Abstract
Hepatoerythropoietic porphyria (HEP) is an inherited metabolic disorder characterized by the accumulation of porphyrins resulting from a deficiency in uroporphyrinogen decarboxylase (UROD). This autosomal recessive disorder is severe, starting early in infancy with no specific treatment. Gene therapy would represent a great therapeutic improvement. Because hematopoietic cells are the target for somatic gene therapy in this porphyria, Epstein-Barr virus-transformed B-cell lines from patients with HEP provide a model system for the disease. Thus, retrovirus-mediated expression of UROD was used to restore enzymatic activity in B-cell lines from 3 HEP patients. The potential of gene therapy for the metabolic correction of the disease was demonstrated by a reduction of porphyrin accumulation to the normal level in deficient transduced cells. Mixed culture experiments demonstrated that there is no metabolic cross-correction of deficient cells by normal cells. However, the observation of cellular expansion in vitro and in vivo in immunodeficient mice suggested that genetically corrected cells have a competitive advantage. Finally, to facilitate future human gene therapy trials, we have developed a selection system based on the expression of the therapeutic gene. Genetically corrected cells are easily separated from deficient ones by the absence of fluorescence when illuminated under UV light.
PORPHYRIAS ARE INHERITED metabolic diseases characterized by specific enzyme defects along the heme biosynthetic pathway.1 Uroporphyrinogen decarboxylase (UROD; EC. 4.1.1.37) is the fifth enzyme of this pathway and catalyzes the sequential decarboxylation of uroporphyrinogen to yield coproporphyrinogen.2 A deficiency of this enzyme is responsible for porphyria cutanea tarda (PCT), the commonest form of porphyria, and for hepatoerythropoietic porphyria (HEP). HEP, the homozygous form of familial PCT, is clinically similar to congenital erythropoietic porphyria (CEP) and is characterized by severe dermal photosensitivity leading to scarring and mutilation of sun-exposed areas of skin, sclerodermoid changes, hypertrichosis, erythrodontia, anemia, and hepatosplenomegaly occuring very early in infancy or childhood.1 The human enzyme has been purified to homogeneity from erythrocytes3 and the human UROD gene has been isolated and sequenced.4 The molecular lesions responsible for HEP were investigated.4-6 The available treatments are mostly unsatisfactory and often limited to supportive care. The enzymatic deficiency is responsible for the accumulation of large amounts of uroporphyrin and heptacarboxylic porphyrin and has been found in all of the tissues investigated, especially bone marrow and liver. This type of porphyria is a good candidate for somatic gene therapy, because the genetic defect is well characterized at the molecular level and inherited as a recessive trait. The ideal therapy should concern two tissues: bone marrow and liver. One approach to somatic gene therapy involves the use of recombinant retroviral vectors to transduce cells ex vivo, followed by autologous transplantation of genetically engineered cells.7 To facilitate the development of ex vivo gene therapy, efficient selection procedures are required for the isolation of genetically corrected cells before autologous transplantation.
The aim of the present study is the transfer of functional UROD cDNA into HEP-deficient cells by means of a retroviral vector. The feasibility of the correction of the enzymatic defect is tested in B-lymphocyte cell lines established from HEP patients. The efficiency of gene transfer was analyzed in terms of retroviral integration, enzymatic restoration, metabolic correction, and cellular expansion. The ability of genetically corrected cells to transform the excess of uroporphyrinogen produced by deficient cells was also analyzed. Finally, a fluorescence-based selection procedure using flow cytometry analysis and sorting was developed to isolate directly metabolically corrected cells.
MATERIALS AND METHODS
Cell lines.
Epstein-Barr virus-transformed B lymphocytes were used because they can be readily obtained from patients and because the cells are easy to grow in culture. Peripheral blood lymphocytes (PBL) were isolated from patients and controls. Cell lines (LB) were established from PBL after Epstein-Barr virus infection at Laboratoire Genethon (Evry, France). Deficient PBL were obtained from three HEP patients carrying two different point mutations of the UROD gene. Two deficient (LBHEP) cell lines, LB44 and LB45, harbored the G281E mutation (substitution of glycine by glutamic acid at codon 281) leading to an unstable protein.5 The third deficient cell line, LB86, had an unpublished mutation F46L (substitution of phenylalanine by leucine at codon 46). The impact of this mutation on UROD protein function is under investigation. Normal (LBN) and deficient (LBHEP) lines are maintained at 0.2 to 2 × 106 cells/mL in RPMI 1640 medium supplemented with 20% fetal calf serum (FCS; Boehringer Mannhein, Meylan, France), 100 U of penicillin per milliliter, and 1 mg/mL of streptomycin (GIBCO BRL, Grand Island, NY) at 37°C in 5% CO2 atmosphere. The ecotropic Gp+env86 and the amphotropic ΨCRIP packaging cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM), supplemented with 10% FCS, 100 U of penicillin per milliliter, and 1 mg/mL of streptomycin at 37°C in 5% CO2 atmosphere.
Construction of retroviral vector LUDSN and obtaining different packaging cell lines.
The pLXSN vector8 has been widely used for gene transfer in hematopoietic cells. The vector contains the NeoR gene that allows the selection of transduced cells by G418 (Geneticin; GIBCO BRL). NeoR gene is controlled by SV40 early promoter sequences. The gene of interest is driven by the Moloney murine leukemia virus (MoMLV)/Moloney murine sarcoma virus (MoMSV) long terminal repeat (LTR). Normal full-length human UROD cDNA (1.2 kb) was obtained from pG7UD vector.5 cDNA was cleaved by EcoRI and Xho I and cloned into the pLXSN vector to obtain the pLUDSN construct. The recombinant amphotropic particles were obtained by a three cell-line transfection process (ping-pong method).9 First, pLUDSN construct was introduced by DOTAP transfection reagent (Boehringer Mannheim) into a first packaging cell line ΨCRIP. Second, filtered supernatants from virus-producing cells were used to infect Gp+env86 cells in the presence of polybrene (Sigma, St Louis, MO) during 6 hours for 3 consecutive days. Two days after the last infection, cells were selected in the presence of 1 mg/mL G418 (Geneticin; GIBCO BRL) for 2 weeks. Resistant clones were isolated and the clones producing the highest viral titer were selected using the NIH3T3 cell line as a target. The Gp+env86/LUDSN8 clone was selected for subsequent transduction experiments because of a high viral titer (1.2 × 106 cfu/mL) and a UROD enzymatic activity 14-fold the normal value. Third, the ΨCRIP packaging cell line was transduced by filtered retroviral supernatant in the presence of protamine sulfate (Sigma). The protocol of infection and selection was as described for the Gp+env86 cells. G418-resistant clones (ΨCRIP/LUDSN) were isolated and screened for those producing the highest titer using NIH3T3 cells as targets. Supernatants were tested for the presence of replication competent virus.10
Transduction of target cells.
Retroviral supernatant was obtained from the producer cell lines (ΨCRIP/LUDSN7, 11, and 16 or ΨCRIP/LXSN) maintained at 32° C in 5% CO2 atmosphere for 24 to 48 hours. Target cells were transduced by retroviral supernatant infection under two different conditions. For the first transduction protocol, 2 mL of the filtered supernatant was added to 4 × 105 target cells (8:1 colony-forming units/cell ratio) in the presence of 8 μg/mL polybrene. Cells and retroviral supernatant were incubated in a 6-well plate and were centrifuged at 1,000g at 32°C for 60 minutes followed by incubation for 6 hours at 32°C in a 5% CO2incubator. Medium from producer cells was then replaced by RPMI 1640 medium supplemented with 20% FCS for 17 hours. Cells were transduced for 3 consecutive days at 24-hour intervals. After the last infection, LB were maintained without selection for 4 days to allow expansion and NeoR expression. After expansion, LBHEP were subjected to an initial period of selection in 0.2 mg/mL G418 for 4 days and then in 0.5 mg/mL for 1 week. Finally, LBHEP were maintained in medium containing 1 mg/mL G418 for 2 to 3 weeks. In a second procedure, wells (of a 6-well plate) were precoated with human recombinant CH296 fibronectin fragment (Retronectin, Takara; Boehringer Ingelheim, Gagny, France) as recommended by the manufacturer. Two milliliters of filtered virus supernatant (2.4 × 106 viral particles) was added for 20 minutes five times and then 4 × 105normal or deficient LB cells were incubated in the wells for 6 hours at 37°C in 5% CO2 atmosphere for 3 consecutive days. After each infection period, the medium was replaced by RPMI 1640 supplemented with 20% FCS. Selection was performed as described above. Supernatants from these three clones were tested for the presence of replication competent virus and were negative.
Southern blot analysis.
Southern blot analysis allows the quantification of integrated viral copies in the target cell using known amounts of the vector as a standard. High molecular weight DNA was extracted from LB cells by proteinase K digestion and phenol/chlorophorm extraction. Ten micrograms of genomic DNA was digested with Sac I; this enzyme cut both LTR regions in our construct and produced a 3-kb band from the LUDSN vector. The resulting fragments were separated on a 0.8% agarose gel and transferred onto nylon membrane (Hybond N+; Amersham, Les Ulis, France). Known amounts of plasmid DNA mixed with control LB DNA were run on the same gel as standard. Filters were hybridized with a 1.2-kb human UROD cDNA fragment that was labeled with 32P dCTP by random priming (Multiprime DNA labeling system; Amersham Pharmacia, Orsay, France). The vector copy number per cell was estimated by comparison of the amount of signal generated by standards. The signal obtained with 10 pg of plasmid DNA corresponds to the presence of one copy of the transgene per cell when 10 μg of genomic DNA is loaded on the gel.
UROD enzymatic activity and porphyrin level.
UROD activity and porphyrin level were determined in normal and LBHEP cell lines to assess the phenotypic expression of the transferred gene. Determination of UROD activity was performed according to de Verneuil et al.3 UROD activity was expressed as picomoles of coproporphyrinogen formed per hour per milligram of protein (units per milligram).
LB cell lines were seeded at 2 × 105 cells per milliliter in the 6-well plate and maintained for 72 hours in RPMI 1640 medium containing 20% FCS. The medium was then kept, the cells were washed with phosphate-buffered saline (PBS) and counted, and porphyrins were extracted from cell lysates and from the medium with 1 mol/L HClO4/CH3OH (1:1, vol/vol). Total porphyrin level was quantitated by spectrofluorimetry (excitation, 405 nm; emission, 595 nm). Uroporphyrin I solution (10 nmol/L) was used as a standard. Porphyrin level was expressed as femtomoles of porphyrins per 103 cells. Individual carboxylated porphyrins were identified by high performance liquid chromatography (HPLC) assay.11
In vitro cell proliferation assays.
The MTS test12 (CellTiter 96rAQueous Solution Cell Proliferation Assay; Promega Corp, Madison, WI) was used to analyze the consequence of the metabolic deficiency on the cell proliferation rate. LB cells were seeded at 2 × 105cells per milliliter in the 6-well plate in RPMI 1640 medium containing 20% FCS. These cells were grown for 2 days before the MTS test. Viable cells (104) were resuspended in 100 μL of fresh medium per well of the 96-well plate reader. Assays were performed at 12, 24, 48, and 72 hours of culture at 37°C in a humidified 5% CO2 atmosphere. Twenty microliters of the CellTiter Solution Reagent was added directly to the wells and the plates were incubated for 4 hours. Absorbance was measured at 490 nm.
The cellular expansion of LBN, LB44, LB44-LUDSN11, and a mixture of LB44/LB44-LUDSN11 (80/20) was assessed at 0, 5, and 10 days by measuring the proliferation index (number of cells at t = x days divided by the number of cells at t = 0 day). Cells were suspended and their viability was assessed with trypan blue. Viable cells were counted in duplicate in a Malassez’s count slide. Cell doubling time was calculated on the first 5 days of culture. UROD enzymatic activity was measured in the mixture at different times of culture.
In vivo cell proliferation in immunodeficient mice.
Injection of LB cells in immunodeficient mice results in the development of a lymphoproliferative disorder characterized by B-cell tumors.13 We used a double mutant, the RAG2−/−/γc− mouse, characterized by a complete absence of B and T lymphocytes and NK cells in peripheral blood.14 Twenty-eight double homozygous mutant male mice between 8 to 10 weeks of age were randomly selected, divided into four groups, and inoculated subcutaneously with 2 × 106 LBN, LB44, or LB44-LUDSN11 cells or by the mixture of LB44/LB44-LUDSN11 (80/20) LB cells as described above. Animals were kept under sterile conditions in microisolators or air-filtered cages and provided with autoclaved food and water. Most of the animals developed subcutaneous tumors at the site of inoculation. Animals were killed when tumors became clinically apparent (30 to 45 days after injection), and tumors were resected for analysis of UROD enzymatic activity.
Fluorescence-based selection of retrovirally transduced cells.
Deficient and corrected HEP LB cell lines were subjected to preparative fluorescence-activated cell sorting (FACS). Culture cells were seeded at 2 × 105 cells/mL into T-75 culture flasks in RPMI 1640 medium supplemented with 7% FCS for 24 hours before flow cytometry analysis. Accumulation of porphyrins in the deficient, transduced, and control cells was monitored by fluorescence microscopy using a 100 W mercury lamp and a near UV band filter. Cells were harvested and concentrated at 106 cells/mL and analyzed by flow cytometry using an Elite cytometer (Coulter, Coultronics, Margency, France) at a rate of 1,500 events/s using saline as a sheath fluid. Excitation was performed by an argon laser (Spectra Physics, Les Ulis, France) tuned to emit 100 mW in UV light (340 to 360 nm). Fluorescence was measured through a 675-nm band filter. For the cell sorting experiments, the cells with either the lowest or the highest fluorescence intensities were sorted in sterile tubes containing 0.5 mL FCS. FACS efficiency was analyzed in terms of UROD enzymatic activity and the percentage of transduced cells estimated by Southern blot.
We then tested a different strategy based on an increase in uroporphyrin accumulation in untransduced deficient LB cells by exposure to δ-aminolevulinic acid (ALA). Melatonin was also added to the medium because of a protective role against oxidative damage induced by ALA.15 Culture cells were seeded at 2.5 × 105 cells/mL into T-75 culture flasks. ALA, melatonin, and FeSO4 (Sigma, Saint-Quentin, France) were added to a final concentration of 1 mmol/L, 2 mmol/L, and 50 μmol/L, respectively, in RPMI 1640 medium containing 7% FCS at pH 7.4. After 12 hours, cells were harvested, washed with PBS, and reseeded at the same dilution in fresh RPMI 1640 medium supplemented with 7% FCS for 4 hours. Porphyrin accumulation in cell lysates was quantitated by spectrofluorimetry, as described above. The fluorescence of intact cells was estimated by cytofluorimetry (Coulter Elite, Coultronics).
RESULTS
Transduction of target cells.
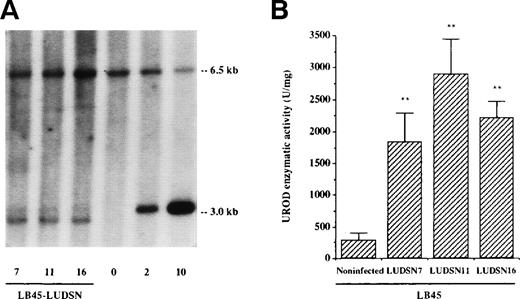
Three clones (ΨCRIP/LUDSN7, ΨCRIP/LUDSN11, and ΨCRIP/LUDSN16) characterized by a retroviral titer greater than 106 cfu/mL were chosen to transduce the deficient LB45 cell line. UROD activity was measured in the nontransduced and in the different transduced and selected cell lines to find the best clone in terms of enzymatic correction. Results are shown in Fig 1. The number of integrated proviral copies was approximately one copy per target cell in the three transduced and selected cell lines, as estimated by Southern blot analysis (Fig 1A). ΨCRIP/LUDSN11 clone, which demonstrated a 10-fold increase in enzymatic activity compared with the deficient nontransduced cells (Fig 1B), was chosen for the following experiments.
Identification of the LUDSN provirus and expression of the transgene in HEP LB cells. Cells were infected with LUDSN7, LUDSN11, and LUDSN16 by centrifugation in the retroviral supernatant at 1,000g and selected for 3 weeks with G418 (see Materials and Methods). (A) Provirus integration into the genomic DNA of LB deficient cells (Southern blot analysis). Ten micrograms of genomic DNA extracted from transduced and selected LB45/LUDSN7, LB45/LUDSN11, and LB45/LUDSN16 cell lines was digested with Sac I, blotted, and probed with the 1.2-kb human UROD cDNA. Lane 0, 10 μg of DNA from untransduced LB cells. Lanes 2 and 10 contained plasmid DNA (20 and 100 pg, respectively) mixed with 10 μg of noninfected DNA corresponding to 2 and 10 copies per cell. The 3.0-kb band corresponds to the integrated provirus and the 6.5-kb band corresponds to the endogenous UROD gene. Note that the migration of the 3.0-kb band for LB45 samples were artefactually sligthly different from the control plasmid DNA. (B) UROD activity in transduced and deficient LB45 cell lines. Values are the means ± SD of three to six determinations.** P < .01v noninfected LB45 cells.
Identification of the LUDSN provirus and expression of the transgene in HEP LB cells. Cells were infected with LUDSN7, LUDSN11, and LUDSN16 by centrifugation in the retroviral supernatant at 1,000g and selected for 3 weeks with G418 (see Materials and Methods). (A) Provirus integration into the genomic DNA of LB deficient cells (Southern blot analysis). Ten micrograms of genomic DNA extracted from transduced and selected LB45/LUDSN7, LB45/LUDSN11, and LB45/LUDSN16 cell lines was digested with Sac I, blotted, and probed with the 1.2-kb human UROD cDNA. Lane 0, 10 μg of DNA from untransduced LB cells. Lanes 2 and 10 contained plasmid DNA (20 and 100 pg, respectively) mixed with 10 μg of noninfected DNA corresponding to 2 and 10 copies per cell. The 3.0-kb band corresponds to the integrated provirus and the 6.5-kb band corresponds to the endogenous UROD gene. Note that the migration of the 3.0-kb band for LB45 samples were artefactually sligthly different from the control plasmid DNA. (B) UROD activity in transduced and deficient LB45 cell lines. Values are the means ± SD of three to six determinations.** P < .01v noninfected LB45 cells.
Results of UROD enzymatic activity in untransduced and transduced normal, LB44, LB45, and LB86 cells are shown in Table 1. UROD-specific activity of unselected LB cells was higher when transduction was performed in plates coated by Retronectin than when cells were centrifuged in the retroviral supernatant. Consequently, all of the following experiments were performed with selected LB cells transduced by Retronectin.
UROD Enzymatic Activity in Untransduced and Transduced LB Cells
| Cell Type . | UROD Activity (U/mg) . | |||
|---|---|---|---|---|
| Mean ± SD . | Range . | % of Normal Values . | n . | |
| Untransduced LB cells | ||||
| LBN | 2,728 ± 238 | 2,278-3,029 | 100 | 14 |
| LB44 | 219 ± 123 | 46.2-443 | 8 | 8 |
| LB45 | 288 ± 108 | 155-422 | 11 | 5 |
| LB86 | 396 ± 129 | 302-585 | 15 | 4 |
| Transduced LB cells | ||||
| LBN-LUDSN (S + C) | 3,063 ± 83 | 3,001-3,157 | 112 | 3 |
| (Retro) | 3,274 ± 156 | 3,161-3,440 | 120 | 3 |
| (Retro) + G418 | 5,850 ± 71 | 5,784-5,925 | 214 | 3 |
| LB44-LUDSN (S + C) | 305 ± 116 | 229-438 | 11 | 4 |
| (Retro) | 570 ± 186 | 420-778 | 21 | 4 |
| (Retro) + G418 | 1,927 ± 266 | 1,595-2,402 | 71 | 9 |
| LB45-LUDSN (S + C) | 318 ± 139 | 127-439 | 12 | 4 |
| (Retro) | 791 ± 201 | 582-983 | 29 | 4 |
| (Retro) + G418 | 2,901 ± 543 | 2,274-3,720 | 106 | 6 |
| LB86-LUDSN (S + C) | 412 ± 157 | 330-642 | 15 | 4 |
| (Retro) | 749 ± 264 | 552-1,138 | 27 | 4 |
| (Retro) + G418 | 2,440 ± 366 | 2,068-2,799 | 89 | 4 |
| Cell Type . | UROD Activity (U/mg) . | |||
|---|---|---|---|---|
| Mean ± SD . | Range . | % of Normal Values . | n . | |
| Untransduced LB cells | ||||
| LBN | 2,728 ± 238 | 2,278-3,029 | 100 | 14 |
| LB44 | 219 ± 123 | 46.2-443 | 8 | 8 |
| LB45 | 288 ± 108 | 155-422 | 11 | 5 |
| LB86 | 396 ± 129 | 302-585 | 15 | 4 |
| Transduced LB cells | ||||
| LBN-LUDSN (S + C) | 3,063 ± 83 | 3,001-3,157 | 112 | 3 |
| (Retro) | 3,274 ± 156 | 3,161-3,440 | 120 | 3 |
| (Retro) + G418 | 5,850 ± 71 | 5,784-5,925 | 214 | 3 |
| LB44-LUDSN (S + C) | 305 ± 116 | 229-438 | 11 | 4 |
| (Retro) | 570 ± 186 | 420-778 | 21 | 4 |
| (Retro) + G418 | 1,927 ± 266 | 1,595-2,402 | 71 | 9 |
| LB45-LUDSN (S + C) | 318 ± 139 | 127-439 | 12 | 4 |
| (Retro) | 791 ± 201 | 582-983 | 29 | 4 |
| (Retro) + G418 | 2,901 ± 543 | 2,274-3,720 | 106 | 6 |
| LB86-LUDSN (S + C) | 412 ± 157 | 330-642 | 15 | 4 |
| (Retro) | 749 ± 264 | 552-1,138 | 27 | 4 |
| (Retro) + G418 | 2,440 ± 366 | 2,068-2,799 | 89 | 4 |
Deficient LB cells were transduced by supernatant infection and centrifugation (S + C) or by supernatant infection on plates coated with Retronectin (Retro) and selected or not by G418.
Deficient cell lines were transduced with LXSN (control virus) to investigate a possible effect of the virus itself. The level of UROD activity in LBHEP cell lines was not modified when compared with selected LBHEP-LXSN cell lines (195 ± 76 U/mg for LB44-LXSNv 219 ± 123 U/mg for LB44; 251 ± 159 U/mg for LB45-LXSNv 288 ± 108 U/mg for LB45; and 369 ± 195 U/mg for LB86-LXSN v 396 ± 129 U/mg for LB86). After transduction of the LBHEP cells by LUDSN11 and selection with G418, UROD activity was increased to nearly normal levels (71%, 106%, and 89% for LB44, LB45, and LB86 cells, respectively; Table 1). Transduced LB cells expressed the same level of UROD activity for 2 months after the end of selection, demonstrating that the UROD transgene introduced by the retroviral vector was stably integrated and expressed (data not shown).
To evaluate the ability of the transgene to correct the metabolic defect, the porphyrin level was measured in the cells and in the medium for untransduced and transduced cells (Fig2A). In the normal cells, a very low amount of porphyrins was found (0.12 ± 0.04 fmol per 103 cells). In deficient cells, a significant increase in porphyrin level was found (from 4.5 to 24 fmol per 103 cells) with the predominance of the highly carboxylated forms, uroporphyrin, and heptacarboxylic porphyrin, which are specific of the disease. Normalization of porphyrin level, documented in Fig 2A, demonstrated the ability of the transduction to correct the metabolic defect.
Porphyrin level in normal, deficient, and transduced deficient LB cells. (A) LB cells were maintained in exponential phase of culture and then placed in culture at 2 × 105cells/mL. After 48 hours, porphyrins were measured ([▧] Cell porphyrin level, [□] Supernatant porphyrin level) as described in Materials and Methods. Data are the mean ± SD of four to six determinations. ** P < .01 v noninfected LBN cells. (B) Porphyrin level in LBN cells, deficient LB45 cells, and a mixture (50/50) of LB45 and LBN cells. Cells were placed in culture at 2 × 105 cells/mL. Porphyrin level was measured by spectrofluorimetry immediatly ([▨] 0 hour) and after 24 (□), 48 (▧), and 72 hours (▨), as described in Materials and Methods. Data are the means ± SD of six independent experiments.
Porphyrin level in normal, deficient, and transduced deficient LB cells. (A) LB cells were maintained in exponential phase of culture and then placed in culture at 2 × 105cells/mL. After 48 hours, porphyrins were measured ([▧] Cell porphyrin level, [□] Supernatant porphyrin level) as described in Materials and Methods. Data are the mean ± SD of four to six determinations. ** P < .01 v noninfected LBN cells. (B) Porphyrin level in LBN cells, deficient LB45 cells, and a mixture (50/50) of LB45 and LBN cells. Cells were placed in culture at 2 × 105 cells/mL. Porphyrin level was measured by spectrofluorimetry immediatly ([▨] 0 hour) and after 24 (□), 48 (▧), and 72 hours (▨), as described in Materials and Methods. Data are the means ± SD of six independent experiments.
Absence of metabolic cross-correction between deficient and transduced cells.
Porphyrin levels in normal, deficient, and a mixture (50/50) of normal LB and LB45 cells at 0, 24, 48, and 72 hours are shown in Fig 2B. The porphyrin level in the mixed cells was approximately half that in the LB45 cell line. These in vitro results show that the UROD enzyme from normal LB cells was not able to metabolize the excess of uroporphyrinogens produced by the nontransduced deficient LB cells.
In vitro cell proliferation.
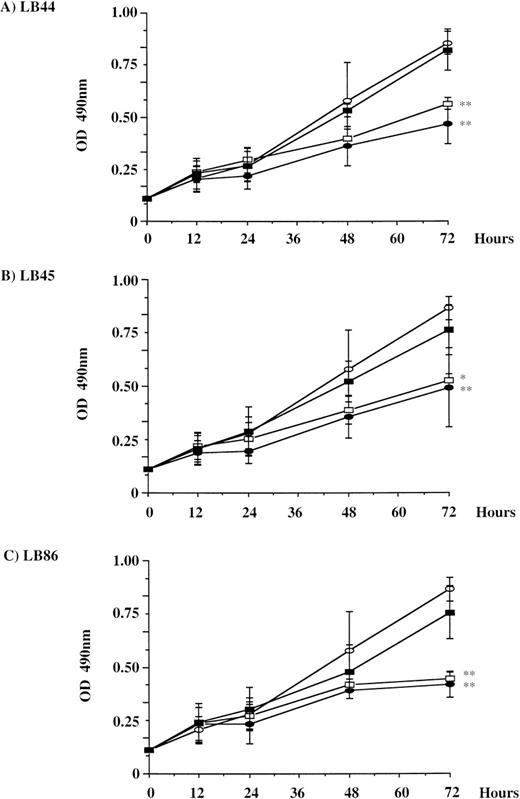
Two different tests were performed to analyze the influence of metabolic correction on the cell proliferation rate. The MTS test was performed to determine the number of metabolically active LB cells. For these experiments, transduction with LXSN was used to rule out a putative effect of the transduction itself. At 72 hours, cell numbers in normal and LBHEP/LUDSN11 cells were significantly higher than in noninfected and LB HEP/LXSN cells in the three LB HEP cell lines investigated (Fig 3). The cell doubling time was 34.5 hours for LB44 versus 19.4 hours for LB44-LUDSN11, 32.5 hours for LB45 versus 21 hours for LB45-LUDSN11, and 38.25 hours for LB86 versus 21.1 hours for LB86-LUDSN11. As a reference, LBN doubling time was 18.7 hours. The second test determined the proliferation index by counting the cells at different time intervals (Table 2). In these experimental conditions, the proliferation index was 10.3 versus 21.1 after 10 days of culture for the LB44 versus LB44-LUDSN11 cells, respectively, corresponding to a doubling time of 46.7 hours versus 22.7 hours. As a reference, the proliferation index was 21.9 for the LBN cells and the doubling time was 21.9 hours.
Cell proliferation in normal, deficient, and deficient transduced (LXSN or LUDSN11) and selected LB cell lines. Assays were performed at different time intervals (0 to 72 hours) with the LB44 (A), LB45 (B), and LB86 (C) cell lines. For each cell line, results are shown for nontransduced (•), LXSN-transduced (□), and LUDSN-transduced cells (▪). As a reference, a normal LB cell line is also shown (○). Determination was performed by the CellTiter 96rAQueous Solution Cell Proliferation Assay (Promega Corp; MTS test). Results of experiments for each cell line were the mean of five determinations from three independent experiments. * P < .05, **P < .01 v the LBHEP/LUDSN11 cells.
Cell proliferation in normal, deficient, and deficient transduced (LXSN or LUDSN11) and selected LB cell lines. Assays were performed at different time intervals (0 to 72 hours) with the LB44 (A), LB45 (B), and LB86 (C) cell lines. For each cell line, results are shown for nontransduced (•), LXSN-transduced (□), and LUDSN-transduced cells (▪). As a reference, a normal LB cell line is also shown (○). Determination was performed by the CellTiter 96rAQueous Solution Cell Proliferation Assay (Promega Corp; MTS test). Results of experiments for each cell line were the mean of five determinations from three independent experiments. * P < .05, **P < .01 v the LBHEP/LUDSN11 cells.
Proliferation Index in LBN and LB44 Cells, in Transduced and Selected LB44-LUDSN11 Cells, and in a Mixture (80/20) of LB44 and Selected LB44-LUDSN11 Cells
| . | Day 5/Day 0 . | Day 10/Day 0 . |
|---|---|---|
| LBN | 7.50 ± 1.15* | 21.9 ± 5.2* |
| LB44 | 3.55 ± 1.10 | 10.3 ± 4.8 |
| LB44-LUDSN11 | 4.55 ± 1.04† | 21.1 ± 2.6* |
| LB44/LB44-LUDSN11 (80/20) | 4.41 ± 1.01† | 17.3 ± 4.6* |
| . | Day 5/Day 0 . | Day 10/Day 0 . |
|---|---|---|
| LBN | 7.50 ± 1.15* | 21.9 ± 5.2* |
| LB44 | 3.55 ± 1.10 | 10.3 ± 4.8 |
| LB44-LUDSN11 | 4.55 ± 1.04† | 21.1 ± 2.6* |
| LB44/LB44-LUDSN11 (80/20) | 4.41 ± 1.01† | 17.3 ± 4.6* |
The proliferation index was determined by dividing the number of cells at t = x days by the number of cells at t = 0 day. Data are the mean ± SD. The mean proliferation index is shown for five determinations from three independent experiments.
P < .01 v LB44 cells.
P < .05 v LB44 cells.
Comparison of UROD activity in LB44/LB44-LUDSN11 mixed-cell experiments between different time intervals indicated that there was a 1.3- and 2.3-fold enrichment of enzymatically corrected cells at days 5 and 10 of coculture, respectively (Fig 4).
Enrichment of enzymatically corrected cells in vitro and in vivo in mixed-cell culture experiments evidenced by the increase in enzymatic activity. LB44 (□), LB44-LUDSN (▧), and a mixture of LB44 (80%) and LB44-LUDSN (20%) (▨) was cultured in vitro for 5 and 10 days or injected into immunodeficient mice and analyzed after 30 to 45 days. LB cells or B-cell tumors were resected and UROD activity was measured. * P < .05, **P < .01 v the LB44/LB44-LUDSN11 cells at day 0.
Enrichment of enzymatically corrected cells in vitro and in vivo in mixed-cell culture experiments evidenced by the increase in enzymatic activity. LB44 (□), LB44-LUDSN (▧), and a mixture of LB44 (80%) and LB44-LUDSN (20%) (▨) was cultured in vitro for 5 and 10 days or injected into immunodeficient mice and analyzed after 30 to 45 days. LB cells or B-cell tumors were resected and UROD activity was measured. * P < .05, **P < .01 v the LB44/LB44-LUDSN11 cells at day 0.
In vivo cell proliferation in immunodeficient mice.
The subcutaneous injection of LB cells into immunodeficient mice resulted in the development of a B-cell tumor (0.1 to 1.5 g in weight for the tumor compared with a mean weight of 28 g for the whole mouse) at the site of inoculation between 30 and 45 days after injection. Cell proliferation was estimated in terms of increase in UROD enzymatic activity in a mixture of deficient (80%) and transduced (20%) cells.
Control experiments with LB44 and LB44-LUDSN11 cells (Fig 4) showed that UROD activity was not modified during engraftment. By contrast, UROD activity in the mixture of deficient (80%) and transduced (20%) cells was increased 1.9-fold compared with the initial UROD activity. Mice injected with LB44-deficient cells evidenced higher plasma porphyrin concentrations at the day of death (24.7 nmol/L; range, 7.4 to 73.8 nmol/L) than did noninjected control mice (4.6 nmol/L; range, 3.9 to 5.6 nmol/L) or injected mice with LBN (5.1 nmol/L; range, 2.9 to 5.9 nmol/L) or with LB44-LUDSN11 (7.2 nmol/L; range, 5.4 to 10.7 nmol/L).
Fluorescence-based selection of retrovirally transduced cells.
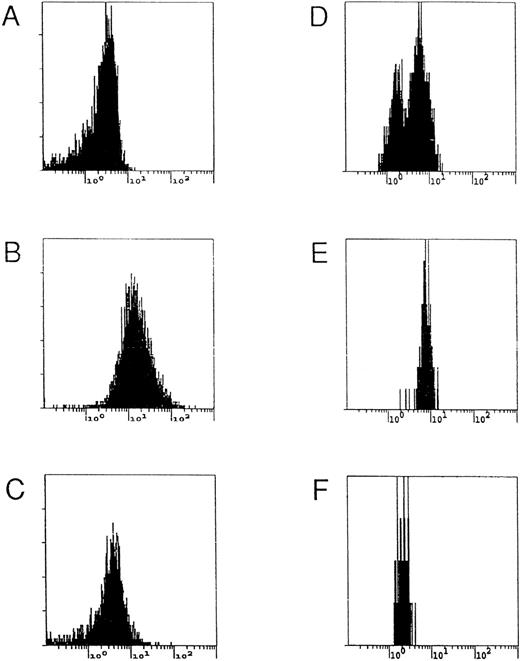
Deficient LB cells are characterized by an accumulation of porphyrins resulting from the deficient UROD activity. In genetically modified cells, the metabolic flow from uroporphyrinogen to protoporphyrin and heme was restored and a decrease in porphyrin accumulation was observed (Table 1). A mixture of deficient (80%) and corrected (20%) LBHEP cells were cocultured and analyzed by flow cytometry (Fig 5). Two overlapping cell populations were identified, which correspond to the enzymatically corrected (ie, low fluorescent) and noncorrected (ie, high fluorescent) cells (Fig5D). After sorting, the cell population with low fluorescence (Fig 5F) had a normal UROD activity and normal porphyrin accumulation (Table 3). Southern blot analysis demonstrated that the low fluorescence fraction (Fig 5F) of cells had one copy of retroviral vector per cell, the mixed cells had 0.2 copy per cell, and the high fluorescent fraction (Fig 5E) had no band corresponding to a retroviral vector sequence (data not shown).
Flow cytometry and FACS analyses of different populations of LB cells. LBN (A), LB45 (B), selected LB45-LUDSN11 (C), and a mixture of LB45/LB45-LUDSN11 (80/20) (D) cells were analyzed by flow cytometry using a UV light excitation to characterize porphyrin fluorescence in the cells. FACS analysis was then performed on mixed cells. After sorting, the cell populations corresponding to high (E) and low fluorescence (F) were reanalyzed.
Flow cytometry and FACS analyses of different populations of LB cells. LBN (A), LB45 (B), selected LB45-LUDSN11 (C), and a mixture of LB45/LB45-LUDSN11 (80/20) (D) cells were analyzed by flow cytometry using a UV light excitation to characterize porphyrin fluorescence in the cells. FACS analysis was then performed on mixed cells. After sorting, the cell populations corresponding to high (E) and low fluorescence (F) were reanalyzed.
UROD Enzymatic Activity and Porphyrin Level in Untransduced and Transduced HEP LB Cells Before and After FACS Selection
| Cell Population . | UROD Activity (U/mg) . | %v LBHEPLUDSN11 . | Porphyrin Level (fmol/1,000 cells) . | % v LBHEP . |
|---|---|---|---|---|
| Experiment no. 1 | ||||
| LB44 | 219 ± 123 | 11 | 9.29 ± 1.92 | 100 |
| LB44-LUDSN11 | 1,927 ± 266 | 100 | 0.29 ± 0.34 | 3 |
| LB44/LB44-LUDSN11 (80/20) | 647 ± 187 | 34 | 7.71 ± 0.09 | 83 |
| FACS sorted | ||||
| High fluorescence | 539 ± 101 | 28 | 7.88 ± 2.24 | 85 |
| Low fluorescence | 1,459 ± 148 | 76 | 0.95 ± 0.13 | 10 |
| Experiment no. 2 | ||||
| LB45 | 288 ± 108 | 10 | 13.28 ± 2.53 | 100 |
| LB45-LUDSN11 | 2,901 ± 543 | 100 | 0.06 ± 0.02 | 0.5 |
| LB45/LB45-LUDSN11 (80/20) | 1,100 ± 308 | 38 | 9.94 ± 0.18 | 75 |
| FACS sorted | ||||
| High fluorescence | 880 ± 236 | 30 | 6.53 ± 1.62 | 49 |
| Low fluorescence | 3,521 ± 1,635 | 121 | 0.64 ± 0.58 | 5 |
| Cell Population . | UROD Activity (U/mg) . | %v LBHEPLUDSN11 . | Porphyrin Level (fmol/1,000 cells) . | % v LBHEP . |
|---|---|---|---|---|
| Experiment no. 1 | ||||
| LB44 | 219 ± 123 | 11 | 9.29 ± 1.92 | 100 |
| LB44-LUDSN11 | 1,927 ± 266 | 100 | 0.29 ± 0.34 | 3 |
| LB44/LB44-LUDSN11 (80/20) | 647 ± 187 | 34 | 7.71 ± 0.09 | 83 |
| FACS sorted | ||||
| High fluorescence | 539 ± 101 | 28 | 7.88 ± 2.24 | 85 |
| Low fluorescence | 1,459 ± 148 | 76 | 0.95 ± 0.13 | 10 |
| Experiment no. 2 | ||||
| LB45 | 288 ± 108 | 10 | 13.28 ± 2.53 | 100 |
| LB45-LUDSN11 | 2,901 ± 543 | 100 | 0.06 ± 0.02 | 0.5 |
| LB45/LB45-LUDSN11 (80/20) | 1,100 ± 308 | 38 | 9.94 ± 0.18 | 75 |
| FACS sorted | ||||
| High fluorescence | 880 ± 236 | 30 | 6.53 ± 1.62 | 49 |
| Low fluorescence | 3,521 ± 1,635 | 121 | 0.64 ± 0.58 | 5 |
The mean activity is shown for three determinations in one experiment for the mixture LB44/LB44-LUDSN11 (80/20) and in three independent experiments for the mixture LB45/LB45-LUDSN11 (80/20). Theoretical values of UROD activity were 561 U/mg for the mixture in experiment no. 1 and 811 U/gm for the mixture in experiment no. 2. Data are the mean ± SD.
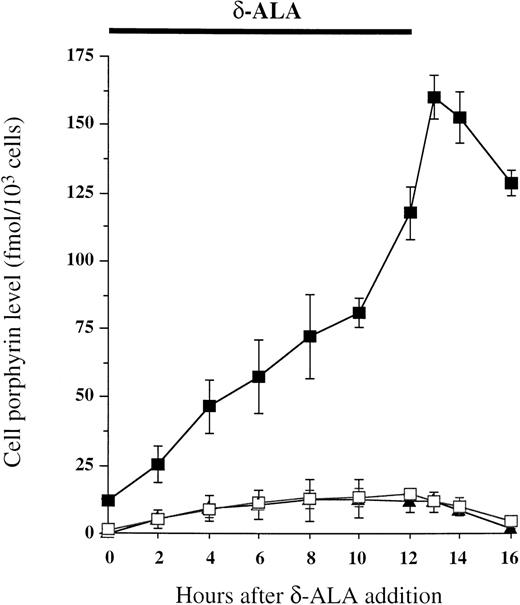
A similar experiment was performed with another selected cell line, LB45-LUDSN7. In these experiments, transduced cells had only 67% of normal UROD activity (1,836 ± 460 U/mg in LB45-LUDSN7 v2,728 ± 238 U/mg in LBN; P < .01) and the porphyrin level was 0.6 ± 0.2 fmol/103 cells in LB45-LUDSN7 versus 0.12 ± 0.04 fmol/103 cells in LBN cells (P < .05). The mixture of transduced (20%) and nontransduced cells (80%) showed a single peak, ie, the two populations were not discriminated by flow cytometry, in relation to the weak residual fluorescence of transduced cells. Thus, we used a different strategy, based on an increase in porphyrin accumulation in untransduced deficient LB cells by exposure to ALA (Fig 6). After 12 hours of ALA exposure, normal or genetically corrected cells were able to metabolize porphyrinogens accumulated in the cell within 4 hours. By contrast, deficient cells still presented a high accumulation of porphyrin(ogen)s. The maximum difference of porphyrin accumulation, determined by the LB45/LB45-LUDSN7 ratio, was observed 4 hours after ALA incubation (39.1 ± 8 or 27.6 ± 2 when estimated in intact cell by flow cytometry or in cell lysate, respectively, v 18.2 ± 5.1 before ALA exposure, P < .05). After ALA exposure, flow cytometry analysis showed the mixture of LB45/LB45-LUDSN7 cells (80/20) to have two nonoverlapping cell populations. Sorted cell population with low fluorescence showed the same UROD activity (2,101 ± 310 U/mg) and porphyrin level (0.65 ± 0.31 fmol/103 cells) than the transduced cells (UROD activity, 1,836 ± 460 U/mg; porphyrin level, 0.6 ± 0.2 fmol/103 cells) with one copy of retroviral vector per cell (data not shown).
In vitro cell porphyrin accumulation during and after ALA exposure. LB45 (▪), LBN (▴), and LB45-LUDSN7 (□) cells were cultured in RPMI 1640 and 7% FCS in the presence of 1 mmol/L ALA, 2 mmol/L melatonin, and 50 μmol/L FeSO4 for 12 hours as indicated at the top of the figure. The medium was then removed, and cells were washed with PBS and then resuspended with regular medium (RPMI 1640 and 7% FCS). At regular time intervals, the porphyrin level was measured in the cells.
In vitro cell porphyrin accumulation during and after ALA exposure. LB45 (▪), LBN (▴), and LB45-LUDSN7 (□) cells were cultured in RPMI 1640 and 7% FCS in the presence of 1 mmol/L ALA, 2 mmol/L melatonin, and 50 μmol/L FeSO4 for 12 hours as indicated at the top of the figure. The medium was then removed, and cells were washed with PBS and then resuspended with regular medium (RPMI 1640 and 7% FCS). At regular time intervals, the porphyrin level was measured in the cells.
In a separate experiment, cell survival was quantitated in untransduced cells without (90% ± 1.4%) or with ALA exposure (73.9% ± 4.2%, P < .05) and also in transduced cells (91.8% ± 2.4% without ALA v 91.1% ± 2.1% after ALA exposure).
DISCUSSION
Porphyrias are inherited metabolic diseases characterized by specific enzyme defects along the heme biosynthetic pathway.1Therapy is often limited to supportive care that is only partially successful, especially in the severe forms, ie, CEP, HEP, some cases of erythropoietic protoporphyria (EPP) inherited either as a dominant or a recessive trait, and homozygous forms of acute hepatic porphyrias. These are good candidates for gene therapy, because the genetic defect is well characterized at the molecular level and inherited as a recessive trait. We have previously documented sufficient gene transfer rate and metabolic correction in different cells to indicate that CEP is a good candidate for treatment by gene therapy in hematopoietic stem cells (HSC).16,17 We also showed a persistance of the expression of the transgene during erythroid differentiation by using the in vitro model of K562 cells.18
In HEP, the UROD enzymatic defect is responsible for the accumulation of large amounts of uroporphyrin in the different tissues investigated. Both erythropoietic cells and liver are involved in the disease. The available treatments are mostly unsatisfactory, and gene therapy by retrovirus-mediated insertion of the normal UROD gene into hematopoietic or/and hepatic cells seems to be the most appropriate treatment in the future.19
We constructed a recombinant retroviral vector LUDSN to mediate insertion and expression of human UROD cDNA in three lymphopoietic target cell populations. We documented sufficient metabolic correction of the porphyric phenotype in three different deficient LB cell lines from HEP after gene transfer of the UROD cDNA. A complete correction of enzymatic activity was found in the first two deficient lymphoblastoid cell lines and only 71% of normal UROD activity in the third one. This difference in expression is probably related to the site of insertion of the provirus in the genome of the different cell lines. Nevertheless, the porphyrin level was normal in all of the transduced LB cells. Therefore, it may not be necessary to achieve a 100% normal UROD activity to have a normal porphyrin level and a disappearence of the photosensitivity for the patient.
One approach for the treatment by gene therapy of hematologic genetic disease, such as HEP and other erythropoietic porphyrias, is to introduce ex vivo a normal counterpart of the defective gene into HSC from the affected patient and then to return them as an autologous bone marrow transplant.7 19
Restoration of the functionality of all the HSC of a patient will necessitate a number of consecutive autologous transplantations of genetically modified cells. To avoid correcting all the HSC, we wondered whether transformation of uroporphyrinogen from a deficient cell by a UROD enzyme synthesized in a normal cell was possible. Metabolic cross-correction is well-established for human lysosomal storage diseases such as mucopolysaccharidosis type I (Hurler syndrome),20 type II (Hunter syndrome),21 type VII (Sly disease),22-24 or metachromatic leukodystrophy.25 For adenosine deaminase (ADA) deficiency, the toxic circulating substrate can be reduced by repeated intramuscular injections of bovine ADA cross-linked to polyethylene glycol26 or by inserting a normal ADA allele into peripheral lymphocytes and circulating stem cells27-30 or into epidermal keratinocytes.31 Our in vitro experiments with deficient cells showed a high level of porphyrins in the supernatant, implying that uroporphyrinogen or uroporphyrin are able to cross the cell membrane of deficient cells, but these porphyrin(ogen)s cannot be metabolized further because a cross-correction was not observed. This is probably due to the oxidation of uroporphyrinogen into uroporphyrin that becomes a nonmetabolisable substrate and/or due to the high affinity of porphyrins to serum proteins, such as albumin or hemopexin.32 These results suggest that a high percentage of transduced HSC must be corrected for a benefit in gene therapy trials involving HEP. However, in the case of a selective advantage of corrected cells, a benefit could be obtained even if a small percentage of genetically corrected cells are reinfused to the patient. A striking observation in this study was the growth inhibition of LB cells in the presence of the metabolic defect. Transformation of uroporphyrinogen by UROD enzyme encoded by the transgene reduced porphyrin accumulation and increased the half-life of the cell. Growth inhibition can be due to a heme defect and/or prolonged exposure to the porphyrins. Heme synthesis is necesary for the electron-transport chain, and a defective activity of one of the heme pathway enzymes could affect cell proliferation. Koningsberger et al33demonstrated that an elevated protoporphyrin level causes high intracellular damage and inhibits HepG2 cell proliferation. How protoporphyrin exerts its toxic dark effect has not been well clarified. Nevertheless, it has been demonstrated that both protoporphyrin and uroporphyrin, in the presence of H2O2, can potentiate the peroxidase-catalyzed oxidation of NADPH.34 35
In our cell proliferation studies, the selective advantage in in vivo experiments was lesser than in vitro. This could be due to much more prolonged exposure to uroporphyrin in in vitro cultures. In vivo, uroporphyrin excreted from LB cells is bound to albumin and hemopexin and then excreted in the urine. Growth of transduced cells was higher than in untransduced cells, suggesting a competitive advantage for these corrected cells. Because bone marrow is the target organ for somatic gene therapy of HEP and this is a proliferative organ, corrected HSC could have a selective advantage.
To facilitate future ex vivo gene therapy in human beings, it could be advantageous to design efficient selection procedures to increase the frequency of genetically corrected patient cells before autologous transplantation. The standard approach to selecting transduced cells is the use of vectors encoding genes conferring cellular resistance to potentially toxic drugs such as neomycin, hygromycin, puromycin, and multidrug resistance (MDR-1).7 That study described a rapid and efficient procedure for isolating retrovirally transduced HEP LB cell lines. Because normal metabolic flow from uroporphyrinogen to coproporphyrinogen was restored in transduced cells, the porphyrin level (measured by fluorescence) returned to normal. The difference of fluorescence between transduced and untransduced cells was detected by flow cytometry analysis, and two populations could be isolated differing in their fluorescence. Culture of the sorted LB cells with low fluorescence led to the generation of a homogeneous cell population that was metabolically corrected.
ALA is an immediate precursor in the biosynthetic pathway of heme. Normally, the rate of synthesis of protoporphyrin is determined by the rate of ALA synthesis, which, in turn, is regulated via a feedback control mechanism dependent on the concentration of free heme.1 The presence of exogeneous ALA bypasses the feedback control and thus may induce the intracellular accumulation of porphyrins. When porphyrin levels were not very different, ALA exposure increased the difference in fluorescence between transduced and untransduced cells, thus improving the discriminating power of the method. In genetically modified cells, partially restored metabolic flow from uroporphyrinogen to protoporphyrin led to a lesser accumulation of porphyrins in the cells.
In the current study, LB cells from HEP patients were used as a model to develop methods for the direct selection of genetically corrected cells. Studies must now be performed on the HSC, the ultimate targets for erythropoietic porphyria somatic gene therapy. Analogous selection systems, based solely on the expression of a therapeutic gene, could be used in other metabolic disorders, in which fluorescent substrates are directly accessible.
Finally, HSC gene therapy in this porphyria is not the unique therapy to be considered. Because there are two main target tissues, bone marrow and liver, the ideal therapy should concern both tissues. We do not know whether gene transfer in vivo in HSC could be sufficient to metabolize the excess of uroporphyrinogen produced by the different deficient tissues, especially the liver. Also, the decrease in porphyrin synthesis in the bone marrow should be sufficient to limit the excess of uroporphyrin accumulated in the body and then to suppress the porphyric phenotype. The future availability of a mouse model of the disease will permit ex vivo gene therapy experiments on hematopoietic or/and hepatic cells to solve this problem.
ACKNOWLEDGMENT
The authors thank Prof R. Enriquez de Salamanca and Prof J.M. Mascaro for providing patients specimens and Laboratoire Généthon (Evry, France) for transforming human lymphocytes.
Supported by grants from Association Française Contre les Myopathies, from Institut National de la Santé et de la Recherche Médicale (Grant No. CRI 9508), and from Conseil Régional d’Aquitaine. A.F. was supported by a postdoctoral TMR Marie Curie research fellowship.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Hubert de Verneuil, MD, PhD, Laboratoire de Pathologie Moléculaire et Thérapie Génique, Université Victor Segalen Bordeaux 2, 146 rue Léo Saignat, 33076 Bordeaux Cédex, France; e-mail: verneuil@u-bordeaux2.fr.

![Fig. 2. Porphyrin level in normal, deficient, and transduced deficient LB cells. (A) LB cells were maintained in exponential phase of culture and then placed in culture at 2 × 105cells/mL. After 48 hours, porphyrins were measured ([▧] Cell porphyrin level, [□] Supernatant porphyrin level) as described in Materials and Methods. Data are the mean ± SD of four to six determinations. ** P < .01 v noninfected LBN cells. (B) Porphyrin level in LBN cells, deficient LB45 cells, and a mixture (50/50) of LB45 and LBN cells. Cells were placed in culture at 2 × 105 cells/mL. Porphyrin level was measured by spectrofluorimetry immediatly ([▨] 0 hour) and after 24 (□), 48 (▧), and 72 hours (▨), as described in Materials and Methods. Data are the means ± SD of six independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/94/2/10.1182_blood.v94.2.465/5/m_blod41414002x.jpeg?Expires=1769104101&Signature=gjqE58SeRpAIkDE7muriPt3q7FA221UzLKE5XLrWTeTxEEGK2xjjrXkglxMvbsW0AjfynqTSAo9~WdB-qc~6L7kbFhpfpiEsC7XWnfSTBQcj9BkNCU6Knh8~NB1lGIU7LC8QbOzR6KcMila2cM1ybnm7ht1U6cptjuJI7Qjc1ctR9vzoccTBlwPpHn-0IASVSGfIZ58kUbyLJV8YxFQGaWH7zfox-nWm6P5zOnQWJiMpoWB38mt8THNZE5-8wrjSXeV~6sQz75RmYVCxZZ4TFI7-EuPlp3cQcMC2brAGeLPyZhtIin8jlBwi7hhCB7BAMTEX3v7PVM04AEXDgHozGA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal