In this study, we examined in detail the interaction of platelet factor-4 (PF-4) with fibroblast growth factor-2 (FGF-2) and vascular endothelial growth factor (VEGF) and the effect of PF-4–derived synthetic peptides. We show that a peptide between amino acids 47 and 70 that contains the heparin-binding lysine-rich site inhibits FGF-2 or VEGF function. This is based on the following observations: PF-4 peptide 47-70 inhibited FGF-2 or VEGF binding to endothelial cells; it inhibited FGF-2 or VEGF binding to FGFRs or VEGFRs in heparan sulfate–deficient CHO cells transfected with FGFR1 (CHOFGFR1) or VEGFR2 (CHOmVEGFR2) cDNA; it blocked proliferation or tube formation in three-dimensional angiogenesis assays; and, finally, it competed with the direct association of 125I-PF-4 with FGF-2 or VEGF, respectively, and inhibited heparin-induced FGF-2 dimerization. A shorter C-terminal peptide (peptide 58-70), which still contained the heparin-binding lysin-rich site, had no effect. Peptide 17-58, which is located in the central part of the molecule, although it does not inhibit FGF-2 or VEGF binding or biologic activity in endothelial cells, inhibited heparin-dependent binding of125I-FGF-2 or 125I-VEGF to CHOmFGFR1 or CHOmVEGFR2 cells, respectively. Shorter peptides (peptides 34-58 and 47-58) did not show any of these effects.
ANGIOGENESIS IS CONTROLLED by a balance of positive and negative regulators. Fibroblast growth factor-2 (FGF-2) and vascular endothelial growth factor (VEGF) are among the principal positive regulators.1,2 Negative regulators include inhibitory molecules, such as angiostatin,3,4endostatin,5 thrombospondin-1 (TSP-1),6 the 16-kD human prolactin fragment (16-kD PRL),7 and platelet factor-4 (PF-4) 8.
FGF-2 and VEGF mediate their biologic activity by binding to specific cell surface receptors. The interaction of these ligands with their receptors is modulated by heparan sulfate proteoglycans (HSPGs). For example, HSPG stabilizes the FGF-2/FGF receptor complex, protects FGF-2 from degradation, and facilitates FGF-2 dimerization.1HSPGs also modulate the binding of VEGF to VEGFR1 or VEGFR2.9 This indicates that angiogenesis inhibitors may possibly interfere with FGF-2 or VEGF activity by disrupting HSPG/FGF-2/FGF receptor or HSPG/VEGF/VEGF receptor interactions.
PF-4 is a member of the C-X-C chemokine family.10 PF-4 is a 7.8-kD protein of 70–amino acid length11 that shares homologies in particular with β-thromboglobulin and interleukin-8 (IL-8).10,12 The crystal structure of human PF-4 has been solved to a resolution of 2.4 A.13 The major heparin-binding site forms a α helicoidal structure and is located at the C-terminus between 61 and 70. In contrast, the central core forms antiparallel β-sheet-like structures. Furthermore, additonal potential heparin-binding sites exist as a positively charged ring of lysine and arginine side chains that encircles the PF-4 molecule.14
A number of observations indicate that PF-4 is an inhibitor of angiogenesis. First, PF-4 inhibits endothelial cell proliferation, migration, and angiogenesis in vitro and in vivo.8,15Second, PF-4 is targeted in vivo to endothelial cells that undergo active angiogenesis.16 Third, tumor growth in vivo is inhibited by PF-4 through an angiogenesis-dependent mechanism.17,18 Recombinant human PF-4 inhibits tumor angiogenesis and the growth of melanoma cells or HCT 116 colon carcinoma cells.17 In addition, human glioma cells infected with a secretable PF-4 cDNA grew slowly in vivo and formed only hypovascular tumors in vivo.19 Megakaryocytes and platelets are the major source of PF-4. Indeed, activated platelets release an inhibitor of FGF-2 activity that was shown to be identical to PF-4.20 Thus, PF-4 may counteract excessive angiogenic factor activity at sites of platelet activation.
In a series of systematic studies, we have recently partially elucidated the mechanism of inhibition by which PF-4 inhibits FGF-2 activity.21 We have demonstrated that PF-4 inhibits FGF-2 binding to high-affinity receptors and inhibits FGF-2 internalization. Furthermore, PF-4 is able to bind surface-immobilized or soluble FGF-2 and inhibited endogenous and heparin-induced FGF-2 dimerization. Similarily, PF-4 also binds surface-immobilized VEGF.22
In the present study, we sought to determine the inhibitory capacity of various peptides that correspond to different PF-4 domains.
MATERIALS AND METHODS
Synthetic peptides.
Peptides were synthesized using standard solid-phase methodology and purified by high-performance liquid chromatography (HPLC) using a C18 column and a 0% to 80% linear acetonitrile gradient in 0.1% trifluorouracetic acid. The following synthetic peptides were used for the study. Peptide 47-70: NGRKICLDLQAPLYKKIIKKLLES; peptide 58-70: PLYKKIIKKLLES; peptide 47-58: NGRKICLDLQAP; peptide 34-58: PHCPTAQLIATLKNGRKICLDLQAP; peptide 17-58: SQVRPRHITSLEVIKAGPHCPTAQLIATLKNG RKICLDLQAP. Recombinant human PF-4 or platelet-purified PF-4 (Serbio, Gennevilliers, France) was used as control in the different experiments.
Cells.
Murine lung microvascular endothelial cells (LEII cells; (kindly donated by Dr Thomas Maciag, American Red Cross, Rockville, MD) were grown in Dulbecco’s modifed Eagle’s medium (DMEM; GIBCO, Life Technologies, Gaithersburg, MD) containing 10 % fetal calf serum (FCS; GIBCO), 1 g/L glucose, 1% glutamin, and 50 ΙU/mL penicillin-50 μg/mL streptomycin at 37°C in a 5% CO 2 atmosphere. Adrenal cortex capillary endothelial cells (ACE cells; kindly donated by Dr Jean-Jacques Feige, CENG, Grenoble, France) were grown in the same medium as LEII cells, except that FCS newborn calf serum (NCS) was used instead of FCS. Heparan sulfate–deficient chinese hamster ovary cells (CHOm-FGFR1 cells, 745-flg; kindly donated by Dr Avner Yayon, The Weizmann Institute, Rehovot, Israel) were grown in DMEM containing 10% FCS, 1 g/L glucose, and 1% nonessential amino acids at 37°C in a 5% CO 2 atmosphere. Heparan sulfate–deficient CHO cells were transfected with a human VEGFR2 cDNA (CHOmVEGFR2 cells) in a pSV7d vector as already described.23
Cell proliferation experiments.
Proliferation assays were performed as described.24Briefly, cells were seeded at 20,000 cells on 3.5-cm2dishes in DMEM containing 10% FCS, 1% glutamin, and antibiotics, or into wells of 24-well plates at 7,000 cells/well. After overnight attachment, the cells were washed once with serum-free DMEM and test medium that contained 1% FCS or 1% NCS and subsequently incubated with the indicated concentrations of recombinant human FGF-2, recombinant human baculovirus-derived VEGF 165 (165–amino acid splicing variant of VEGF), PF-4, or PF-4 peptides. Cells were counted at specified days with a Coulter counter (Coultronics, Margency, France).
In vitro angiogenesis assays.
In vitro angiogenesis assays were performed according to Montesano et al25 using collagen type I as the three-dimensional matrix. Briefly, 20,000 cells were plated on the top of collagen type I gels into wells of 96-well plates. Twenty-four hours after plating, 10 ng/mL FGF-2 or VEGF was added with or without the indicated peptide concentrations. The experiments were performed over a period of 7 days, after which photographs were taken from each well using a 10X objective (Zeiss, Wetzlar, Germany). Alternatively, the cells were fixed with methanol, stained with methylene blue, and photographed. Endothelial tube length was quantified in each well using a Biocom image analyzer (Biocom, les Ulis, France). The results were expressed as the mean ± SD of the total tube length from experiments run in duplicates.
Binding studies and cross-linking to receptors.
FGF-2 and PF-4 were labeled with 125I-Na using iodogen (Pierce, Rockford, IL) as coupling agent according to the manufacturer’s indications and according to Moscatelli.26VEGF was labeled with 125I-Na using iodobeads (Pierce). The specific activities of 125I-FGF-2 , 125I-PF-4 and 125I-VEGF were 80,000 to 200,000 cpm/ng, 35,000 to 100,000 cpm/ng, and 150,000 to 200,000 cpm/ng, respectively. FGF-2 binding experiments to high- and low-affinity sites were performed essentially as described by Moscatelli.26 Cells were seeded at 2.5 × 105/cm2 and cultured in complete medium into 3.5-cm diameter dishes for 2 days. Cells were washed twice with ice-cold phosphate-buffered saline (PBS) and incubated with the indicated concentrations of 125I-FGF-2 in DMEM, which contained 20 mmol/L HEPES (pH 7.4), 0.15% gelatin, for 2 hours at 4°C. At the end of the incubation period, cells were washed three times with ice-cold PBS. 125I-FGF-2 was dissociated from its cellular low-affinity binding sites by two 20-second washes with ice-cold 20 mmol/L HEPES (pH 7.4), 2 mol/L NaCl, and from its high-affinity sites by two 20-second washes with ice-cold 20 mmol/L NaAc (pH 4.0), 2 mol/L NaCl. Bound 125I-FGF-2 was quantified using a Kontron MR 250 γ-counter (Saint-Quentin-Yvelines, France). Nonspecific binding was determined by incubating separate dishes with 125I-FGF-2 and a 100-fold excess of unlabeled ligand. Specific binding was determined by substracting nonspecific binding from total binding.
Cross-linking of 125I-FGF-2 or 125I-VEGF to receptors was performed and analyzed as described by Bikfalvi et al,21 using 0.2 mmol/L Bis(sulfosuccinyl) suberate (BS3; Pierce) in PBS as a coupling agent. The quantity of protein used in each experiment was normalized according to cell number or protein. Samples were run on a 7% sodium dodecyl sulfate (SDS)-polyacrylamide gel. Autoradiographies were performed with X-OMAT AR films (Eastman Kodak, Rochester, NY) at −80°C in the presence of an intensifying screen or analyzed by PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
125I-PF-4 binding experiments with surface-immobilized FGF-2 or VEGF.
Binding of 125I-PF-4 to FGF-2 or VEGF 165–coated wells was performed as follows. Ninety-six–well enzyme-linked immunosorbent assay (ELISA) plates were coated with FGF-2 or VEGF in buffer A (1 mmol/L EDTA, 20 mmol/L K2HPO4, 10 mmol/L KH2PO4, 150 mmol/L NaCl) in a volume of 50 μL. After a 2-hour incubation at room temperature, the plates were washed five times with buffer B (10 mmol/L Tris-HCl pH 7.2, 150 mmol/L NaCl, 0.1% Tween 20). The wells were subsequently incubated with buffer A with an additional 0.1% gelatin and washed five times after 1 hour with buffer B. The FGF-2– or VEGF-coated wells were then incubated in buffer A with 10 ng/well 125I-PF-4, competitors, or polyclonal anti–PF-4 antibodies at 37°C for 1 hour. Rabbit polyclonal anti–PF-4 antibodies were prepared as previously reported.27 At the end of the incubation period, the wells were washed five times with buffer B. Surface-associated125I-PF-4 was then extracted with 200 μL of 0.2 mol/L NaOH and counted in a γ-counter. At the end of the preincubation period, the wells were washed twice before the addition of125I-PF-4. To determine nonspecific binding, separate wells were not coated with FGF-2 and only preincubated with buffer A before the initiation of binding. Specific binding was determined by substracting nonspecific binding from total binding.
FGF-2 dimerization experiments.
FGF-2 dimerization was studied according to the method described by Ornitz et al.28 Briefly, 5 ng 125I-FGF-2 and 500 ng unlabeled FGF-2 were incubated for 1 hour at room temperature with or without the indicated concentrations of heparin and PF-4 or peptides in PBS in a final volume of 45 μL. At the end of the incubation period, 5 μL BS3 (0.1 mmol/L final concentration) was added for another 30-minute incubation. The reaction was stopped by adding SDS-sample buffer from a five-times concentrated stock solution. The samples were boiled and run on a 12% or 15% SDS-polyacrylamide gel. The gels were analyzed by PhosphorImager (Molecular Dynamics, Bondoufle, France) or autoradiography. For all of the experiments outlined here, autoradiograms or PhosphoImager results were analyzed by a public domain National Institutes of Health (NIH) Image Program developed at the US NIH and available over the internet by anonymous FTP fromzippy.nimh.nih.gov or by using a Bio Profil V 6.0 scanner with Bio 1 D software (Vilber Lourmat, Paris, France).
RESULTS
Effect of PF-4 peptides on FGF-2 or VEGF-induced endothelial cell proliferation or angiogenesis in vitro.
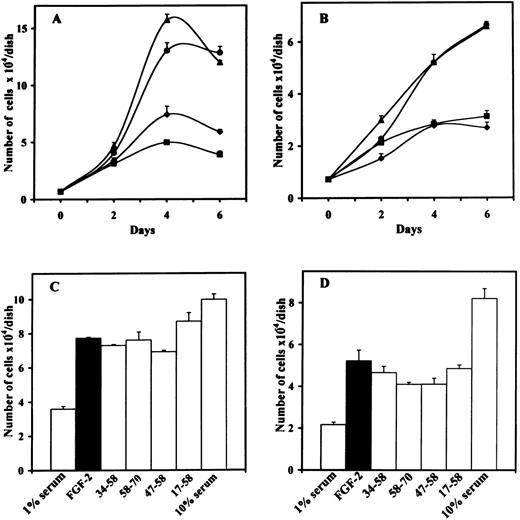
We examined the effect of the different peptides on the biologic activity induced by FGF-2 or VEGF. We first performed proliferation assays using bovine ACE cells or murine LEII cells. Inhibition of FGF-2–induced endothelial cell proliferation was only observed with 10 μmol/L of peptide 47-70, but not with peptide 47-70, 58-70, 17-58, 34-58, or 47-58 (Fig 1A through D). Inhibition of endothelial cell proliferation was observed for both ACE cells and LEII cells and was consistent during the whole time course of incubation.
Effect of PF-4–derived peptides on endothelial cell proliferation. (A) ACE cells or (B) LEII cells are incubated in DMEM containing 1% bovine serum (FCS for LEII cells and NCS for ACE cells) with or without 10 ng/mL FGF-2 and 10 μmol/L PF-4–derived peptides. 1% serum without FGF-2 (⧫), 1% serum with FGF-2 (•), 1% serum with FGF-2 and peptide 47-70 (▪), 1% serum with FGF-2 and peptide 34-58 (▴). Cells are counted every other day. (C) ACE cells or (D) LEII cells are incubated with 10 ng/mL FGF-2 and 10 μmol/L of various peptides and counted after 6 days.
Effect of PF-4–derived peptides on endothelial cell proliferation. (A) ACE cells or (B) LEII cells are incubated in DMEM containing 1% bovine serum (FCS for LEII cells and NCS for ACE cells) with or without 10 ng/mL FGF-2 and 10 μmol/L PF-4–derived peptides. 1% serum without FGF-2 (⧫), 1% serum with FGF-2 (•), 1% serum with FGF-2 and peptide 47-70 (▪), 1% serum with FGF-2 and peptide 34-58 (▴). Cells are counted every other day. (C) ACE cells or (D) LEII cells are incubated with 10 ng/mL FGF-2 and 10 μmol/L of various peptides and counted after 6 days.
To ensure that sprouting and tubulogenesis were also impaired by peptide 47-70, we performed in vitro angiogenesis assays according to Montesano et al.25 In the absence of peptide, FGF-2 or VEGF induced a total tube length of 3.5 mm ± 0.25/well (mean ± SD) and 2.1 mm ± 0.65/well. respectively. A 10-μmol/L quantity of peptide 47-70 completely inhibited tube formation induced by FGF-2 or VEGF (Fig 2A and B). On the contrary, peptides 17-58, 47-58, 34-58, or 58-70 were unable to inhibit tube formation induced by FGF-2 or VEGF (Fig 2A and B). However, peptide 47-58 was able to increase FGF-2–stimulated tube formation to 3.5 mm ± 0.25/well to 6 mm ± 0.65/well (mean ± SD; 175% in comparison to control). This effect was not seen when tube formation was induced with VEGF.
Effect of PF-4–derived peptides on angiogenesis in vitro. ACE cells are grown onto 3-dimensional collagen type I gels in the presence of 10 ng/mL FGF-2 (A) or VEGF (B) in the presence or absence of 10 μmol/L of the different peptides. Photomicrographs were taken after 7 days incubation. Results were quantified as indicated in Materials and Methods and expressed as the mean ± SD of experiments done in duplicates.
Effect of PF-4–derived peptides on angiogenesis in vitro. ACE cells are grown onto 3-dimensional collagen type I gels in the presence of 10 ng/mL FGF-2 (A) or VEGF (B) in the presence or absence of 10 μmol/L of the different peptides. Photomicrographs were taken after 7 days incubation. Results were quantified as indicated in Materials and Methods and expressed as the mean ± SD of experiments done in duplicates.
These experiments indicate that FGF-2 or VEGF activities are inhibited by peptide 47-70, but not by peptides 58-70, 17-58, 47-58, or 34-58. In addition, peptide 47-58 appears to increase FGF-2–stimulated tube formation.
Effect of PF-4 peptides on 125I-FGF-2 or125I-VEGF binding on endothelial cells.
We next investigated the effects of peptides 17-58, 34-58, 47-58, 47-70, or 58-70 on the inhibition of 125I-FGF-2 or125I-VEGF binding to endothelial cells. We used LEII cells and ACE cells for these studies, because both cell types express FGF or VEGF receptors. PF-4 inhibited 125I-FGF-2 binding to low- and high-affinity binding sites in endothelial cells with a half-maximum inhibition (IC50) of 0.6 μg/mL for the inhibition of FGF-2 binding to the high-affinity binding sites.21 Binding of 125I-VEGF to endothelial cells was also inhibited by PF-4 with an IC50 of 0.25 μmol/L (data not shown). When the peptides indicated above were used in these binding experiments, inhibition was only observed with peptide 47-70 (Fig 3A through C). High-affinity binding of 125I-FGF-2 to FGF receptors on LEII or ACE cells was inhibited maximally at 5 μmol/L and half maximally at 1 μmol/L peptide concentration (Fig 3A and data not shown). Peptides 58-70, 17-58, 34-58, and 47-58 were without effect (Fig 3B and C).
Effect of PF-4–derived peptides on binding of125I-FGF-2 to endothelial cells. LEII cells or ACE cells are incubated with 10 ng/mL 125I-FGF-2 in the presence or absence of 2 μg/mL unlabeled ligand (nonspecific binding, NS), 10 μg/mL PF-4 or PF-4–derived peptides. Binding experiments are performed as indicated in Materials and Methods. Binding experiments on LEII cells with increasing concentrations (A) of peptides 47-70 (•) or 34-58 (▪) or fixed concentrations (B, 10 μmol/L) of the indicated peptides; binding experiments on ACE cells with fixed concentrations (C, 10 μmol/L) of indicated peptides. Results are expressed as the mean± SD of experiments done in duplicates.
Effect of PF-4–derived peptides on binding of125I-FGF-2 to endothelial cells. LEII cells or ACE cells are incubated with 10 ng/mL 125I-FGF-2 in the presence or absence of 2 μg/mL unlabeled ligand (nonspecific binding, NS), 10 μg/mL PF-4 or PF-4–derived peptides. Binding experiments are performed as indicated in Materials and Methods. Binding experiments on LEII cells with increasing concentrations (A) of peptides 47-70 (•) or 34-58 (▪) or fixed concentrations (B, 10 μmol/L) of the indicated peptides; binding experiments on ACE cells with fixed concentrations (C, 10 μmol/L) of indicated peptides. Results are expressed as the mean± SD of experiments done in duplicates.
We then examined the effect of the different peptides on the binding of125I-VEGF to LEII or ACE cells (Fig4A through D). Peptide 47-70 at 5 to 10 μmol/L completely inhibited 125I-VEGF binding to both endothelial cell types with a IC50 of 1.5 and 2 μmol/L for LEII and ACE cells, respectively (Fig 4B and D). Peptides 58-70, 17-58, 34-58, and 47-58 did not show inhibitory activity (Fig 4 A and C).
Effect of PF-4±derived peptides on binding of125I-VEGF to endothelial cells. LEII cells or ACE cells are incubated with 5 ng/mL 125I-VEGF in the presence or absence of 500 ng/mL unlabeled ligand (nonspecific binding, NS), or PF-4–derived peptides. Binding experiments are performed as indicated in Materials and Methods. Binding experiments on LEII cells with 10 μmol/L of indicated peptides (A) or increasing concentrations (B) of peptide 47-70; binding experiments on ACE cells with fixed (10 μmol/L) concentrations (C) of the different peptides or increasing concentrations (D) of peptide 47-70. Results are expressed as the mean ± SD of experiments done in duplicates.
Effect of PF-4±derived peptides on binding of125I-VEGF to endothelial cells. LEII cells or ACE cells are incubated with 5 ng/mL 125I-VEGF in the presence or absence of 500 ng/mL unlabeled ligand (nonspecific binding, NS), or PF-4–derived peptides. Binding experiments are performed as indicated in Materials and Methods. Binding experiments on LEII cells with 10 μmol/L of indicated peptides (A) or increasing concentrations (B) of peptide 47-70; binding experiments on ACE cells with fixed (10 μmol/L) concentrations (C) of the different peptides or increasing concentrations (D) of peptide 47-70. Results are expressed as the mean ± SD of experiments done in duplicates.
These results indicate that peptide 47-70 not only interferes with the binding of FGF-2, but also with the binding of VEGF to endothelial cells.
Effect of PF-4–derived peptides on 125I-FGF-2 or125I-VEGF binding on heparan sulfate–deficient CHO cells expressing FGFR1 or VEGFR2.
We next set out to examine the inhibitory activity of the various peptides by using heparan sulfate–deficient CHO cells that were transfected with either expression vectors that contained FGFR1(CHOmFGFR1) or VEGFR2 (CHOmVEGFR2) cDNAs. We have previously shown that PF-4 inhibits the heparin-dependent and -independent binding of125I-FGF-2 in CHOmFGFR1.21 As depicted in Fig5A, peptide 47-70 did only slightly inhibit125I-FGF-2 binding to CHOmFGFR1 cells in the absence of heparin (≈20% at 10 μmol/L in comparison to control). However, in the presence of heparin (100 ng/mL), heparin-induced binding was strongly impaired by peptide 47-70 (Fig 5B and C). Maximum inhibition of 125I-FGF-2 binding by peptide 47-70 was attained at 2 μmol/L (IC50 at 0.35 μmol/L). This is also reinforced by cross-linking of 125I-FGF-2 to FGFR1 in the presence of peptide 47-70 (Fig 5D). These experiments show that increasing concentrations of peptide 47-70 (0.6 to 4 μmol/L) progressively decreased the intensity of the signal that corresponds to125I-FGF-2/FGFR1 complexes (Fig 5D). None of the other peptides inhibited 125I-FGF-2 binding in the absence of heparin (Fig 5A and data not shown). However, in the presence of heparin, peptide 17-58, like peptide 47-70, inhibited125I-FGF-2 binding (maximum inhibition of ≈50% to 60% of binding, Fig 5C). Maximum inhibition was reached at 2 μmol/L and the IC50 at 0.7 μmol/L. Peptides 34-58, 47-58, and 58-70 did not inhibit heparin-induced 125I-FGF-2 binding to CHOmFGFR1 cells.
Effect of PF-4–derived peptides on binding and cross-linking of 125I-FGF-2 to CHOmFGFR1 cells. CHOmFGFR1 cells are incubated with 10 ng/mL 125I-FGF-2 with or without 2 μg/mL unlabeled ligand, 10 μg/mL PF-4 or PF-4–derived peptides in the absence (A) or presence (B, C) of 100 ng/mL heparin. 47-70 (▴); 34-58 (•); 17-58 (⧫). (A and C) Concentration dependency; (B) 10 μmol/L PF-4–derived peptides. Cross-linking (D) is done with the different peptides in the presence of 100 ng/mL heparin. Peptide 47-58 (5.3 μmol/L), 34-58 (5.3 μmol/L), 47-70 (1, 0.65 μmol/L; 2, 1.3 μmol/L; 3, 2.6 μmol/L; 4, 4.1 μmol/L). Binding or cross-linking experiments are performed as indicated in Materials and Methods. Results (A-C) are expressed as the mean± SD of experiments done in duplicates.
Effect of PF-4–derived peptides on binding and cross-linking of 125I-FGF-2 to CHOmFGFR1 cells. CHOmFGFR1 cells are incubated with 10 ng/mL 125I-FGF-2 with or without 2 μg/mL unlabeled ligand, 10 μg/mL PF-4 or PF-4–derived peptides in the absence (A) or presence (B, C) of 100 ng/mL heparin. 47-70 (▴); 34-58 (•); 17-58 (⧫). (A and C) Concentration dependency; (B) 10 μmol/L PF-4–derived peptides. Cross-linking (D) is done with the different peptides in the presence of 100 ng/mL heparin. Peptide 47-58 (5.3 μmol/L), 34-58 (5.3 μmol/L), 47-70 (1, 0.65 μmol/L; 2, 1.3 μmol/L; 3, 2.6 μmol/L; 4, 4.1 μmol/L). Binding or cross-linking experiments are performed as indicated in Materials and Methods. Results (A-C) are expressed as the mean± SD of experiments done in duplicates.
In the absence of heparin, 125I-VEGF only weakly bound CHOmVEGFR2 (data not shown). Heparin increased the binding of125I-VEGF to these cells by fourfold to fivefold (data not shown). We therefore investigated the effect of the different peptides on 125I-VEGF binding to CHOmVEGFR2 cells in the presence of 100 ng/mL heparin (Fig 6).125I-VEGF binding to CHOmVEGFR2 was also inhibited by peptides 47-70 and 17-58. Peptide 47-70 inhibited maximally and half maximally 125I-VEGF binding at 1 and 0.5 μmol/L, respectively. For peptide 17-58, maximum inhibition was attained at 1 μmol/L and the IC50 at 0.17 μmol/L. None of the other peptides interfered with 125I-VEGF binding to CHOmVEGFR2 cells.
Effect of PF-4–derived peptides on binding of125I-VEGF to CHOmVEGFR2 cells. CHOmVEGFR2 cells are incubated with 5 ng/mL 125I-VEGF with or without 500 ng/mL unlabeled ligand (NS) or PF-4–derived peptides in the presence of 100 ng/mL heparin. Binding experiments are performed as indicated in Materials and Methods. Peptide 47-70 (▴), 34-58 (▪); 17-58 (⧫), 47-58 (□). Results are expressed as the mean ± SD of experiments done in duplicates.
Effect of PF-4–derived peptides on binding of125I-VEGF to CHOmVEGFR2 cells. CHOmVEGFR2 cells are incubated with 5 ng/mL 125I-VEGF with or without 500 ng/mL unlabeled ligand (NS) or PF-4–derived peptides in the presence of 100 ng/mL heparin. Binding experiments are performed as indicated in Materials and Methods. Peptide 47-70 (▴), 34-58 (▪); 17-58 (⧫), 47-58 (□). Results are expressed as the mean ± SD of experiments done in duplicates.
Effect of PF-4 peptides on the interaction between PF-4 and FGF-2 or VEGF.
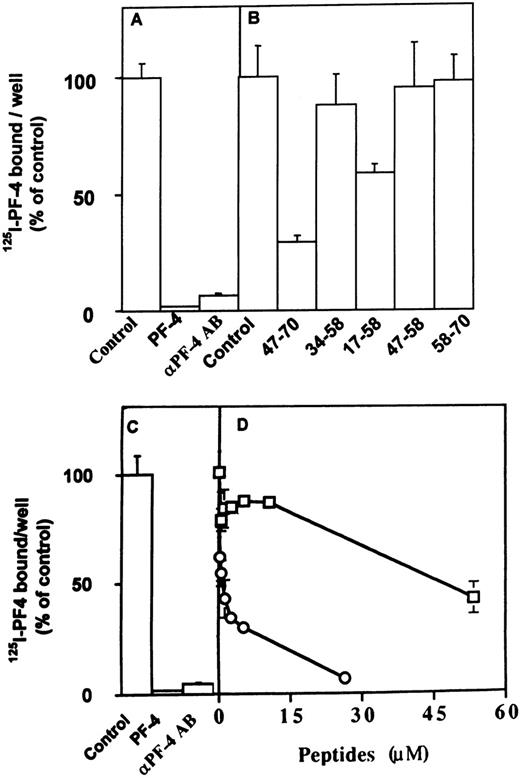
We next analyzed the effect of the different peptides on the direct interaction of PF-4 with FGF-2 or VEGF. These experiments were performed using a solid-phase binding assay. We have previously demonstrated that PF-4 directly binds to FGF-2 and inhibits FGF-2 dimerization.21 In the solid-phase binding assay, unlabeled PF-4 competed with 125I-PF-4 binding to FGF-2 or VEGF nearly completely at 5 μmol/L and half maximally at 0.1 μmol/L (Fig7A and C; and data not shown). Furthermore,125I-PF-4 binding to FGF-2 or VEGF was also nearly completely blocked by 10 μg/mL anti–PF-4 antibodies (Fig 7A and C). At 10 μmol/L, peptide 47-70 inhibited the association of125I-PF-4 to FGF-2 or VEGF by 75% in comparison to control (Fig 7B and D; and data not shown). When increasing concentrations of peptide 47-70 were added, maximum inhibition (95% inhibition) was reached between 20 to 30 μmol/L and the IC50 at approximately 0.5 μmol/L (Fig 7D and data not shown). Peptide 17-58 was also able to compete with the association of 125I-PF-4 to surface-immobilized growth factors (≈42% at 10 μmol/L of peptide 17-58), but the other peptides were unable to do so (Fig 7B and data not shown).
Effect of PF-4–derived peptides on the direct association of 125I-PF-4 with FGF-2 or VEGF. Wells of 96-well plates are coated with 15 ng FGF-2 (A, B) or 35 ng VEGF (C, D) and incubated with 10 ng 125I-PF-4 with or without 2 μg unlabeled PF-4, 10 μg anti–PF-4 antibodies, or peptides in 50 μL incubation volume. Solid-phase binding experiments are performed as indicated Materials and Methods with fixed (10 μmol/L) or increasing peptide concentrations. Peptide 47-70 (○), 34-58 (□). Results are expressed as the mean ± SD of experiments done in triplicates.
Effect of PF-4–derived peptides on the direct association of 125I-PF-4 with FGF-2 or VEGF. Wells of 96-well plates are coated with 15 ng FGF-2 (A, B) or 35 ng VEGF (C, D) and incubated with 10 ng 125I-PF-4 with or without 2 μg unlabeled PF-4, 10 μg anti–PF-4 antibodies, or peptides in 50 μL incubation volume. Solid-phase binding experiments are performed as indicated Materials and Methods with fixed (10 μmol/L) or increasing peptide concentrations. Peptide 47-70 (○), 34-58 (□). Results are expressed as the mean ± SD of experiments done in triplicates.
We then tested the effect of the different PF-4 peptides on FGF-2 dimerization (Fig 8A through C). PF-4 by itself severely decreased multimerization and simultaneously promoted the appearance of a intermediate band of 25 kD corresponding to PF-4/FGF-2 complexes.21 Five nanograms of125I-FGF-2 and 500 ng unlabeled FGF-2 were incubated with various concentrations of peptides in a 50-μL incubation volume. The experiments were performed in the absence and presence of heparin. When PF-4 peptide 47-70 was used in these experiments, a modulation of FGF-2 dimer formation was observed. Surprisingly, in the absence of heparin, FGF-2 dimers were increased by peptide 47-70 with a maximum increase at 10.8 μmol/L (Fig 8A). At higher peptide concentrations (21 to 134 μmol/L), FGF-2 dimers progressively decreased. To the contrary, in the presence of 50 ng/50 μL heparin, dimer formation was already severely inhibited at 5.4 μmol/L of peptide 47-70 and remained low with increasing peptide concentrations (Fig 8B). We also investigated the effects of peptides 17-58, 34-58, 47-58, and 58-70 on FGF-2 dimerization (Fig 8C). A 10-μmol/L quantity of each peptide was added to the incubation mixture. In the presence of 50 ng/50 μL heparin, FGF-2 dimerization was slightly increased by peptide 17-58 and an additional band of approximately 20 kD was observed. This signal may correspond to cross-linked 125I-FGF-2–peptide 17-58 complexes. Peptides 34-58, 47-58, and 58-70 were unable to modulate FGF-2 dimer formation (Fig 8C).
Effect of PF-4–derived peptides on FGF-2 dimerization. 10 ng 125I-FGF-2, 500 ng unlabeled FGF-2 are incubated in the presence or absence of 50 ng heparin with fixed (10 μmol/L) or increasing concentrations of the different PF-4–derived peptides. Dimerization experiments were performed as indicated in Materials and Methods. (A) Increasing concentrations of peptide 47-70 in the absence of heparin (lane 1: control, lane 2: 2.6 μmol/L, lane 3: 5.4 μmol/L, lane 4: 10.8 μmol/L, lane 5: 21.4 μmol/L, lane 6: 42.7 μmol/L, lane 7: 87 μmol/L, lane 8: 134.5 μmol/L); (B) increasing concentrations of peptide 47-70 in the presence of heparin (lane 1: control with heparin, lane 2: 5.4 μmol/L, lane 3: 10.8 μmol/L, lane 4: 21.4 μmol/L, lane 5: 42.7 μmol/L, lane 6: 87 μmol/L, lane 7: 134.5 μmol/L) ; C, fixed concentrations (10 μmol/L) of various PF-4–derived peptides (lane 1: control without heparin, lane 2: control with heparin, lane 3: peptide 34-58, lane 4: peptide 47-58, lane 5: peptide 58-70, lane 6: peptide 17-58).
Effect of PF-4–derived peptides on FGF-2 dimerization. 10 ng 125I-FGF-2, 500 ng unlabeled FGF-2 are incubated in the presence or absence of 50 ng heparin with fixed (10 μmol/L) or increasing concentrations of the different PF-4–derived peptides. Dimerization experiments were performed as indicated in Materials and Methods. (A) Increasing concentrations of peptide 47-70 in the absence of heparin (lane 1: control, lane 2: 2.6 μmol/L, lane 3: 5.4 μmol/L, lane 4: 10.8 μmol/L, lane 5: 21.4 μmol/L, lane 6: 42.7 μmol/L, lane 7: 87 μmol/L, lane 8: 134.5 μmol/L); (B) increasing concentrations of peptide 47-70 in the presence of heparin (lane 1: control with heparin, lane 2: 5.4 μmol/L, lane 3: 10.8 μmol/L, lane 4: 21.4 μmol/L, lane 5: 42.7 μmol/L, lane 6: 87 μmol/L, lane 7: 134.5 μmol/L) ; C, fixed concentrations (10 μmol/L) of various PF-4–derived peptides (lane 1: control without heparin, lane 2: control with heparin, lane 3: peptide 34-58, lane 4: peptide 47-58, lane 5: peptide 58-70, lane 6: peptide 17-58).
DISCUSSION
PF-4 has several interesting structural features. Three major clusters of basic amino acids are found within the PF-4 sequence. One cluster is localized near the N-terminus (Arg20, Arg22, His23), another in the middle of the molecule (Arg49, Lys50), and a third at the C-terminus (Lys61, Lys62, Lys65, Lys66). The cluster found at the C-terminus constitutes the major heparin-binding domain of PF-4. This sequence is located within an α-helicoidal structure that protrudes from the whole PF-4 molecule. Furthermore, PF-4 has two motifs with the amino acids AspLeuGln (DLQ) at position 7 to 9 and 54 to 56. This sequence seems to be implicated in the inhibitory activity of PF-4 on colony-forming unit–granulocyte, macrophage (CFU-GM) progenitor cells.29 Finally, four cystein residues are found in this chemokine at position 10, 12, 36, and 52 that form disulfite bridges between Cys10 and Cys 36 and between Cys 12 and Cys 52. To identify antiangiogenic PF-4–derived peptides, we synthesized a number of overlaping peptides that corresponded to different domains of PF-4 and investigated their effect in different biological assays. These comprise two peptides that map PF-4 from position 17 at the N-terminus to two thirds of the molecule (peptide 17-58 and peptide 34-58) and three other peptides that map PF-4 from the middle of the molecule to the carboxy terminus (peptides 47-58, 47-70, and 58-70). Peptide 17-58 has two clusters of basic amino acids and peptide 34-58 contains only one cluster of basic amino acids. However, both peptides contain a DLQ motif. Therefore, peptide 17-58 but not peptide 34-58 contains the first cluster of basic amino acids. Peptide 47-70 contains two clusters of basic amino acids (including the major heparin-binding domain) and the DLQ motif. Peptide 47-58 only possess the DLQ motive and one cluster of basic amino acids, but not the major heparin-binding domain. Peptide 58-70 only contains the major heparin-binding domain, but not the DLQ sequence or the second cluster of basic amino acids at position 49 and 50.
We chose to compare the activity of the different PF-4–derived peptides towards FGF-2 and VEGF for the following reasons. FGF-2 exist as monomers and dimerizes for receptor activation.30 VEGF, on the contrary, exists already as a natural dimer.2,9Dimerization seems not to be required for VEGF receptor activation.9 Nevertheless, HSPGs regulate the activity of both growth factors.9,30 Furthermore, FGF-2 and VEGF are inhibited by PF-4.20 21 It was therefore of interest to examine whether similar PF-4–derived peptides are involved in the inhibition of the activity of both growth factors.
Peptide 47-70, but not the other peptides, completely inhibited proliferation and angiogenesis in vitro induced by FGF-2 or VEGF. This is in agreement with Sato et al,31 who have reported that a carboxyl-terminal peptide that corresponds to the heparin-binding fragment of PF-4 retained the blocking effect on FGF-2 activity in endothelial cells. Peptides 47-70 and 47-58 have also been shown to inhibit the proliferation of leukemic cell lines and megakaryocytopoiesis.32, 33 However, myelopoiesis (CFU-GM, burst-forming unit–erthrocyte [BFU-E], and CFU-megakaryocyte [CFU-MK]) is also inhibited by peptide 34-58 and this at much lower concentrations when compared with native PF-4 or peptide 47-70.27 The inhibition of megakaryocytopoiesis by peptide 34-58 does not exceed 30% to 40% at 20 nmol/L peptide concentration. To the contrary, peptide 47-70 nearly completely inhibited endothelial cell function, albeit at higher peptide concentrations.
Binding of 125I-FGF-2 or 125I-VEGF to endothelial cell FGF or VEGF receptors was inhibited by peptide 47-70, but not by the other peptides. Peptide 47-70 inhibited binding of125I-FGF-2 to FGF receptors and to proteoheparan sulfates. Proteoheparan sulfates also modulate VEGF binding.9 This may indicate that peptide 47-70 interferes directly with FGF-2/FGF or VEGF/VEGF receptor interactions or indirectly via proteoheparan sulfates.
To address the requirement of proteoheparan sulfates in the effect of peptide 47-70 on FGF-2 or VEGF binding, binding studies were performed with heparan sulfate–deficient CHO cells transfected with a vector that contained a FGFR1 or VEGFR2 cDNA. Peptide 47-70 inhibited heparin-induced binding of 125I-FGF-2 or125I-VEGF to FGFR1 or VEGFR2 in CHOm cells, respectively. However, binding was not inhibited in the absence of heparin. We have previously shown that the whole PF-4 molecule inhibits FGF-2 binding to CHOmFGFR1 in the absence of heparin by 50%.21 This would indicate that other PF-4 domains are required for full inhibitory activity. We also found that a peptide derived from the central part of PF-4 between amino acids 17 to 58 interfered with the heparin-induced binding of FGF-2 or VEGF to CHOmFGFR1 or CHOmVEGFR2 cells. Peptide 17-58 contains at position 20, 22, 23 49, and 50 basic amino acids with potential heparin-binding activity. This might explain the effect of the peptide on heparin-dependent binding of FGF-2 or VEGF to CHOmFGFR1 or CHOmVEGFR2 cells.
PF-4 associates directly with FGF-2 or VEGF.21,22 We therefore examined whether the different peptides were able to compete with the direct binding of PF-4 to FGF-2 or VEGF. As expected, peptide 47-70 competed with the association of PF-4 with FGF-2 or VEGF. This may indicates that the PF-4 domain 47-70 is essential for direct association of PF-4 to FGF-2 or VEGF. We investigated the effect of peptides on the formation of FGF-2 dimers because PF-4 inhibits endogenous or heparin-induced FGF-2 dimerization.21 In the absence of heparin, peptide 47-70 modulated FGF-2 dimer formation. With increasing concentrations of peptide, FGF-2 dimers increased first and subsequently declined. However, in the presence of heparin, FGF-2 dimerization is severely inhibited. What is the explanation for these findings? The stimulation of FGF-2 dimer formation may be explained by direct binding of peptide 47-70 to FGF-2, thus inducing inactive FGF-2 dimers. Through an elegant series of structural studies, it has recently been shown that FGF-2 undergoes two types of dimerizations that lead to active or inactive FGF-2 dimers.34 Active FGF-2 dimers are “side by side” dimers and induced with heparan sulfate oligomers, dodecamers, dimers, or trimers, but not by hexamers or septamers. Inactive dimers are “head to head“ dimers and are induced by compounds such as sucralsulfate. These studies suggest that peptide 47-70 behaves to a certain extent similarily to sucralsulfate and induces head to head dimers in a heparin-free context. Structural biology studies should establish the type of FGF-2 dimer that occurs in the presence of peptide 47-70. Peptide 17-58 was also able to interfere to some extent with the association of PF-4 to the angiogenic growth factors. However, peptide 17-58 did not inhibit heparin-dependent dimer formation of FGF-2. This may account for the lack of inhibitory activity of this peptide in the biological assays and in the binding experiments of FGF-2 or VEGF to endothelial cells.
It was surprising that peptide 17-58 inhibited to some extent the heparin-dependent binding of FGF-2 or VEGF to heparan sulfate–deficient cells that expressed FGF or VEGF receptors, but did not inhibit the binding of these growth factors to endothelial cells. The reasons for these differences are not understood. Peptide 17-58 is alone perhaps insufficient to disturb the interaction of FGF-2 or VEGF and their receptors with heparan sulfates bound to proteoglycans at the cell surface.
Another observation is the stimulatory activity of peptide 47-58 on FGF-2 induced tube formation in in vitro angiogenesis, but not in any other assays we performed. The reasons for this stimulatory activity remain to be elucidated.
What are the implications of these results for the structure and function of PF-4? The fact that peptide 47-70 but not 47-58 or 58-70 showed inhibitory activity would indicate that the heparin-binding domain is not sufficient for full antiangiogenic activity. An additional sequence that contains the second basic amino acid cluster and/or the DLQ motif (Asp54, Leu55, Gln56) is possibly required. On the other hand, peptide 17-58, but not peptide 34-58, showed inhibitory activity in CHOm cells transfected with FGFR1 or VEGFR2 cDNA. This indicates that a sequence that comprises amino acid 17-34 is required for this inhibitory activity. As mentioned earlier, a cluster of basic amino acid residues is localized within this sequence. It is possible that this sequence is implicated in the inhibitory activity of peptide 17-58.
Our results are in apparent conflict with those reported by Maione et al.35 Maione et al35 have reported that a mutated PF-4 molecule that had the C-terminal heparin-binding domain mutated was still able to inhibit angiogenesis. The reasons for this difference are not known. In our hands, it is unlikely that PF-4 domains found outside the 47-70 sequence are essential for the inhibitory activity of PF-4 in angiogenesis. However, other domains may contribute to enhance the inhibitory effect of domain 47-70 in vivo. This may also be supported by the intriguing finding that a peptide derived from the central part of PF-4 that contains a potential heparin-binding motif at positions 20, 22, and 23 only showed inhibitory activity on heparin-induced FGF-2 or VEGF binding to FGF or VEGF receptors to heparan sulfate–deficient cells, but not in binding assays performed with capillary endothelial cells and not on biologic activity.
Taken together, our results indicate that peptide 47-70 derived from the C-terminus of PF-4 interferes with both FGF-2 and VEGF function. Furthermore, a peptide derived from the central part of PF-4 that contains a potential heparin-binding domain interferes with heparin-dependent binding of FGF-2 or VEGF to FGF or VEGF receptors in heparan sulfate–deficient cells. These observations are of significance for PF-4’s mechanisms of angiogenesis inhibition.
ACKNOWLEDGMENT
The authors thank Dr H. Prats and Dr D.B Rifkin for providing recombinant human FGF-2, Dr Avner Yayon for providing CHOmFGFR1 cells, and Dr I. Kramer (Growth Factor and Cell Differentiation Laboratory) for critically reading the manuscript.
Supported by grants from the Fondation de France, The Association de la Recherche sur le Cancer (ARC), and the Ministère de la Science et de la Recherche (MSR) to A.B. and from the Del Duca foundation to M.A. and J.P.C.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Andreas Bikfalvi, MD, PhD, Laboratoire des Facteurs de Croissance et de la Differenciation Cellulaire, Bâtiment de Recherche Biologie Animale, Avenue des Facultés, 33405 Talence, France; e-mail:a.bikfalvi@croissance.u-bordeaux.fr.







This feature is available to Subscribers Only
Sign In or Create an Account Close Modal