacI and BglII and ligated into the SacI and BglII sites of pGl3basic (Promega, Mannheim, Germany) (pGl3PAI-766, see Fig 3).
In construct pGl3PAI-766ΔM, the -175/-159 fragment was omitted by deletion mutagenesis with the ExSite Mutagenesis kit (Stratagene, La Jolla, CA) by using the pGl3PAI-766 as template and the oligonucleotides 5′-tcccagcaagttactgggaggga-3′ (-158/-136) as forward primer and 5′-gtgagagggcatgggggataattg-3′ (-176/-199) as reverse primer, respectively. The constructs pGl3PAI-766M1 (see Fig 3) and pGl3PAI-766M2 (see Fig 3) were constructed by the Altered Sites in vitro Mutagenesis kit (Promega) using the mutagenized oligonucleotides M1 and M2 shown in Table 1.
Oligonucleotides Used for Mutagenesis
| −191/−166 | 5′-cccatgccctctcacaCACGTACAca-3′ WT; HRE-1, −175/−168 underlined | WT; HRE-1, −175/−168 underlined |
| 5′-cccatgccctctcacacaattaatga-3′ | M1, alterations boldface | |
| −165/−142 | 5′-ccagcaagttactggg-3′ | WT; HRE-2, −165/−158 underlined |
| 5′-aagcattcccagcaagttactggg-3′ | M2, alterations boldface |
| −191/−166 | 5′-cccatgccctctcacaCACGTACAca-3′ WT; HRE-1, −175/−168 underlined | WT; HRE-1, −175/−168 underlined |
| 5′-cccatgccctctcacacaattaatga-3′ | M1, alterations boldface | |
| −165/−142 | 5′-ccagcaagttactggg-3′ | WT; HRE-2, −165/−158 underlined |
| 5′-aagcattcccagcaagttactggg-3′ | M2, alterations boldface |
The plasmid pcDNAHIF-1 containing the full-length rat HIF-1α cDNA under the control of the cytomegalovirus (CMV) promoter was constructed by excision of the HIF-1α cDNA (Gene bank Y09507) from pCRII withBamHI and EcoRI and subsequent ligation into theBamHI, EcoRI sites of pcDNAI-Amp (Invitrogen, Groningen, The Netherlands). All constructs were verified by sequencing in both directions.
Cell transfection and LUC assay.
Freshly isolated rat hepatocytes (about 1 × 106 cells per dish) were transfected for 5 hours with 2.5 μg plasmid DNA containing 500 ng of a Renilla luciferase (RL) reporter plasmid (pRL-SV40, Promega) to control transfection efficiency and 2 μg of the appropriate PAI-1 promoter Firefly luciferase (FL) construct essentially as described.28 In HIF-1α cotransfection assays, 2 μg of the appropriate PAI-1 promoter FL construct were transfected together with 200 ng pcDNAHIF or in the controls with pcDNA, thereby reducing the amount of pRL-SV40 to 300 ng. After 5 hours, the medium was changed and the cells were cultured under normoxia for 19 hours. The medium was changed again and the cells were further cultured for 24 hours under normoxia or mild hypoxia.
Western blot analysis.
Western blot analysis was performed as described.29 In brief, media from primary cultured hepatocytes were collected and the protein content was determined using the Bradford method. A total of 50 μg of protein was loaded onto a 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel and after electrophoresis blotted onto nitrocellulose membranes. The primary rabbit antibody against rat PAI-1 (American Diagnostics, Pfungstadt, Germany) was used in a 1:200 dilution. The primary rabbit aryl hydrocarbon nuclear translocator (ARNT) antibody (Affinity Bioreagents, Grünwald, Germany) was used in a 1:500 dilution. The secondary antibody was a goat antirabbit IgG (Santa Cruz Biotech, Santa Cruz, CA) and used in a 1:2000 dilution. The ECL Western blotting system (Amersham, Arlington Heights, IL) was then used for detection. Under these conditions, PAI-1 was seen as a double band, the major 49 kD band and the minor 46 kD band.29
Preparation of nuclear extracts.
Nuclear extracts were prepared by modification of a standard protocol30 31 with buffers A and C containing 0.5 mmol/L dithioerythritol (DTE; Sigma, St Louis, MO), 0.4 mmol/L phenylmethylsulfonyl fluoride (Serva, Heidelberg, Germany), 2 μg of leupeptin per mL (Boehringer), 2 μg of pepstatin per mL (Boehringer), 2 μg of aprotinin per mL (Bayer, Leverkusen, Germany), 1 mmol/L sodium vanadate (Sigma), and the “complete” protease inhibitor cocktail tablets (Boehringer). Briefly, confluent HepG2 cells in 10-cm dishes were cultured at 8% O2 or 16% O2 for 24 hours, the cells were then washed twice with ice cold 0.9% NaCl, scraped into 300 μL NaCl, and pelleted by centrifugation at 2000 rpm for 2 minutes at 4°C in an Sorvall high-speed centrifuge RC 5B (SS 34 rotor). The cell pellet was resuspended in 4 packed cell volumes (PCV) of buffer A (10 mmol/L Hepes-KOH [pH 7.9], 1.5 mmol/L MgCl2, 10 mmol/L KCl), and incubated on ice for 10 minutes. The cell suspension was Dounce homogenized with a type-B pestle, and the nuclei were pelleted by centrifugation at 2300 rpm for 10 minutes, resuspended in 3 PCV of buffer A, and pelleted once again by centrifugation at 14,500 rpm for 20 minutes. The nuclei were resuspended in 3 PCV of buffer C (20 mmol/L HEPES-KOH [pH 7.9], 1.5 mmol/L MgCl2, 0.42 mmol/L NaCl, 0.2 mmol/L EDTA, 20% glycerol), and incubated at 4°C with gentle agitation for 30 minutes. Nuclear debris was pelleted by centrifugation for 30 minutes at 14,500 rpm. The supernatant was dialyzed against a 50-fold volume of buffer D (20 mmol/L Hepes-KOH [pH 7.9], 0.1 mmol/L KCl, 0.2 mmol/L EDTA, 20% glycerol) for 4 hours. The dialysate was centrifuged for 10 minutes at 14,500 rpm, and aliquots of the supernatant were frozen in liquid N2 and stored at −70°C. Protein concentration was determined by the Bradford method with bovine serum albumin standards.
Electrophoretic mobility shift assay (EMSA).
The sequences of the PAI-1 oligonucleotides used for the EMSA are shown (see Fig 4). Equal amounts of complementary oligonucleotides were annealed and labeled by 5′-end-labelling with [γ-32P]ATP (Amersham) and T4 polynucleotide kinase (MBI). They were purified with the Nucleotide Removal Kit (Quiagen, Hilden, Germany). Binding reactions were performed in a total volume of 20 μL containing 250 mmol/L KCl, 5 mmol/L MgCl2, 5.5 mmol/L EDTA, 25% glycerol, 10 μg of nuclear extract, 250 ng poly d(I-C), and 5 mmol/L DTE. After preincubation for 5 minutes at room temperature, 1 μL of the labeled probe (104 cpm) was added and the incubation was continued for an additional 10 minutes. For supershift analysis, 1 μL preimmune serum or a HIF-1α antibody (1 μL or 0.1 μL of a 1:5 dilution) was added to the EMSA reaction and then incubated at 4°C for 2 hours. The antibodies against rat-HIF–1α were raised in rabbits against a 223 amino acid peptide (rat HIF aa 166-388).
The electrophoresis was then performed with a 5% nondenaturing polyacrylamide gel in TBE buffer (89 mmol/L Tris, 89 mmol/L boric acid, 5 mmol/L EDTA) at 200 V. After electrophoresis, the gels were dried and exposed to a phosphorimager screen.
RESULTS
Induction of PAI-1 mRNA and protein by mild hypoxia.
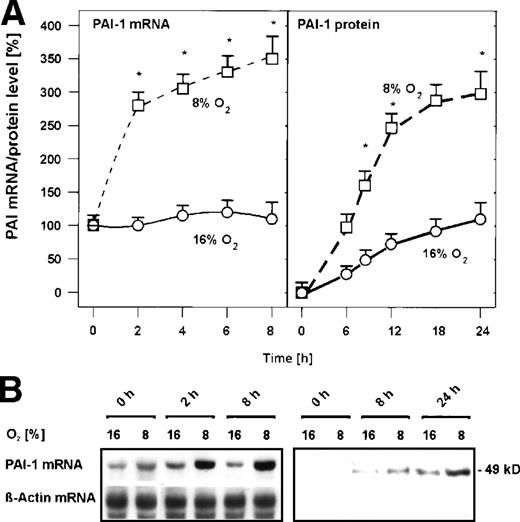
In primary cultured rat hepatocytes, mild hypoxia (8% O2) induced PAI-1 mRNA within 2 hours by about 2.5-fold (Fig 1) and within 8 hours by about 3.5-fold (Fig 1). Induced PAI-1 mRNA then remained almost stable at this level up to 24 hours (data not shown). The increase of PAI-1 mRNA by mild hypoxia was followed by an increase of PAI-1 protein. PAI-1 protein was first detectable in the medium of the cells after 8 hours; under mild hypoxia, the amount of PAI-1 protein was increased by about 2-fold compared with normoxia (Fig 1). After 24 hours exposure to mild hypoxia, the amount of PAI-1 protein was further increased by about 3-fold (Fig 1). Thus, PAI-1 mRNA and protein expression in primary rat hepatocytes were induced by mild hypoxia.
Hypoxia-dependent induction of PAI-1 mRNA and protein expression in primary rat hepatocyte cultures. Hepatocytes were cultured for 24 hours under arterial pO2. At 24 hours, the medium was changed and cells were further cultured for the times indicated under normoxic (16% O2) and hypoxic (8% O2) conditions. (A) In each experiment, the PAI-1 mRNA level measured by Northern blotting (cf B) at 0 hour under normoxia (16% O2) was set equal to 100%. The PAI-1 protein level measured by Western blotting (cf B) at 0 hour under normoxia (16% O2) was set equal to 1%. Values are means ± standard error of mean (SEM) of 3 independent culture experiments. Statistics, Student's t-test for paired values: * significant differences 16% O2 versus 8% O2,P ≤ .05. (B) Representative Northern and Western blot. For Northern analysis, 15 μg total RNA prepared from the cultured hepatocytes were hybridized to DIG-labelled PAI-1 and β-actin antisense RNA probes (cf, Materials and Methods). A total of 50 μg of protein from the medium of the cultured hepatocytes was subjected to Western analysis with an antibody against rat PAI-1 (cf, Materials and Methods). Autoradiographic signals were obtained by chemiluminescence and scanned by videodensitometry.
Hypoxia-dependent induction of PAI-1 mRNA and protein expression in primary rat hepatocyte cultures. Hepatocytes were cultured for 24 hours under arterial pO2. At 24 hours, the medium was changed and cells were further cultured for the times indicated under normoxic (16% O2) and hypoxic (8% O2) conditions. (A) In each experiment, the PAI-1 mRNA level measured by Northern blotting (cf B) at 0 hour under normoxia (16% O2) was set equal to 100%. The PAI-1 protein level measured by Western blotting (cf B) at 0 hour under normoxia (16% O2) was set equal to 1%. Values are means ± standard error of mean (SEM) of 3 independent culture experiments. Statistics, Student's t-test for paired values: * significant differences 16% O2 versus 8% O2,P ≤ .05. (B) Representative Northern and Western blot. For Northern analysis, 15 μg total RNA prepared from the cultured hepatocytes were hybridized to DIG-labelled PAI-1 and β-actin antisense RNA probes (cf, Materials and Methods). A total of 50 μg of protein from the medium of the cultured hepatocytes was subjected to Western analysis with an antibody against rat PAI-1 (cf, Materials and Methods). Autoradiographic signals were obtained by chemiluminescence and scanned by videodensitometry.
Transcriptional and translational regulation of the mild hypoxia-dependent PAI-1 mRNA induction.
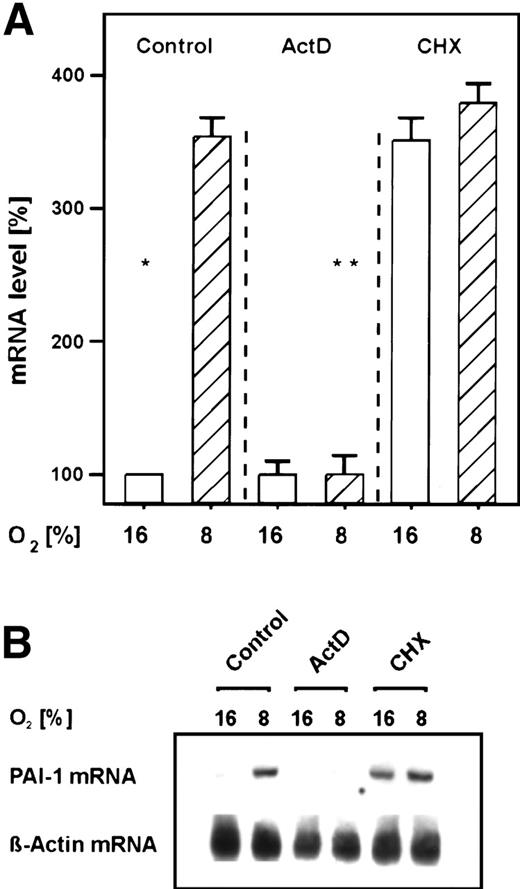
In the primary rat hepatocyte cultures, the transcriptional inhibitor ActD prevented the mild hypoxia-dependent PAI-1 mRNA induction within 3 hours (Fig 2). Thus, PAI-1 gene activation by mild hypoxia occurred mainly on the transcriptional level. The protein synthesis inhibitor cycloheximide caused an induction of PAI-1 mRNA under normoxia to levels normally induced by mild hypoxia, but it did not increase any further the induction elicited by mild hypoxia (Fig 2). The induction by cycloheximide under normoxia was probably due to a stabilization of PAI-1 mRNA11; the lack of an additivity of induction by cycloheximide and mild hypoxia may indicate that the mild hypoxia-dependent induction of PAI-1 mRNA was dependent on ongoing protein synthesis of, eg, HIF-1. Thus, the present finding would be in line with previous reports showing that cycloheximide (CHX) blocked the hypoxic induction of HIF-dependent genes.31
Inhibition of the hypoxia-dependent PAI-1 mRNA induction by ActD and cycloheximide in primary rat hepatocyte cultures. Hepatocytes were cultured as in Fig 1. (A) The cells were then treated for 30 minutes with ActD (1 μg/mL) or cycloheximide (CHX; 10 μg/mL) before they were placed under hypoxia for 3 hours. A total of 15 μg total RNA was subjected to Northern analysis. In each experiment, the PAI-1 mRNA level measured by Northern blotting (cf B) under normoxia (16% O2) was set equal to 100%. The induction of PAI-1 mRNA by cycloheximide under normoxia was due to a stabilization of PAI-1 mRNA.11 Values ± SEM represent the fold induction of PAI-1 mRNA of 3 independent experiments. Statistics, Student'st-test for paired values: significant differences * 16% O2 versus 8% O2, ** 8% O2 versus ActD, P ≤ .05. (B) Representative Northern blot.
Inhibition of the hypoxia-dependent PAI-1 mRNA induction by ActD and cycloheximide in primary rat hepatocyte cultures. Hepatocytes were cultured as in Fig 1. (A) The cells were then treated for 30 minutes with ActD (1 μg/mL) or cycloheximide (CHX; 10 μg/mL) before they were placed under hypoxia for 3 hours. A total of 15 μg total RNA was subjected to Northern analysis. In each experiment, the PAI-1 mRNA level measured by Northern blotting (cf B) under normoxia (16% O2) was set equal to 100%. The induction of PAI-1 mRNA by cycloheximide under normoxia was due to a stabilization of PAI-1 mRNA.11 Values ± SEM represent the fold induction of PAI-1 mRNA of 3 independent experiments. Statistics, Student'st-test for paired values: significant differences * 16% O2 versus 8% O2, ** 8% O2 versus ActD, P ≤ .05. (B) Representative Northern blot.
Identification of sequences responsible for the mild hypoxia-dependent transcription from the PAI-1 promoter.
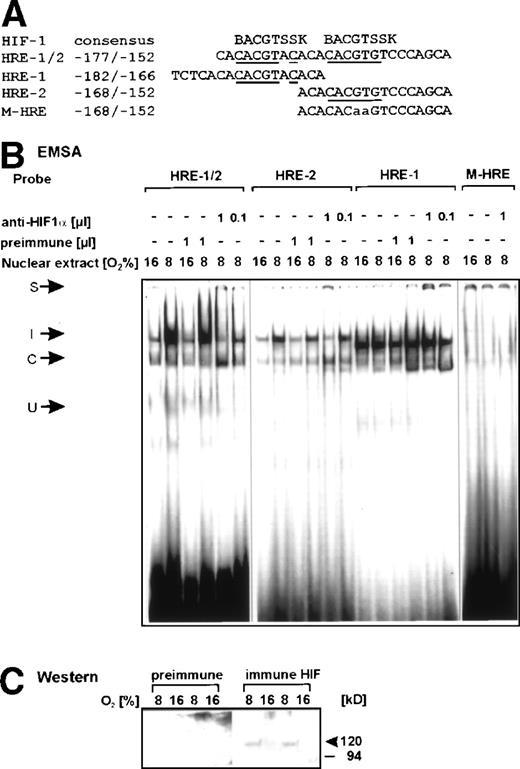
Sequence analysis of the rat PAI-1 promoter showed that 2 potential hypoxia response elements (HRE), which might bind HIF-1, are present within the first 200 bp of the promoter. Both are located inside the “C”-site identified by footprint analysis with liver nuclear extracts.27 The first potential HIF-1 binding element (HRE-1) inside the “C-site” -175/-168 5′-CACGTACA-3′ matches the HIF-1 binding consensus sequence BACGTSSK32(with B=G/C/T, S=G/C, and K=G/T) in 6 of 8 bp. The second potential HIF-1 binding element (HRE-2) -165/-158 5′-CACGTGTC-3′ also matches the HIF-1 consensus site in 6 of 8 bp (Fig 3A).
Regulation of transfected rat PAI-1 promoter LUC gene constructs by hypoxia and cotransfected HIF-1 in transiently transfected rat hepatocyte cultures. Enhancement of PAI-1 production by HIF-1. (A) The 5′-flanking region of the rat PAI-1 gene with its footprinted sites [A-G]. In the “C-site”, the 2 potential HIF-1 binding elements (HRE) matching the HIF-1 consensus sequence BACGTSSK32 were B=G/C/T, S=G/C and K=G/T are underlined. B, S, or K are shown only if the actual PAI-1 sequence does not match any of the bases allowed by the consensus. (B) Hepatocytes were transiently cotransfected with pcDNAHIF-1 containing the full-length rat HIF-1 cDNA and LUC gene constructs driven by a wild-type 766 bp rat PAI-1 promoter (pGl3PAI-766), or the 766 bp promoter mutated at either the HRE-1 (pGl3PAI-766M1) or HRE-2 (pGl3PAI-766M2) site. After 24 hours, the transfected cells were cultured for 18 hours under hypoxia, additionally pGl3PAI-766-transfected cells were treated with CHX (10 μg/mL). In each experiment, the fold stimulation of LUC activity by hypoxia (8% O2) was determined relative to the normoxic (16% O2) control, which was set equal to 1. The values represent means ± SEM of 3 independent experiments. Statistics, Student'st-test for paired values: * significant differences 16% O2 versus 8% O2, P ≤ .05; ** significant differences 16% O2 versus 16% O2+ HIF-1, P ≤ .05. In pGl3PAI-766M1 and pGl3PAI-766M2, the wild-type PAI-1 sequence is shown on the upper strand, mutated bases are indicated by *, and are shown in lower case letters. (C) Western blot analysis of PAI-1, HIF-1, and ARNT in cultured hepatocytes transfected with a HIF-1 vector. A total of 50 μg of protein from the medium and 100 μg of total cellular protein of the transfected hepatocytes were subjected to Western analysis (cf, Materials and Methods) with antibodies against rat PAI-1, rat HIF-1, and ARNT, respectively. Blots were scanned by videodensitometry, and in each experiment, the PAI-1 level and HIF-1 level measured under normoxia (16% O2) were set equal to 1. Values represent the fold induction of PAI-1 and HIF-1 of 3 independent experiments.
Regulation of transfected rat PAI-1 promoter LUC gene constructs by hypoxia and cotransfected HIF-1 in transiently transfected rat hepatocyte cultures. Enhancement of PAI-1 production by HIF-1. (A) The 5′-flanking region of the rat PAI-1 gene with its footprinted sites [A-G]. In the “C-site”, the 2 potential HIF-1 binding elements (HRE) matching the HIF-1 consensus sequence BACGTSSK32 were B=G/C/T, S=G/C and K=G/T are underlined. B, S, or K are shown only if the actual PAI-1 sequence does not match any of the bases allowed by the consensus. (B) Hepatocytes were transiently cotransfected with pcDNAHIF-1 containing the full-length rat HIF-1 cDNA and LUC gene constructs driven by a wild-type 766 bp rat PAI-1 promoter (pGl3PAI-766), or the 766 bp promoter mutated at either the HRE-1 (pGl3PAI-766M1) or HRE-2 (pGl3PAI-766M2) site. After 24 hours, the transfected cells were cultured for 18 hours under hypoxia, additionally pGl3PAI-766-transfected cells were treated with CHX (10 μg/mL). In each experiment, the fold stimulation of LUC activity by hypoxia (8% O2) was determined relative to the normoxic (16% O2) control, which was set equal to 1. The values represent means ± SEM of 3 independent experiments. Statistics, Student'st-test for paired values: * significant differences 16% O2 versus 8% O2, P ≤ .05; ** significant differences 16% O2 versus 16% O2+ HIF-1, P ≤ .05. In pGl3PAI-766M1 and pGl3PAI-766M2, the wild-type PAI-1 sequence is shown on the upper strand, mutated bases are indicated by *, and are shown in lower case letters. (C) Western blot analysis of PAI-1, HIF-1, and ARNT in cultured hepatocytes transfected with a HIF-1 vector. A total of 50 μg of protein from the medium and 100 μg of total cellular protein of the transfected hepatocytes were subjected to Western analysis (cf, Materials and Methods) with antibodies against rat PAI-1, rat HIF-1, and ARNT, respectively. Blots were scanned by videodensitometry, and in each experiment, the PAI-1 level and HIF-1 level measured under normoxia (16% O2) were set equal to 1. Values represent the fold induction of PAI-1 and HIF-1 of 3 independent experiments.
A 766-bp fragment of the 5′-flanking region of rat PAI-1 gene27 was cloned in front of the LUC gene in pGl3-basic (Promega) to generate pGl3PAI-766 (Fig 3). In hepatocytes transfected with pGl3PAI-766, LUC activity was induced by about 3-fold under mild hypoxia (Fig 3B). To verify that the mild hypoxia-dependent induction of pGl3PAI-766 was dependent on ongoing protein synthesis of, eg, HIF-1, pGl3PAI-766–transfected hepatocytes were treated with cycloheximide while cultured under mild hypoxia. It was found that cycloheximide abolished the hypoxia-dependent induction of LUC activity in transfected hepatocytes (Fig 3B). These results further support the involvement of HIF-1 in the hypoxia-dependent PAI-1 gene activation.
Deletion of the “C”-site containing the 2 PAI-1-HREs in pGl3PAI-766ΔM abolished the hypoxia-dependent induction of LUC activity in transfected hepatocytes indicating that these elements are necessary for the enhanced transcription of the PAI-1 gene under mild hypoxia (data not shown). Mutation of the nucleotides -173/-167 encompassing the HRE-1 site in the construct pGl3PAI-766M1 resulted again in a loss of the enhancement of LUC activity by mild hypoxia in transfected hepatocytes. When the nucleotides -165/-160 inside HRE-2 were mutated in the construct pGl3PAI-766M2, the mild hypoxia-elicited enhancement of LUC activity was also abolished (Fig 3B). These data indicate that both HREs in the PAI-1 promoter contributed to the mild hypoxia-dependent induction of PAI-1 gene transcription.
Activation of transfected rat PAI-1 promoter LUC gene constructs by HIF-1α.
To identify the direct regulation of the PAI-1 HREs by HIF-1, the wild-type PAI-1 promoter LUC constructs and the mutated PAI-1 promoter constructs were cotransfected into rat hepatocytes with an expression vector encoding rat HIF-1α. In hepatocytes transfected with pGl3PAI-766 and the HIF-1α vector, the LUC activity was increased by about 12-fold under 16% O2 (Fig 3B). Mild hypoxia and cotransfected HIF-1α were synergistic in pGl3PAI-766–transfected hepatocytes; LUC activity was induced to about 18-fold (Fig 3B).
Cotransfection of pGl3PAI-766M1 mutated at HRE-1 and the HIF-1α vector resulted in an enhanced LUC activity by about 5-fold under normoxia and hypoxia indicating that the remaining complete HRE-2 may be sufficient for a HIF-1–dependent submaximal induction (Fig 3B).
In hepatocytes cotransfected with pGl3PAI-766M2 mutated in the HRE-2 and the HIF-1α vector, LUC activity under both O2tensions was comparable to that obtained by transfection of the wild-type PAI-1 promoter construct under normoxia and could not be enhanced by mild hypoxia or HIF-1α (Fig 3B) despite the high presence of HIF-1α in the cotransfected cultures (Fig 3C). This suggests that HRE-2 seems to be the main or even only HIF-1 target sequence in the PAI-1 promoter.
Enhancement of PAI-1 protein production by transfection of rat HIF-1α.
Because the PAI-1 protein was also induced by mild hypoxia and this response was mediated by HIF-1, it should be tested whether the PAI-1 production by rat hepatocytes could be increased by transfection of HIF-1α. We found that HIF-1α–transfected hepatocytes secreted about 2-fold more PAI-1 protein into the medium under 16% O2 compared with the nontransfected control cells (Fig 3C). Again, mild hypoxia and transfected HIF-1α were synergistic; the amount of PAI-1 was increased by about 5-fold compared with the normoxic untransfected control and by about 2-fold compared with the normoxic transfected control (Fig 3C). Thus, transfection of HIF-1α can mimic the response to mild hypoxia and enhance the production of PAI-1 protein.
Binding of HIF-1 to sequences in the rat PAI-1 promoter.
The binding of nuclear proteins to oligonucleotide probes spanning the putative HRE-1 and HRE-2 sites of the PAI-1 promoter was examined by EMSA.
The oligonucleotide -177/-152, which enclosed both the HRE-1 and HRE-2 site (Fig 4A), was able to bind, besides a constitutive protein complex, a hypoxia-inducible nuclear protein complex (Fig 4B, left panel). Thus, it seemed likely that the hypoxia-induced DNA-protein complex contained HIF-1. To ensure this, several dilutions of a specific antiserum against HIF-1α was included in the binding reaction. The HIF-1α antibody was raised against a peptide from amino acid 166 to 388 of the rat HIF-1α protein. This region contains parts of the per-arnt-sim (PAS)-domain, which is involved in protein dimerization. Thus, the addition of the HIF-1α antibody should prevent the mild hypoxia-dependent DNA-protein complex formation. With the -177/-152 PAI-1 (HRE-1/2) oligonucleotide, this was indeed found (Fig 4B, left panel). Addition of 1 μL and 0.1 μL HIF-1α antibody to the EMSA reaction inhibited the formation of the mild hypoxia-dependent DNA complex and led to a supershifted complex, which could not be further resolved on the gel. Inhibition of the hypoxia-inducible complex was best seen with 1 μL of the HIF-1α antibody (Fig 4B, left panel). At this HIF-1α antibody concentration, binding of the so-called constitutive complex to the HRE1/2 oligonucleotide was en- hanced. With a further reduction of the HIF-1α antibody (1:10), the formation of the hypoxia-inducible complex returned (Fig 4B, left panel). Specificity of the HIF-1α antiserum was ensured by using the preimmune serum in the EMSA and in the Western blot analysis (Fig 4C). When the preimmune serum was used, the formation of the mild hypoxia-dependent DNA-protein complex was not impaired (Fig 4B), and the presence of the HIF-1α protein in the nuclear extracts could not be detected, whereas with the immune serum, it was found that only under mild hypoxia the HIF-1α protein was present in the nuclear extracts (Fig 4C).
Binding of HIF-1 to the rat PAI-1 promoter region. (A) Oligonucleotides: the HIF-1 consensus sequence BACGTSSK32were B=G/C/T, S=G/C, and K=G/T and the sense strand of the rat PAI-1 promoter sequence -177/-152 containing the HRE-1 and HRE-2 site, the sense strands of the oligonucleotides containing HRE-1, HRE-2, and the oligonucleotide with a mutation in the HRE-2 site M-HRE are shown. Bases matching the consensus sequences are underlined, mutated bases are in lower case letters. (B) EMSA, left panel: the32P-labeled PAI-1 HRE-1/2 oligonucleotide was incubated with either 10 μg protein of nuclear extracts from normoxic (16% O2) or hypoxic (8% O2) cells in the absence or presence of preimmune serum or rat HIF-1 antibody as indicated (cf, Materials and Methods). In EMSA with antibodies, the nuclear extracts were preincubated with 1 μL of the HIF-1 antibody for 2 hours at 4°C before adding the labeled probe. Middle panel: the32P-labeled HRE-2 (-168/-152) oligonucleotide and the PAI-1 HRE-1 (-182/-166) oligonucleotide were incubated with either 10 μg of nuclear extracts from normoxic (16% O2) or hypoxic (8% O2) cells in the absence or presence of preimmune serum or rat HIF-1 antibody as indicated. The inducible (I) HIF-1 complex was seen with the HRE-2, but not with the HRE-1 oligonucleotide. Right panel: the 32P-labeled mutated HRE-2 (-168/-152) oligonucleotide (M-HRE) was incubated with either 10 μg of nuclear extracts from normoxic (16% O2) or hypoxic (8% O2) cells or with extracts from hypoxic cells in the presence of rat HIF-1 antibody as indicated. The image had to be overexposed to visualize the weak constitutive binding. The DNA-protein binding was analyzed by electrophoresis on 5% native polyacrylamide gels. I, induced HIF-1 complex; C, constitutive complex; S, supershifted complex. (C) Western analysis of nuclear extracts from normoxic (16% O2) or hypoxic (8% O2) cells. A total of 20 μg nuclear extract from 2 different preparations was resolved on a 10% SDS gel and probed with preimmune serum or the HIF-1 antibody.
Binding of HIF-1 to the rat PAI-1 promoter region. (A) Oligonucleotides: the HIF-1 consensus sequence BACGTSSK32were B=G/C/T, S=G/C, and K=G/T and the sense strand of the rat PAI-1 promoter sequence -177/-152 containing the HRE-1 and HRE-2 site, the sense strands of the oligonucleotides containing HRE-1, HRE-2, and the oligonucleotide with a mutation in the HRE-2 site M-HRE are shown. Bases matching the consensus sequences are underlined, mutated bases are in lower case letters. (B) EMSA, left panel: the32P-labeled PAI-1 HRE-1/2 oligonucleotide was incubated with either 10 μg protein of nuclear extracts from normoxic (16% O2) or hypoxic (8% O2) cells in the absence or presence of preimmune serum or rat HIF-1 antibody as indicated (cf, Materials and Methods). In EMSA with antibodies, the nuclear extracts were preincubated with 1 μL of the HIF-1 antibody for 2 hours at 4°C before adding the labeled probe. Middle panel: the32P-labeled HRE-2 (-168/-152) oligonucleotide and the PAI-1 HRE-1 (-182/-166) oligonucleotide were incubated with either 10 μg of nuclear extracts from normoxic (16% O2) or hypoxic (8% O2) cells in the absence or presence of preimmune serum or rat HIF-1 antibody as indicated. The inducible (I) HIF-1 complex was seen with the HRE-2, but not with the HRE-1 oligonucleotide. Right panel: the 32P-labeled mutated HRE-2 (-168/-152) oligonucleotide (M-HRE) was incubated with either 10 μg of nuclear extracts from normoxic (16% O2) or hypoxic (8% O2) cells or with extracts from hypoxic cells in the presence of rat HIF-1 antibody as indicated. The image had to be overexposed to visualize the weak constitutive binding. The DNA-protein binding was analyzed by electrophoresis on 5% native polyacrylamide gels. I, induced HIF-1 complex; C, constitutive complex; S, supershifted complex. (C) Western analysis of nuclear extracts from normoxic (16% O2) or hypoxic (8% O2) cells. A total of 20 μg nuclear extract from 2 different preparations was resolved on a 10% SDS gel and probed with preimmune serum or the HIF-1 antibody.
When the oligonucleotide -168/-152 containing HRE-2 was used as a probe in the EMSA, the HIF-1 complex was formed (Fig 4B, middle panel), but the complex with the oligonucleotide containing HRE-1 and HRE-2 -177/-152 was stronger (Fig 4B, left panel). Again, the addition of 1 μL of the HIF-1α antibody prevented the HIF-1 complex formation, enhanced formation of the constitutive complex, and formed the same supershifted complex as with the HRE-1/2 probe (Fig 4B, middle panel). The HIF-1 complex was also no longer detectabel when a mutation was introduced in the HRE-2 site as with the M-HRE oligunucleotide (Fig 4B, right panel). In contrast, with the PAI-1 promoter oligonucleotide -182/-166 containing HRE-1, a very strong complex smaller than the HIF-1 complex was detectable (Fig 4B, middle panel). The formation of this DNA-protein complex was independent from the pO2, and addition of the HIF-1α antibody did not interfere with the formation of this strong complex. However, the HIF-1α antibody addition resulted in the detection of a supershifted complex (Fig 4B, middle panel).
These data indicate that mainly the HRE-2 site in the PAI-1 promoter was necessary for the interaction with HIF-1, and that the sequences in the region -182/-166 had supporting functions in forming a stronger HIF-1-DNA complex.
DISCUSSION
In this study, we show that PAI-1 mRNA and protein expression were activated by hypoxia in primary rat hepatocytes (Figs 1and 2), and that this induction was mediated by HIF-1 (Figs 3 and 4). Transfection assays with rat PAI-1 promoter LUC gene constructs provided evidence that hypoxia activated PAI-1 via 2 HREs matching the HIF-1 consensus sequence (Fig 3). Cotransfection of PAI-1 promoter LUC gene constructs and full-length HIF-1α cDNA (Fig 3) and EMSAs (Fig 4) showed that HIF-1 bound to HRE-2 and that HRE-1 had supportive functions in the response of the rat PAI-1 promoter to hypoxia.
Functional and nonfunctional HIF-1 binding sites.
A variety of O2 responsive genes has been identified.33 In the response to hypoxia, the transcription factor HIF-1 appears to be a key regulator. HIF-1 is a heterodimer consisting of HIF-1α and HIF-1β, the latter being identical to the ARNT protein.34 In the response to hypoxia, HIF-1 binds to HREs, which can be located in the 5′-flanking or 3′-flanking regions of a gene. HIF-1 binding sites were identified in a variety of hypoxia-inducible genes encoding erythropoietin (EPO), vascular endothelial growth factor (VEGF), and glycolytic enzymes.31,35 From these genes, the HRE-consensus sequence 5′-BACGTSSK-3′ (B=G/C/T, S=C/G, K=G/T) was deduced.36 Furthermore, in a detailed analysis of HREs in the human aldolase A (ALDA), enolase 1, and mouse lactate dehydrogenase A gene promoters with transfection assays in Hep3B cells and EMSAs, it was found that a HIF-1 sensitive DNA sequence should consist of a pair of transcription factor binding sites, which contained the HIF-1 core sequence 5′-RCGTG-3′ (R=A/G).37 Both the HRE-1 (-175/-168) and the HRE-2 (-165/-158) in the hypoxia-responsive DNA sequence of the rat PAI-1 promoter, -175/-157, identified in this study matched the HIF-1 consensus sequence 5′-BACGTSSK-3′ in 6 of 8 bp (Fig 3A). The HRE-2 contained the HIF-1 core sequence 5′-ACGTG-3′, whereas HRE-1 with 5′-ACGTA-3′ did not. However, inspection of the antisense strand showed that the antisense HRE-1 sequence -170/-177 5′-TACGTGTG-3′ and the antisense HRE-2 sequence -160/-167 5′-CACGTGTG-3′ matches the HIF-1 binding consensus sequence in 7 of 8 bp containing also the core sequence 5′-RCGTG-3′. This existence of 2 adjacent HIF-1 sites, arranged as either direct or inverted repeats, has been noted in other hypoxically regulated genes including human enolase 1,37mouse lactate dehydrogenase A,37 human phosphoglycerate kinase 1,38 and human transferrin.39
In hepatocytes transfected with either the PAI-1 promoter LUC constructs pGl3PAI-766M1 or pGl3PAI-766M2, in which the HRE-1 and HRE-2, respectively, were mutated (Fig 3B), the hypoxia-dependent increase in LUC activity was abolished (Fig 3B). Thus, both the HRE-1 and HRE-2 were involved in the hypoxia-dependent activation of the PAI-1 promoter.
Cotransfection of hepatocytes with full-length HIF-1α cDNA and pGl3PAI-766M2, in which the HRE-1, but not the HRE-2 was intact, did not result in any induction of Luciferase (Fig 3). Gel shift analysis with the 17-mer oligonucleotide -182/-166 spanning the HRE-1 did not provide evidence for the same HIF-1 oligonucleotide complex as with the HRE-1/2 or the HRE-2 probe (Fig 4). However, the presence of the supershifted complex implicates that a small amount of HIF-1α was present in this complex. This would mean that other partners of the bHLH-protein family, such as USF or MYC, may interact with HIF-1α at the HRE-1 of the PAI-1 promoter. Studies to clarify this question are under way. However, these results clearly indicated that HRE-1 did not bind HIF-1 with the same affinity as HRE-2 or HRE-1/2. In contrast, cotransfection of hepatocytes with pGl3PAI-766M1 in which the HRE-2, but not the HRE-1, was intact and a vector encoding full-length HIF-1α cDNA caused induction of LUC activity (Fig 3). However, this induction was not as strong as with the wild-type pGl3PAI-766 construct (Fig 3). The 17-mer oligonucleotide -168/-152 containing the HRE-2 formed the HIF-1 complex (Fig 4). These findings showed that HRE-2 bound HIF-1. Thus, it seems likely that HRE-1 is a low affinity or a nonfunctional HIF-1 binding site. Such nonfunctional binding sites were also identified in the human ALDA gene and in the mouse HIF-1α gene. In the case with the ALDA gene, a part of the intervening sequence IV (IVS-IV; intron D) 5′-CACGTGCG-3′ from +128/+13537 and in the promoter of HIF-1α exon I1 (E-I1),40 the region -82/-75 5′-TACGTGGT-3′ also matched the HRE-consensus sequence. Transfection assays with ALDA-CAT gene constructs containing IVS-IV (intron D) sequences and HIF-1α E-I1 promoter LUC gene constructs in Hep3B cells in concordance with EMSAs showed that neither the ALDA IVS-IV-HRE nor the HRE of the HIF-1α E-I1promoter was functionally active.37 40
Indeed, a more detailed inspection of the PAI-1 promoter HRE-1 and sequences immediately flanking its 5′-region showed that it contained 3 CA-repeats, which were present also in the 17-mer HRE-1 oligonucleotide used for EMSA (Fig 4A). These CA-repeats 4-6 nt upstream or downstream from the HRE have been shown to be of importance in conjunction with the HIF-1 binding sites of the EPO and VEGF genes. Mutation of the CACA-sequences 3′ of the EPO-HRE and also 5′ and 3′ of VEGF-HRE resulted in a loss of hypoxia-dependent expression of CAT reporter genes.31,41,35In addition, such CA repeats in the form of a CACAT-sequence have also been found 3′ from the HRE -204/-181 sequence in the human ALDA gene promoter37 and in the form of a CACAG sequence 161 bp in front of the HRE -164/-151 of the rat α1B-adrenergic receptor gene. Thus, in the PAI-1 promoter, HRE-2 served as the major HIF-1 binding site, and HRE-1 in conjunction with the CA-repeat was necessary for the full response of the PAI-1 gene promoter under hypoxia.
Physiological role of the HIF-1–mediated PAI-1 induction.
Hypoxia is an important regulator for vascular homeostasis and a major stimulus for vascular development. Local hypoxia shifts the balance between fibrinogenesis and fibrinolysis towards thrombus formation and stabilization42,43 and elicits neovascularization.44 A key event in these processes is the local increase in HIF-1. HIF-1, in turn, stimulates the formation of the fibrinolysis inhibitor or more general extracellular proteolysis inhibitor PAI-1 as shown in this study and of the angiogenic growth factor VEGF.45 Inhibition of fibrinolysis by PAI-1 is important for thrombus formation and stabilization, and stimulation of growth by VEGF in conjunction with inhibition of extracellular proteolysis by PAI-1 are important for vessel formation.43,46 47
PAI-1 in atherosclerosis and thrombosis.
The importance of PAI-1 for atherosclerosis and thrombosis is indicated by the observation that increased plasma levels of PAI-1 paralleled reduced fibrinolytic activity of aortic tissue48 in rats with O2-deprivation (5% O2) and caused fibrin deposition in the lung of mice exposed to a hypoxic environment (5% to 6% O2),49 and that PAI-1 expression was enhanced in human atherosclerotic lesions.50-52 Thus, the HIF-1–mediated activation by hypoxia of the PAI-1 gene, shown here, may help to explain progression of ischemic coronary heart disease and fibrin deposition on venous valves during thromboembotic events.
PAI-1 in tumor progression and vascular development.
Hypoxia is a major stimulus for vascular development. The local increase in HIF-1 in hypoxic areas elicits the formation of the angiogenic growth factor VEGF45 and as shown in this study of the extracellular proteolysis inhibitor PAI-1. Stimulation of growth by VEGF and control of proteolysis are important for angiogenesis,53 because low VEGF levels resulted in reduced blood vessel density46 and low levels of PAI-1 in excessive proteolysis preventing coordinated endothelial sprouting.54 55
The formation of tumors or metastases requires the regulation of adhesive, proteolytic, as well as migrating events, and finally neovascularization of the tumor or metastasis. Tumors derived from embryonic stem (ES) cells with inactivated HIF-1α genes (HIF-1α−/−) did not express VEGF in response to hypoxia resulting in poor blood vessel supply.56,57 In athymic mice, tumors derived from injected HepaC4 cells with a defective ARNT (HIF-1β) and thus with a lack of the HIF-1 (HIF-1α/HIF-1β) heterodimer had reduced vascularity and grew more slowly than tumors derived from the wild-type Hepa-1 cells.58,59 Moreover, after implantation of malignant murine keratinocytes onto the dorsal muscle fascia of wild-type and PAI-1−/− mice, only the tissues of wild-type mice were infiltrated by the tumor cells. The failure of tumor cell invasion in PAI-1−/− mice was due to absent vascularization of the tumor, which was restored after rescue of systemic PAI-1 expression with an adenoviral vector containing the human PAI-1 gene.47 In line with these observations, PAI-1 was found to be expressed at higher levels in tumor than in other cells; it may be necessary for optimal metastasis formation as shown for cultured lung cancer cells.60 Finally, in a number of clinical studies, it was shown that high PAI-1 levels in patients suffering from various types of cancer indicated a poor prognosis.61-63
Thus, it appears that HIF-1 controls production of angiogenic growth factors, such as VEGF, and as shown in this study, inhibitors of proteolysis, such as PAI-1, which both contribute to neovascularization and tumor growth.
ACKNOWLEDGMENT
The authors thank Dr T.D. Gelehrter (Department of Human Genetics, University Michigan Medical School, Ann Arbor, MI) for the kind gift of PAI-1 cDNA.
Supported by the Deutsche Forschungsgemeinschaft SFB 402 Teilprojekt A1.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Thomas Kietzmann, MD, Institut für Biochemie und Molekulare Zellbiologie, Humboldtallee 23, D-37073 Göttingen, Germany; e-mail: tkietzm@gwdg.de.

![Fig. 3. Regulation of transfected rat PAI-1 promoter LUC gene constructs by hypoxia and cotransfected HIF-1 in transiently transfected rat hepatocyte cultures. Enhancement of PAI-1 production by HIF-1. (A) The 5′-flanking region of the rat PAI-1 gene with its footprinted sites [A-G]. In the “C-site”, the 2 potential HIF-1 binding elements (HRE) matching the HIF-1 consensus sequence BACGTSSK32 were B=G/C/T, S=G/C and K=G/T are underlined. B, S, or K are shown only if the actual PAI-1 sequence does not match any of the bases allowed by the consensus. (B) Hepatocytes were transiently cotransfected with pcDNAHIF-1 containing the full-length rat HIF-1 cDNA and LUC gene constructs driven by a wild-type 766 bp rat PAI-1 promoter (pGl3PAI-766), or the 766 bp promoter mutated at either the HRE-1 (pGl3PAI-766M1) or HRE-2 (pGl3PAI-766M2) site. After 24 hours, the transfected cells were cultured for 18 hours under hypoxia, additionally pGl3PAI-766-transfected cells were treated with CHX (10 μg/mL). In each experiment, the fold stimulation of LUC activity by hypoxia (8% O2) was determined relative to the normoxic (16% O2) control, which was set equal to 1. The values represent means ± SEM of 3 independent experiments. Statistics, Student'st-test for paired values: * significant differences 16% O2 versus 8% O2, P ≤ .05; ** significant differences 16% O2 versus 16% O2+ HIF-1, P ≤ .05. In pGl3PAI-766M1 and pGl3PAI-766M2, the wild-type PAI-1 sequence is shown on the upper strand, mutated bases are indicated by *, and are shown in lower case letters. (C) Western blot analysis of PAI-1, HIF-1, and ARNT in cultured hepatocytes transfected with a HIF-1 vector. A total of 50 μg of protein from the medium and 100 μg of total cellular protein of the transfected hepatocytes were subjected to Western analysis (cf, Materials and Methods) with antibodies against rat PAI-1, rat HIF-1, and ARNT, respectively. Blots were scanned by videodensitometry, and in each experiment, the PAI-1 level and HIF-1 level measured under normoxia (16% O2) were set equal to 1. Values represent the fold induction of PAI-1 and HIF-1 of 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/94/12/10.1182_blood.v94.12.4177/5/m_blod42414003w.jpeg?Expires=1763700660&Signature=dR4M37oJiwkWpKN4~iZOeWaxjthfSFGx6BBDr5rn4ER3eHE5hcKDfTJu34Utb-WYgfo5E1KVGQOcxXi-FGYLaM1yvOagVLj6SvnAxJxe7b8NOeesf1ocaPylDQKFtQvLxR~zKvixXpl39RIYJn~rZoH3GMRQHwqq-4xqUTx8HtJ96X0NQpqIT~gX9Bh1piehXQFN6-MmZNRmi~2pF0a5rwSX-RhBoG8-QOQkYdHX13EEjStpgKHxQhaM~LAtwUxjoeDQk1afKbPHfehmUzdYeENPaBdzYLKGZXEt6gnfPhLaK~BxvclrQCnuIbr4JJLF64uP9An6cKKbG3FrBuSY5w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal