Abstract
Deletion in chromosome bands 11q22-q23 is one of the most common chromosome aberrations in B-cell chronic lymphocytic leukemia (B-CLL). It is associated with extensive lymph node involvement and poor survival. The minimal consensus deletion comprises a segment, which contains the ATM gene presenting an interesting candidate gene, as mutations in ATM predispose A-T patients to lymphoid malignancies. To investigate a potential pathogenic role of ATM in B-cell tumorigenesis, we performed mutation analysis of ATM in 29 malignant lymphomas of B-cell origin (B-CLL = 27; mantle cell lymphoma, [MCL] = 2). Twenty-three of these carried an 11q22-q23 deletion. In five B-CLLs and one MCL with deletion of one ATMallele, a point mutation in the remaining allele was detected, which resulted in aberrant transcript splicing, alteration, or truncation of the protein. In addition, mutation analysis identified point mutations in three cases without 11q deletion: two B-CLLs with one altered allele and one MCL with both alleles mutated. In four cases analyzed, theATM alterations were not present in the germ line indicating a somatic origin of the mutations. Our study demonstrates somatic disruption of both alleles of the ATM gene by deletion or point mutation and thus its pathogenic role in sporadic B-cell lineage tumors.
MUTATIONS IN THE ATM (ataxia telangiectasia mutated) gene are responsible for the autosomal recessive disorder ataxia telangiectasia (A-T). A-T is characterized by neurologic degeneration, immunodeficiency, infertility, radiation sensitivity, and a marked predisposition to cancer (for review, see Sedgewick and Boder,1 Lavin and Shiloh,2 and Jeggo et al3). The cellular phenotype of A-T includes hypersensitivity to ionizing radiation, genome instability, and dysregulation of the cell cycle checkpoints.2,3 The ATM gene is localized in the chromosomal region 11q22.3-q23.1 and consists of 66 exons spanning 146 kb of genomic DNA. The gene encodes a 370-kD nuclear phosphoprotein sharing homology with phosphatidylinositol 3-kinase (PI-3-K). It is known that PI-3-K–related proteins function in DNA repair, in DNA recombination, and in cell cycle control.4-10
One of the most intriguing features of A-T is the increased predisposition to develop malignancies. The predominant cancers among these are neoplasms of the lymphoid system including both B- and T-cell tumors11 with a risk for leukemia approximately 70 times higher than in the normal population.12 Individuals with heterozygous ATM mutations have also been suggested to be at elevated risk for cancer, particularly for carcinoma of the breast.13 Several studies of breast cancer patients, however, did not find ATM mutations in a frequency higher than expected by chance (eg, Vorechovsky et al,14 FitzGerald et al,15 Bay et al,16 and Chen et al17). Thus, the role of ATM in breast tumors remains unresolved. Recently, biallelic mutations of the ATM gene have been identified in T-prolymphocytic leukemia (T-PLL) of patients without A-T history indicating a tumor-suppressor function of ATM in sporadic T-PLL.18-21 Although loss of heterozygosity in the 11q22-q23 region has been frequently observed in various types of tumors22 (and references therein), to date T-PLL is the only sporadic tumor for which recurrent somatic inactivation of both alleles of the ATM gene has been demonstrated.
In B-cell chronic lymphocytic leukemia (B-CLL), we identified deletion of the chromosomal region 11q22-q23 as a recurrent aberration by using fluorescence in situ hybridization (FISH) in a large series of more than 200 B-CLLs.23,24 Loss of the region was shown to be a prognostic marker predicting poor survival.24 The commonly deleted region was defined as a 2 to 3 megabasepairs (Mbp) segment, which contains the ATM, DDX10, RDX(radixin), and FDX1 (ferredoxin 1) genes.23 24 The frequent monoallelic deletion of the gene in sporadic B-CLL and the association between ATM inactivation and the development of T- and B-cell leukemia in A-T patients had led us to analyze the ATM gene in malignant B-cell lymphoma.
MATERIALS AND METHODS
Patient and control material.
The study comprised 29 patients with malignant lymphoma of B-cell type and 50 unrelated healthy probands of central European origin. Samples were collected after informed consent. Based on morphology and immunophenotype, the tumors were classified as B-CLL (n = 27) and mantle cell lymphoma (MCL, n = 2). Dual-color interphase FISH detected deletions of chromosome bands 11q22-q23 in 22 of the B-CLL cases and one of the MCL cases (MCL-A) and translocations affecting 11q23 in two B-CLLs (B-CLL-F and -H). Many of these cases were contained in the study by Stilgenbauer et al.23 Both MCLs were shown to have a t(11;14)(q13;q32). None of the patients had clinical evidence for A-T. From six B-CLL patients and one MCL patient, skin biopsies were available serving as specimen for the analysis of the ATMgermline status.
Mutation analysis of the ATM gene.
Total RNA and genomic DNA from mononuclear cell preparations of malignant B-cell lymphomas and control persons were extracted with the Trizol reagent (Gibco BRL, Eggenstein, Germany). Genomic DNA from skin biopsy cells was isolated with the QIAamp kit (Qiagen, Hilden, Germany) and from normal healthy controls by the standard phenol-chloroform method.
The following oligonucleotides were used for polymerase chain reaction (PCR) and sequence analysis:
1A, 1B, 2A, 2B, 4A, 4A2, 4B, 5A, 5B, 6A, and 6B are listed in Stilgenbauer et al.18 Additionally used primers were 1A1 (exon 38) 5′-TGGATAAAGACACTGACT-3′; 1A2 (exon 39/40) 5′-GGATTCAGAGTCAGAGCAC-3′; 1B1 (exon 41) 5′-CAGCACAAGACTGAGCTACC-3′; 3A (exon 42/43) 5′-CTGGAATAAGTTTACAGGATCTTC-3′; 3B (exon 51) 5′-GATGATTTCATGTAGTTTTCAATTC-3′; 4A1 (exon 59) 5′-GAATGGTGCACAGGAACTG-3′; 4B3 (exon 61) 5′-TCTGTACATGTCTATCACC-3′; 5B1 (exon 52) 5′-TACCCACATATCATGTTC-3′; 39A (intron 38) 5′-CATTTTTACTCAAACTATTG-3′; 39B (intron 39) 5′-TCTTAAATCCATCTTTCTCTA-3′; 59A (intron 58) 5′-AGGTCAACGGATCATCAAAT-3′; 59B (intron 59) 5′-TTAATTTTGGGTGTCACTC-3′; 65A (intron 64) 5′-TTAAACTGTTCACCTCACTGA-3′; 65B (exon 65) 5′-GTTAAAAATAAAGGCTAAAATA-3′.
1A1, 1A2, 1B1, 4A1↑, 4B3, and 5B1 were derived from cDNA sequence data of the ATM gene (GenBank accession no. U33841). PCR amplification of the exons 39, 59, the coding part of exon 65, and the flanking intronic regions from genomic DNA was performed according to Vorechovsky et al14 using primer pairs 39A/B, 59A/B, and 65A/B.
Reverse-transcription PCR (RT-PCR) and single-strand conformation polymorphism (SSCP) analysis were performed as described previously.18 25 Single-strand cDNA was synthesized using random hexamers (GeneAmp RNA PCR System, Perkin-Elmer, Weiterstadt, Germany). Six overlapping fragments covering 6.2 kb of the coding region of the ATM transcript were amplified with primer sets 1A/B, 2A/B, 3A/B, 4A/B, 5A/B, and 6A/B. For SSCP analysis, RT-PCR products were digested with restriction endonucleases and end-labeled with T4 polynucleotide kinase in the presence of γ-33P deoxyadenosine triphosphate. Fragments were separated by electrophoresis through nondenaturing 6% polyacrylamide gels containing 5% glycerol (exclusively for runs at room temperature) in 0.5 × Tris-borate buffer. Electrophoresis was performed at room temperature or at 4°C at 8 W for at least 10 hours. Gels were dried and subjected to autoradiography.
Direct sequencing of PCR and RT-PCR products was performed by cycle sequencing with dye terminator chemistry (ABI PRISM big dye terminator cycle sequencing ready reaction kit, Perkin-Elmer). Sequencing reactions were run on a Perkin-Elmer ABI-377 automated sequencer.
To test whether the two heterozygote mutations identified in MCL-B were localized on the same ATM transcript, RT-PCR products amplified with primer set 5A/B were cloned into pT-Adv (AdvanTAge PCR Cloning Kit, Clontech, Heidelberg, Germany). Insert DNA from individual clones was amplified by colony PCR using the original PCR primers and PCR conditions. Amplified DNA fragments were subsequently sequenced.
RESULTS
Mutations in malignant B-cell lymphomas with 11q deletions.
Based on FISH analysis, 23 malignant lymphomas of B-cell type (22 B-CLL cases; one MCL case) with monoallelic deletions of chromosomal region 11q22-q23 were selected for mutation screening in the ATM gene.ATM transcript was analyzed by RT-PCR amplification of a set of six overlapping fragments representing exons 22-30, 29-36, 36-43, 42-51, 51-57, 56-65.25 This 6.2-kb part of the coding region includes the protein domains, which are responsible for the kinase function (exons 57-65) and the binding of c-Abl (exon 30), as well as a postulated rad 3 homology domain (exons 32-42) and a leucine zipper motif (exon 27).4,5,26 27 After digestion with restriction endonucleases, the RT-PCR products were subjected to SSCP analysis.
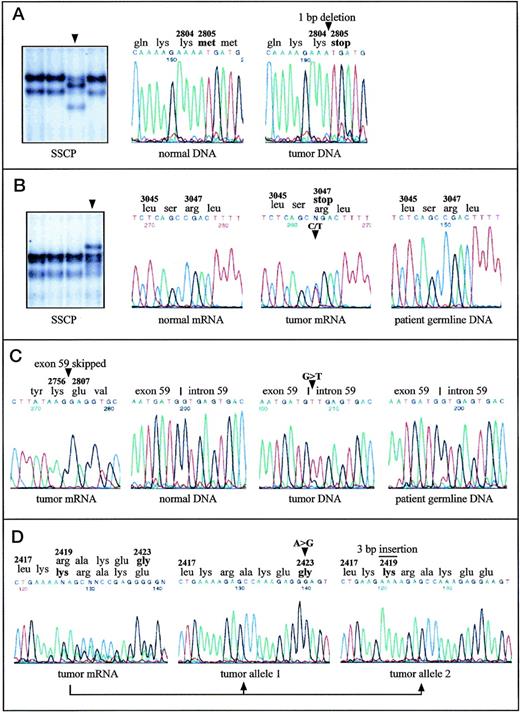
Aberrantly migrating DNA fragments were found in five cases. Direct sequencing of the corresponding RT-PCR fragments detected one single-base deletion and four single-base substitutions as detailed in Table 1. The deletion of one nucleotide in B-CLL-A causes a frameshift after codon 2804 resulting in a truncation of the protein at this position and the loss of about 70% of the kinase domain (Fig 1A). In B-CLL-B, a transition creates a new stop codon at position 3047, which leads to the removal of the last 10 amino acids from the translation product (Fig 1B). In three cases, amino acid substitutions were detected. Two of them are predicted to alter the protein structure in the PI-3-K domain (in B-CLL-D and B-CLL-E), the other one affected the so-called rad 3 homology domain (in B-CLL-C). The nonmalignant cells from B-CLL-B (Fig 1B), B-CLL-D, and B-CLL-E, obtained by skin biopsy, did not harbor the same nucleotide changes as the malignant cells, indicating a somatic, but not a germline origin of the mutations in all three cases.
Alterations Affecting Both ATM Alleles in Malignant B-Cell Lymphomas of Non–A-T Individuals
| B-Cell Lymphoma . | First Allele . | Second Allele . | ||
|---|---|---|---|---|
| DNA*,† . | Gene Product . | DNA† . | Gene Product . | |
| B-CLL-A | del(11q22-q23) | — | 8412delA (exon 59) | Frameshift after codon 2804 + truncation of the protein |
| B-CLL-B1-153 | del(11q22-q23) | — | 9139C > T (exon 65) | Arg3047ter |
| B-CLL-C | del(11q22-q23) | — | 5858C > G (exon 41) | Thr1953Arg |
| B-CLL-D1-153 | del(11q22-q23) | — | 9023G > A (exon 65) | Arg3008His |
| B-CLL-E1-153 | del(11q22-q23) | — | 9054A > C (exon 65) | Lys3018Asn |
| MCL-A1-153 | del(11q22-q23) | — | IVS59 + 1G > T (intron 59) | Skipping of exon 59: Del Val2757-Met2806 |
| B-CLL-F | Nondeleted‡ | Unknown | 5558A > T (exon 39) | Asp1853Val |
| B-CLL-G | Nondeleted | Unknown | 7258G > C (exon 51) | Ala2420Pro |
| MCL-B | 7268A > G (exon 51) | Glu2423Gly | 7250insGAA (exon 51) | Ins Lys2419 |
| B-Cell Lymphoma . | First Allele . | Second Allele . | ||
|---|---|---|---|---|
| DNA*,† . | Gene Product . | DNA† . | Gene Product . | |
| B-CLL-A | del(11q22-q23) | — | 8412delA (exon 59) | Frameshift after codon 2804 + truncation of the protein |
| B-CLL-B1-153 | del(11q22-q23) | — | 9139C > T (exon 65) | Arg3047ter |
| B-CLL-C | del(11q22-q23) | — | 5858C > G (exon 41) | Thr1953Arg |
| B-CLL-D1-153 | del(11q22-q23) | — | 9023G > A (exon 65) | Arg3008His |
| B-CLL-E1-153 | del(11q22-q23) | — | 9054A > C (exon 65) | Lys3018Asn |
| MCL-A1-153 | del(11q22-q23) | — | IVS59 + 1G > T (intron 59) | Skipping of exon 59: Del Val2757-Met2806 |
| B-CLL-F | Nondeleted‡ | Unknown | 5558A > T (exon 39) | Asp1853Val |
| B-CLL-G | Nondeleted | Unknown | 7258G > C (exon 51) | Ala2420Pro |
| MCL-B | 7268A > G (exon 51) | Glu2423Gly | 7250insGAA (exon 51) | Ins Lys2419 |
The deletion mapping by interphase FISH is described in Stilgenbauer et al23; del(11q22-q23) indicates loss of one copy of the chromosomal region 11q22-q23 containing ATM.
The exon numbering is according to Uziel et al6 and Platzer et al7 (GenBank no. U82828); nucleotide positions refer to the sequence of the ATM transcript (GenBank no.U33841). The first codon of the open reading frame was designated +1. The nomenclature of the mutations was used as suggested by Beaudet and Tsui (1993).46
B-CLL-F has a translocation in 11q23, which is localized approximately 2 Mbp distal from the ATM gene.23
For those patients, the analysis of the germline status ofATM was possible.
Point mutations in the ATM gene in malignant B-cell lymphomas with (A, B, C) and without (D) monoallelic deletions of ATM. Mutations are indicated by arrowheads or by a bar. A, B-CLL-A: SSCP analysis of a 242-bp restriction fragment derived from the RT-PCR product containing exons 56-65 identified an aberrantly migrating fragment. DNA sequence analysis of the respective fragment showed 8412delA causing frameshift and truncation of the protein. DNA sequences of PCR amplified exon 59 are shown. (B) B-CLL-B: SSCP analysis of a 307-bp restriction fragment of the RT-PCR product encompassing exons 56-65 and DNA sequence analysis of the corresponding RT-PCR products identified the nonsense mutation 9139C>T (Arg3047ter). Note that the amount of cells with an 11q22-q23 deletion was 58% in the tumor sample, and therefore the signal intensity of the normal allele was unusually high. DNA sequence analysis of exon 65 from patient’s germline DNA showed absence of the mutation in the germ line. (C) MCL-A: The splice-donor site mutation in intron 59 (IVS59+1G>T), which causes skipping of exon 59 from the ATMtranscript and loss of 50 amino acids, was detected by DNA sequencing of the RT-PCR fragment containing exons 56-65 and by sequencing of PCR amplified exon 59 and its flanking intronic regions. The patient’s germ line DNA did not contain the mutation. (D) MCL-B: Cloning and subsequent sequencing of RT-PCR fragments harboring exons 51-57 allowed assignment of the two heterozygous mutations 7268A>G and 7250insGAA to two separate ATM transcripts. Thus, both alleles were affected by two independent mutations.
Point mutations in the ATM gene in malignant B-cell lymphomas with (A, B, C) and without (D) monoallelic deletions of ATM. Mutations are indicated by arrowheads or by a bar. A, B-CLL-A: SSCP analysis of a 242-bp restriction fragment derived from the RT-PCR product containing exons 56-65 identified an aberrantly migrating fragment. DNA sequence analysis of the respective fragment showed 8412delA causing frameshift and truncation of the protein. DNA sequences of PCR amplified exon 59 are shown. (B) B-CLL-B: SSCP analysis of a 307-bp restriction fragment of the RT-PCR product encompassing exons 56-65 and DNA sequence analysis of the corresponding RT-PCR products identified the nonsense mutation 9139C>T (Arg3047ter). Note that the amount of cells with an 11q22-q23 deletion was 58% in the tumor sample, and therefore the signal intensity of the normal allele was unusually high. DNA sequence analysis of exon 65 from patient’s germline DNA showed absence of the mutation in the germ line. (C) MCL-A: The splice-donor site mutation in intron 59 (IVS59+1G>T), which causes skipping of exon 59 from the ATMtranscript and loss of 50 amino acids, was detected by DNA sequencing of the RT-PCR fragment containing exons 56-65 and by sequencing of PCR amplified exon 59 and its flanking intronic regions. The patient’s germ line DNA did not contain the mutation. (D) MCL-B: Cloning and subsequent sequencing of RT-PCR fragments harboring exons 51-57 allowed assignment of the two heterozygous mutations 7268A>G and 7250insGAA to two separate ATM transcripts. Thus, both alleles were affected by two independent mutations.
In MCL-A, RT-PCR fragment 4 containing exons 56-65 showed a significantly smaller size on agarose gels and presented a complete loss of exon 59 when sequenced directly (Fig 1C). Further genomic DNA sequencing showed a G>T transversion at the first nucleotide of intron 59, in the donor-splice junction of exon 59/intron 59, at position 8418+1 (Table 1). According to the RT-PCR sequence data, this nucleotide substitution obviously leads to complete skipping of the in-frame exon 59 from the ATM transcript, which removes 50 amino acids from the kinase domain of the ATM protein. Sequence analysis of genomic DNA from skin cells of this patient showed that this splice-site mutation was not present in nonmalignant cells, indicating a somatic mutation origin (Fig 1C).
Mutations in malignant B-cell lymphomas without 11q deletions.
In addition to B-cell lymphomas with monoallelic 11q deletions, five B-CLL cases and one MCL case without microscopically detectable deletions of chromosome bands 11q22-q23 were searched for ATMmutations (by SSCP and sequence analysis). In one of the two B-CLLs with a translocation approximately 2 Mbp distal from the ATM gene locus, a heterozygous missense mutation in the rad 3 homology domain was identified (B-CLL-F; Table 1). In B-CLL-G, a nucleotide substitution in one allele lead to an amino acid replacement at position 2420 (Table 1). In the same protein region, two heterozygous mutations were detected in MCL-B (Fig 1D): a trinucleotide insertion incorporated an additional lysine at position 2419 and a single-base substitution caused an amino acid exchange at position 2423. Cloning and subsequent sequencing of RT-PCR fragments, which covered both mutational sites allowed the assignment of the mutations to separateATM transcripts, thus indicating two independent mutation events affecting both alleles in MCL-B (Fig 1D).
Identification of a common genetic polymorphism.
In contrast to the aforementioned point mutations, in our series one amino acid substitution was observed more than once: Asp1853Asn (5557G>A) was found in six of the 29 tumor samples analyzed (21%; B-CLL-B, B-CLL-E, B-CLL-H, B-CLL-J, B-CLL-K, B-CLL-L). An estimation of the germline status by sequence analysis of genomic DNA from skin cells was possible in four cases (B-CLL-B, B-CLL-E, B-CLL-J, and B-CLL-K). In B-CLL-J, the 5557A allele was absent in the nonmalignant cells indicating its somatic origin, whereas in the other three patients the 5557A allele was already present in the germ line in a heterozygous form. Interestingly, in one of those cases (B-CLL-B), the 5557A allele was apparently lost during tumorigenesis, as it was only detectable in a low amount in the tumor sample corresponding to the amount of cells without 11q deletion in the specimen (concluded from sequence analysis of both genomic DNA and RT-PCR products). The analysis of 50 samples from healthy controls detected the 5557A allele in 11 samples (22%). Nine samples were heterozygous and two samples were found to be homozygous for 5557A, resulting in an allele frequency of 0.13 in the population studied. Because of the similar frequency of Asp1853Asn positive tumor samples and carriers in the control population, this variant has to be considered a common polymorphism, which seems not to be disease associated. In a recent study of breast cancer patients in Britain, 5557A was also identified as polymorphic allele.28 The single-base substitution 5558A>T in B-CLL-F, which affected the same codon, but caused a different amino acid exchange (Asp1853Val), was not found in the 50 controls and is therefore compatible with the definition of a pathogenic mutation.
DISCUSSION
Two lines of evidence emphasize the pivotal role of a tumor suppressor gene localized in 11q22-q23 for the leukemogenesis of B-CLL: (1) deletion of this chromosomal region is one of the most frequent chromosome aberrations in this disease.24 (2) The 11q22-q23 deletions have a strong clinical impact, as they define a subset of B-CLL characterized by extensive lymph node involvement, rapid disease progression, and poor survival.24 In the current study, a series of malignant B-cell lymphomas, mostly B-CLLs, with deletions of one copy of chromosome bands 11q22-q23 were analyzed. In six of the 23 analyzed cases, the ATM gene was found affected by a point mutation in the nondeleted allele. Besides those, there was one B-cell lymphoma without 11q deletion, but with mutations in both ATM alleles. The inactivation of both gene copies indicates a pathogenic role ofATM in sporadic B-cell tumors. The identified mutations resulted in aberrant transcript splicing, premature truncation of the protein, or alteration of amino acids. Fifty percent of the mutations affected the PI-3-K domain, which is highly conserved among ATM-related proteins4,5 and crucial for the protein kinase activity of ATM.29,30 Interestingly, three of those mutations correspond to disease-causing mutations in A-T patients and/or mutations associated with T-PLL underlining the pathogenic character of the mutations: (1) the truncation mutation Arg3047ter has been described as an A-T allele9,31-33 and as a mutation in T-PLL19 causing considerable impairment of the protein phosphorylation function of ATM29; (2) the splicing defect caused by IVS59+1G>T resembles A-T mutations, which cause skipping of the affected exon 5925,32,34;(3) the missense mutation Arg3008His replaced the same conserved amino acid as Arg3008Cys in two independent T-PLL cases.18 21
The PI-3-K domain was recently shown to mediate phosphorylation of p53 in response to DNA damage.29,30 Therefore, mutational inactivation of the kinase domain likely affects the p53 mediated control of the cell cycle and response to DNA damage. Constitutional ATM deficiency in cells of A-T patients (for review, see Lavin and Shiloh2 and Jeggo et al3) andatm−/- mice35-37 was found to lead to dysregulation of apoptosis and cell-cycle check point control, as well as to defects in DNA recombination. These cellular defects are considered to be causative for the development of T-cell lymphomas inatm−/- mice36,38 and lymphoid tumors of both the T- and B-cell lineage in A-T patients.11It is conceivable that somatic ATM inactivation in lymphocytes may result in similar cellular defects, which may contribute to T- and B-cell leukemogenesis in non–A-T patients.
Here, we demonstrate that somatic disruption of the ATM gene occurs not only in sporadic T-PLL,18-21 but is also common for B-cells of malignant B-cell lymphomas, in particular, B-CLLs. Two very recent studies analyzing ATM gene mutation and ATM protein levels reported similar evidence for the role of ATM in B-CLL.39-41However, Stankovic et al40 could not observe loss of heterozygosity of the respective genomic region in a subset of tumors with impaired ATM expression, a fact that could be explained by the existence of a second B-CLL associated gene in the critical genomic segment in 11q.
The mutation frequency observed in our study exceeds by far the expected value of random mutations taking the A-T heterozygote frequency of about 1% in the general population42,43 into account. Therefore, ATM is probably the tumor-suppressor gene in 11q22-q23, whose inactivation is responsible for an aggressive course of B-CLL. There are several reasons why we did not observeATM point mutations in a higher proportion of the cases analyzed: (1) mutation analysis focused on a 6.2-kb part of the coding region encompassing the kinase domain, the c-Abl binding site, the rad3 homology region, and a leucine zipper motif. Mutations could also reside, however, in the N-terminal region of ATM, which was recently shown to interact with β-adaptin,44 or in the nontranslated regions or the promoter region impairing gene expression. (2) While we used a modified SSCP method, also known as restriction endonuclease fingerprinting (REF), which is of considerably enhanced efficiency,45 it is still possible that we missed mutations. (3) The commonly deleted region in 11q22-q23 is 2 to 3 Mbp in size and contains besides ATM several other genes.23 At this point, it cannot be excluded that a second gene with pathogenic function for B-CLL exists in this region (see also above).
For four patients with ATM point mutations in the tumor cells, an assessment of the ATM germline status was possible. Absence of the mutations in the nonmalignant cells in all four cases indicated a somatic origin of the mutations. However, others reported ATMgermline mutations in some of the B-CLL patients carrying ATMmutation40 41 showing the existence of genetic predisposition to B-CLL.
ACKNOWLEDGMENT
The authors gratefully acknowledge Ralf Klären, Irina Idler, and Markus Scheuermann for their technical assistance.
Supported by the Wilhelm-Sander-Stiftung (97.003.1), the Deutsche Krebshilfe (10-1289-StI), and the Tumorzentrum Heidelberg/Mannheim (I/I.1).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Peter Lichter, PhD, Abteilung “Organisation komplexer Genome” (H0700), Deutsches Krebsforschungszentrum, Im Neuenheimer Feld 280, D-69120 Heidelberg, Germany; e-mail: p.lichter@dkfz-heidelberg.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal