Abstract
Screening of antibodies to hepatitis C virus (HCV) is widely used for monitoring the prevalence of HCV infections and to assess HCV infectivity. Among HCV-infected individuals in the general population, the interval between the detection of HCV RNA and the development of HCV antibodies is usually 5 to 6 weeks, but in rare cases, seroconversion may be prolonged up to 6 to 9 months. In this study, we tested for the presence of HCV RNA during the antibody-undetectable period of 19 drug-injecting HCV seroconverters to gain insight into the antibody-negative carrier status in this population. HCV seroconversion status was determined by testing the first and last serum samples obtained from each subject, using third-generation antibody screening and confirmation assays. Serial samples were tested for HCV-specific antibodies to establish the moment of seroconversion and HCV RNA by single reverse transcriptase-polymerase chain reaction (RT-PCR) and branched DNA assay (bDNA) in serum. Plasma and peripheral blood mononuclear cells (PBMCs) were independently collected and tested for HCV RNA. HCV RNA-positivity was confirmed by Southern blot hybridization and sequencing of serial samples. The 19 HCV seroconverters had a mean follow-up of 5 years (range, 1 to 8 years). Of the 19, 4 were human immunodeficiency virus (HIV)-infected before HCV seroconversion. HCV RNA was detected in serum before seroconversion in 12 (63.2%) of the 19 HCV seroconverters, independent of HIV status. In 7 of these 12, the antibody-undetectable period was relatively short (2 to 10 months). The other 5, who were all HIV-negative before HCV seroconversion, had intermittent low levels of HCV RNA before seroconversion for a period of more than 12 months, with a mean of 40.8 months (range, 13 to 94 months). In all 5 individuals, independent repeats of the experiments confirmed the presence of HCV RNA in serum, and in 3 of these individuals, HCV-positivity was confirmed in independently collected plasma and PBMC samples. Low levels of HCV RNA may be present during prolonged antibody-undetectable periods before seroconversion in a number of injecting drug users. Independent of HIV status, their immune system appears to be unable to respond to these low HCV RNA levels and was sometimes only activated after reinfections with distinct HCV genotypes. These results indicate that primary HCV infection may not always elicit the rapid emergence of HCV antibodies and suggests that persistent low levels of HCV RNA (regardless of the genotype) may not elicit at all or delay antibody responses for prolonged periods of time.
HEPATITIS C VIRUS (HCV), an RNA virus with marked genetic heterogeneity, is the etiological agent of most cases of posttransfusion and community-acquired non-A and non-B hepatitis.1 This blood-borne virus is widespread among injecting drug users (IDUs),2-4 and HCV infections may cause a benign, asymptomatic disorder with an indolent course5 but can eventually cause progressive liver disease, cirrhosis, and liver cancer.6-8 The illness has a complex course, with RNA levels that are often transient.9-12Antibodies to HCV are detected by immunoassays, and their presence is closely related with infectivity, especially in blood donors,13-15 hemodialysis patients,16hemophiliacs,17 and patients with chronic HCV.18,19 Current immunoassays detect most individuals with HCV infections, and HCV RNA has been detected during short antibody-undetectable periods (the window phase from infection to the development of antibodies to HCV) in persons infected by blood transfusions20,21 or surgical procedures22 and in experimentally infected primates.23,24 However, up to half of immunosuppressed patients, such as organ transplant patients, fail to generate antibodies to HCV,25 and IDUs also have a high proportion of delayed antibody responses.2 In 2 other studies, HCV RNA was significantly more detectable in whole blood than in plasma samples among antibody-negative individuals, indicating that a proportion of HCV RNA in peripheral blood might be missed.26,27 Unexpected clinical profiles, with HCV RNA present for a period of 5 years without antibodies to HCV, were observed in experimentally inoculated chimpanzees.28Spontaneous loss of antibodies to HCV, or seroreversion, among immunocompetent individuals has recently been described,29and we have observed a complete loss of antibodies to HCV in individuals both with and without reinfection.30Therefore, the present study was performed to identify and confirm the presence of HCV RNA in serum and other blood compartments before the moment of seroconversion in the high-risk group of IDUs.
PATIENTS AND METHODS
Patients and sample collection.
The study population consists of IDUs participating in the Amsterdam Cohort Studies on human immunodeficiency virus (HIV) and acquired immunodeficiency syndrome (AIDS) among IDUs. From a cohort started in December 1985,31 we selected drug users in March 1996 who were observed for at least 3 years and had at least 7 visits (n = 358). The 19 HCV seroconverters (10 women and 9 men; mean age, 38 years) and the cohort of 358 participants (148 women and 210 men; mean age, 42 years) from which they are recruited are representative of the original cohort and are 99% whites. A previous study3 identified risk factors for a prevalent HCV infection, which included a history of injecting drug use and duration and frequency of injecting drug use. Because our aim was to study incident cases of HCV infection, we therefore selected in March 1996 only drug users with a history of injecting drug use who had sufficient follow-up (>3 years) and study visits (>6). Because of this selection, we included in this study mainly drug users with a relatively long career of injecting drugs, of whom 88.3% (n = 316) were found to be already infected with HCV at intake. Among the remainder of 42 participants, 23 remained seronegative throughout the study period, whereas 19 seroconverters were identified. Their HCV seroconversion status was established by screening the first and the last serum samples for HCV antibodies, and serial samples of the seroconverters were then tested to establish the approximate seroconversion point. The date of HCV seroconversion was determined by calculating the midpoint between the last seronegative and first seropositive sample. Nineteen HCV seroconverters, of which 1 reseroconverted, were identified and studied longitudinally for the presence of HCV RNA. Serum samples were initially used to determine the presence of HCV RNA before seroconversion. The findings of HCV RNA-positive samples before HCV seroconversion were confirmed by detecting HCV RNA in serum and peripheral blood mononuclear cells (PBMCs) or plasma sampled at the same time point. In addition, the branched DNA (bDNA) assay was performed on the same serial serum samples before and after HCV seroconversion. Serum and EDTA-blood samples of the 19 HCV seroconverters were drawn at different Municipal Health stations in Amsterdam. For final handling and storage, all serum samples were sent to the Academic Medical Centre, whereas all EDTA-blood samples were sent to the Central Laboratory of The Netherlands Red Cross Blood Transfusion Service. Serum samples were stored initially at +4°C, centrifuged, aliquoted, frozen at −20°C within 24 hours of collection, and ultimately stored at −70°C. PBMCs and plasma samples, also initially stored at +4°C, were separated from EDTA-blood by density-gradient centrifugation on Ficoll-Hypaque (Pharmacia, Uppsala, Sweden). Cryopreservation on aliquoted PBMCs was performed using computer-automated freezing in liquid nitrogen.
Serological tests.
Samples were tested for the presence of antibodies to HCV by the third-generation enzyme immunoassay (EIA 3.0; Abbott Laboratories, Chicago, IL). All positive EIA 3.0 assays were confirmed by the third-generation strip immunoblot assay (SIA, RIBA; Chiron Corp, Emeryville, CA). Antibodies to HIV-1 were determined with commercial EIA (Abbott Laboratories) and confirmed by Western blot (Diagnostic Biotechnology, Herent, Belgium). Individuals who remained HIV-negative were screened on all consecutive samples by EIA. All serological assays were performed according to the manufacturer’s manual.
Detection of HCV RNA by reverse transcriptase-polymerase chain reaction (RT-PCR) in serum and plasma samples.
HCV RNA was isolated from a 100-μL serum or plasma sample, according to Boom et al,32 and was used immediately in single RT-PCR experiments, using primers located in the 5′ noncoding region, or stored at −70°C as previously described.12Briefly, one fifth (10 μL) of the isolated RNA was subjected to reverse transcription and a single PCR. For reverse transcription, 10 μL RNA was incubated with 25 ng of antisense primer (nt 319-324) 5′-ACCTCC-3′ for 5 minutes at 65°C and then cooled down to 42°C. Finally, 14 μL of the reverse transcription mixture was added, containing 10 mmol/L Tris-HCl, pH 8.3, 50 mmol/L KCl, 0.1% Triton X-100 (Packard Instrument Co, Inc, Downers Grove, IL), 6 mmol/L MgCl2, 0.6 mmol/L of each dNTP, 20 U RNAse inhibitor (Promega, Madison, WI), and 100 U SuperScript II (Life Technologies, Gaithersburg, MD). The mixture was incubated for 30 minutes at 42°C, and 12.5 μL was used for the single PCR in duplicate. For the PCR, the GeneAmp PCR carry-over prevention kit (Perkin Elmer Cetus, Branchburg, NJ) was used to avoid contamination. The PCR was performed in a 50 μL volume containing 100 ng of sense primer (nt 47-68) 5′-GTGAGGAACTACTGTCTTCACG-3′, 100 ng of antisense primer (nt 292-312) 5′-ACTCGCAAGCACCCTATCAGG-3′, 2.5 U Ampli-Taq polymerase (Perkin Elmer Cetus), 50 mmol/L Tris-HCl, pH 8.3, 20 mmol/L KCl, 1.2 mmol/L MgCl 2, 2.5 μmol/L of each dNTP, 25 μmol/L dUTP, and 0.5 U Uracil N-glycosylase (Perkin Elmer Cetus). The thermal cycler (type 480; Perkin Elmer Cetus) was programmed as follows: 5 minutes at 95°C and subsequently 40 cycles of 95°C for 1 minute, 55°C for 1 minute, and 72°C for 2 minutes, and then incubation of samples for 8 minutes at 72°C. PCR products were subjected to electrophoresis in 2% agarose containing ethidiumbromide and visualized under UV light. As positive controls, we used a pool of HCV-positive serum quantified by the bDNA technology (Chiron Corp) to a level of 1.6 × 106 HCV RNA copies/mL and 100-fold dilution. The sensitivity of our single RT-PCR was evaluated by serial 2-fold dilutions of the quantified pool of serum and was found to have a detection limit of approximately 103 HCV RNA copies/mL (results not shown). As negative controls, we used a pool of commercially available serum (seronegative for HIV, HBV, and HCV) and TE (Tris/EDTA). All positive and negative controls were tested in parallel with the test samples throughout the entire procedure, starting from RNA extraction. Serum samples yielded no different PCR results from plasma samples (results not shown). The RT-PCR rendered good duplicates unless its detection limit was reached, in which case it rendered a plus/minus duplicate (Poisson distribution). Samples were randomly tested for the presence of HCV RNA, and all precautions were taken to avoid any possible contamination, using separated locations for extraction of specimen, amplification of HCV RNA, and analyses of PCR products.
Isolation and detection of HCV RNA by RT-PCR in PBMCs.
HCV RNA was isolated from 100 μL PBMCs containing approximately 105 cells/mL using Total RNA Isolation Reagent (TRIzol Reagent; Life Technology), which is designed for the isolation of total RNA from cells and tissue, according to the manufacturer’s manual. HCV RNA was detected in PBMCs with nested PCR, processing one tenth of the initial RT-PCR products under the same conditions as in the single PCR, but with 25 cycles using sense primer (nt 74-91) 5′-AGCGCCTAGCCATGGCGT-3′ and antisense primer (nt 243-260) 5′-TACCACAAGGCCTTTCGC-3′, which were extended at the 5′-end with the −21 M13 primer and the reverse M13 primer, respectively. Samples were randomly tested for the presence of HCV RNA, and all precautions were taken to avoid any possible contamination, using separated locations for extraction of specimen, amplification of HCV RNA, and analyses of PCR products.
HCV RNA quantification.
The HCV RNA load in serum was determined longitudinally by the bDNA signal amplification assay 2.0 (Quantiplex HCV RNA; Chiron Corp) according to the manufacturer’s manual. All samples were tested in duplicate, and the mean value of the duplicate tests was used for data analysis. Viral load, expressed as HCV RNA copies per milliliter, was determined by comparison with an external standard curve, having a quantitation limit of 2.0 × 105 HCV RNA copies/mL.
Specificity and confirmation of HCV PCR products.
The specificity of the PCR products was confirmed by using a 5′-digoxigenine labeled probe33 (nt 264-291), 5′-TTGGGTCGCGAAAGGCCTTGTGGTACTG-3′, in high stringency hybridizations, according to the manual (Boehringer Mannheim GmbH, Mannheim, Germany) and by genotyping. The genotypes were determined either using the HCV LiPa protocol (Line Probe Assay [LiPa]; Innogenetics, Ghent, Belgium),34 according to the manual, or by direct-sequencing the products obtained by nested PCR processing one tenth of the initial RT-PCR products. Nested PCR (encompassing the same region as used in the HCV LiPa protocol) was performed under the same conditions as the single PCR, but for 25 cycles, using sense primer (nt 74-91) 5′-AGCGCCTAGCCATGGCGT-3′ and antisense primer (nt 243-260) 5′-TACCACAAGGCCTTTCGC-3′, which were extended at the 5′-end with the −21 M13 primer and the reverse M13 primer, respectively. The Thermo Sequenase cycle-sequencing reaction was performed according to the manual, using the Dye-primer method (Amersham Life Science, Buckinghamshire, UK). Serum samples were initially used to determine the presence of HCV RNA before seroconversion. The finding of HCV RNA-positive samples before HCV seroconversion were confirmed by detecting and genotyping HCV in samples of serum and PBMCs or plasma drawn at the same time point.
Computer sequence analysis.
The PCR products were directly sequenced with an ABI Automated Sequencer model 373A (Applied Biosystems, Columbia, MD) using the 1.2.0 software. The direct sequences were assembled by the Sequence Navigator program (ABI) and were further optimized manually. P-distances and consensus sequences of part of the 5′UTR (based on at least 10 sequences of the 6 major genotypes as found in Genbank) were calculated with MEGA.35Phylogenetic trees were computed using the neighbour-joining algorithm.36 The resulting tree was assessed by the bootstrap method37 based on 500 replicates.
RESULTS
Determination of HCV seroconverters.
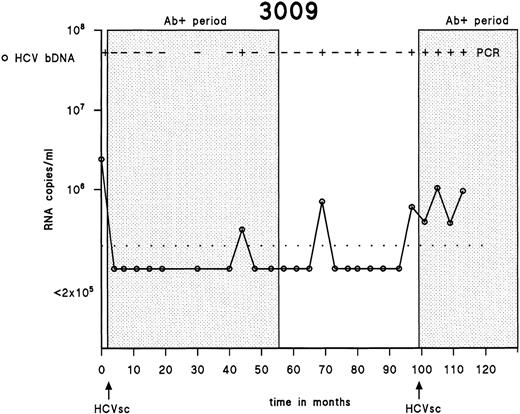
Testing of the first and the last samples drawn from 358 IDUs showed that 316 (88.3%) were positive for antibodies to HCV, 23 (6.4%) were negative for antibodies to HCV, and 19 (5.3%) seroconverted for HCV during the follow-up period. One of 19 HCV seroconverters seroreverted during follow-up, as described earlier.30 Antibodies to HCV disappeared completely after the first 55 months, although HCV RNA was intermittently detectable in serum during that period of 45 months. Subsequently, a new seroconversion occurred at 98 months (Fig 1).
Patterns of HCV viremia and serological response in an individual showing reseroconversion. The detection limit of the bDNA assay is shown as a dotted line.
Patterns of HCV viremia and serological response in an individual showing reseroconversion. The detection limit of the bDNA assay is shown as a dotted line.
Confirmation by hybridization of the initial HCV RNA-positive serum samples in 19 HCV seroconverters.
To confirm all PCR products initially detected in serum samples of the 19 HCV seroconverters after UV illumination, we performed high-stringency Southern blot hybridizations using a digoxigenine-labeled probe that disclosed the specificity of the RT-PCR. The RT-PCR rendered good duplicates unless the detection limit of the system was reached (Fig 2). As a control panel, we included the first serum samples of 6 HCV seronegative subjects who were seronegative with EIA 3.0 at both first and last sample over a mean period of 5 years. RT-PCR and Southern blot hybridization experiments performed on this control panel showed that no detectable HCV RNA was present in any of the first samples (results not shown).
Southern blot hybridization of the duplicate RT-PCR results using an internal 5′-digoxigenine–labeled HCV probe. The initially detectable PCR products in duplicate of the 19 HCV seroconverters were blotted and hybridized under high-stringency conditions. The 19 seroconverters are indicated by an identification number. Positive and negative controls are indicated.
Southern blot hybridization of the duplicate RT-PCR results using an internal 5′-digoxigenine–labeled HCV probe. The initially detectable PCR products in duplicate of the 19 HCV seroconverters were blotted and hybridized under high-stringency conditions. The 19 seroconverters are indicated by an identification number. Positive and negative controls are indicated.
Presence of HCV RNA in different blood compartments among HIV-negative subjects with prolonged antibody-negative periods.
RT-PCR was used to analyze serial blood samples for the presence of HCV RNA before HCV seroconversion in different blood compartments drawn from the 5 HIV-negative subjects (0073, 0146, 1083, 1085, and 3059), when available. In 3 of these 5 individuals (0073, 1083, and 3059), the initial sample was HCV RNA-negative. HCV RNA, as detected by single RT-PCR, fluctuated but was repeatedly found at low levels in serial samplings of all 5 subjects before HCV seroconversion. On some occasions, HCV RNA was found by the bDNA assay in serum more than 1 year before HCV seroconversion. In 3 of 5 subjects (1085, 3059, and 0146), the presence of HCV RNA in serum could be confirmed independently in PBMCs or plasma. The antibody status and presence of HCV RNA in these 5 subjects are summarized in Table 1.
Presence of HCV RNA in 5 Individuals Without Antibodies to HCV More Than 1 Year
| Subject . | Visit . | Time in Months . | EIA 3.0 . | RT-PCR . | bDNA (Meq/mL) . | ||
|---|---|---|---|---|---|---|---|
| Serum . | Plasma . | PBMCs . | Serum . | ||||
| 1085 | 1 | −94 | − | +* | NA | NA | 0.254* |
| 2 | −51 | − | +* | NA | + | <0.2 | |
| 3 | −47 | − | −* | NA | NA | <0.2 | |
| 4 | −43 | − | +* | NA | + | <0.2 | |
| 5 | −39 | − | −* | −* | NA | <0.2 | |
| 6 | −35 | − | −* | −* | NA | <0.2 | |
| 7 | −27 | − | −* | −* | NA | <0.2 | |
| 8 | −23 | − | −* | −* | NA | <0.2 | |
| 9 | −18 | − | −* | −* | NA | <0.2 | |
| 10 | −14 | − | −* | −* | − | <0.2 | |
| 11 | −10 | − | −* | −* | − | <0.2 | |
| 12 | −6 | − | +* | +* | NA | <0.2 | |
| 13 | −2 | − | −* | −* | NA | <0.2 | |
| 14 | +2 | + | +* | NA | NA | <0.2 | |
| 3059 | 1 | −42 | − | − | NA | NA | <0.2 |
| 2 | −38 | − | +* | NA | + | 0.439* | |
| 3 | −34 | − | +* | NA | + | 0.686* | |
| 4 | −31 | − | −* | NA | + | 0.306* | |
| 5 | −27 | − | −* | NA | NA | <0.2 | |
| 6 | −22 | − | −* | NA | NA | <0.2 | |
| 7 | −18 | − | −* | NA | NA | <0.2 | |
| 8 | −14 | − | + | NA | + | <0.2 | |
| 9 | −11 | − | −* | − | NA | <0.2 | |
| 10 | −6 | − | −* | − | NA | <0.2 | |
| 11 | −2 | − | +* | +* | + | 0.641* | |
| 12 | +2 | + | +* | + | NA | 0.390 | |
| 13 | +6 | + | NT | + | NA | 0.211 | |
| 14 | +10 | + | NT | + | NA | 1.337 | |
| 0146 | 1 | −28 | − | +* | NA | NA | <0.2 |
| 2 | −24 | − | −* | NA | NA | <0.2 | |
| 3 | −21 | − | −* | NA | NA | <0.2 | |
| 4 | −15 | − | +* | NA | NA | 0.528* | |
| 5 | −12 | − | −* | NA | NA | <0.2 | |
| 6 | −9 | − | −* | NA | + | <0.2 | |
| 7 | −5 | − | −* | NA | NA | <0.2 | |
| 8 | −2 | − | +* | NA | + | <0.2 | |
| 9 | +2 | + | +* | NA | NA | <0.2 | |
| 10 | +5 | + | −* | NA | NA | <0.2 | |
| 11 | +9 | + | +* | NA | NA | <0.2 | |
| 0073 | 1 | −44 | − | − | NA | NA | <0.2 |
| 2 | −40 | − | − | NA | NA | <0.2 | |
| 3 | −35 | − | − | NA | NA | <0.2 | |
| 4 | −31 | − | +* | NA | NA | <0.2 | |
| 5 | −25 | − | +* | NA | NA | <0.2 | |
| 6 | −16 | − | −* | NA | NA | <0.2 | |
| 7 | −11 | − | −* | NA | NA | <0.2 | |
| 8 | +11 | + | NT | −* | NA | <0.2 | |
| 9 | +16 | + | NT | −* | NA | <0.2 | |
| 10 | +21 | + | NT | +* | NA | <0.2 | |
| 11 | +25 | + | NT | +* | NA | 0.346 | |
| 1083 | 1 | −16 | − | − | NA | NA | <0.2 |
| 2 | −13 | − | +* | NA | NA | <0.2 | |
| 3 | −8 | − | −* | NA | NA | <0.2 | |
| 4 | −6 | − | −* | NA | NA | <0.2 | |
| 5 | −3 | − | +* | NA | NA | 0.698* | |
| 6 | +3 | + | −* | NA | NA | <0.2 | |
| 7 | +12 | + | +* | NT | NA | 0.286 | |
| 8 | +16 | + | NT | + | NA | 0.472 | |
| Subject . | Visit . | Time in Months . | EIA 3.0 . | RT-PCR . | bDNA (Meq/mL) . | ||
|---|---|---|---|---|---|---|---|
| Serum . | Plasma . | PBMCs . | Serum . | ||||
| 1085 | 1 | −94 | − | +* | NA | NA | 0.254* |
| 2 | −51 | − | +* | NA | + | <0.2 | |
| 3 | −47 | − | −* | NA | NA | <0.2 | |
| 4 | −43 | − | +* | NA | + | <0.2 | |
| 5 | −39 | − | −* | −* | NA | <0.2 | |
| 6 | −35 | − | −* | −* | NA | <0.2 | |
| 7 | −27 | − | −* | −* | NA | <0.2 | |
| 8 | −23 | − | −* | −* | NA | <0.2 | |
| 9 | −18 | − | −* | −* | NA | <0.2 | |
| 10 | −14 | − | −* | −* | − | <0.2 | |
| 11 | −10 | − | −* | −* | − | <0.2 | |
| 12 | −6 | − | +* | +* | NA | <0.2 | |
| 13 | −2 | − | −* | −* | NA | <0.2 | |
| 14 | +2 | + | +* | NA | NA | <0.2 | |
| 3059 | 1 | −42 | − | − | NA | NA | <0.2 |
| 2 | −38 | − | +* | NA | + | 0.439* | |
| 3 | −34 | − | +* | NA | + | 0.686* | |
| 4 | −31 | − | −* | NA | + | 0.306* | |
| 5 | −27 | − | −* | NA | NA | <0.2 | |
| 6 | −22 | − | −* | NA | NA | <0.2 | |
| 7 | −18 | − | −* | NA | NA | <0.2 | |
| 8 | −14 | − | + | NA | + | <0.2 | |
| 9 | −11 | − | −* | − | NA | <0.2 | |
| 10 | −6 | − | −* | − | NA | <0.2 | |
| 11 | −2 | − | +* | +* | + | 0.641* | |
| 12 | +2 | + | +* | + | NA | 0.390 | |
| 13 | +6 | + | NT | + | NA | 0.211 | |
| 14 | +10 | + | NT | + | NA | 1.337 | |
| 0146 | 1 | −28 | − | +* | NA | NA | <0.2 |
| 2 | −24 | − | −* | NA | NA | <0.2 | |
| 3 | −21 | − | −* | NA | NA | <0.2 | |
| 4 | −15 | − | +* | NA | NA | 0.528* | |
| 5 | −12 | − | −* | NA | NA | <0.2 | |
| 6 | −9 | − | −* | NA | + | <0.2 | |
| 7 | −5 | − | −* | NA | NA | <0.2 | |
| 8 | −2 | − | +* | NA | + | <0.2 | |
| 9 | +2 | + | +* | NA | NA | <0.2 | |
| 10 | +5 | + | −* | NA | NA | <0.2 | |
| 11 | +9 | + | +* | NA | NA | <0.2 | |
| 0073 | 1 | −44 | − | − | NA | NA | <0.2 |
| 2 | −40 | − | − | NA | NA | <0.2 | |
| 3 | −35 | − | − | NA | NA | <0.2 | |
| 4 | −31 | − | +* | NA | NA | <0.2 | |
| 5 | −25 | − | +* | NA | NA | <0.2 | |
| 6 | −16 | − | −* | NA | NA | <0.2 | |
| 7 | −11 | − | −* | NA | NA | <0.2 | |
| 8 | +11 | + | NT | −* | NA | <0.2 | |
| 9 | +16 | + | NT | −* | NA | <0.2 | |
| 10 | +21 | + | NT | +* | NA | <0.2 | |
| 11 | +25 | + | NT | +* | NA | 0.346 | |
| 1083 | 1 | −16 | − | − | NA | NA | <0.2 |
| 2 | −13 | − | +* | NA | NA | <0.2 | |
| 3 | −8 | − | −* | NA | NA | <0.2 | |
| 4 | −6 | − | −* | NA | NA | <0.2 | |
| 5 | −3 | − | +* | NA | NA | 0.698* | |
| 6 | +3 | + | −* | NA | NA | <0.2 | |
| 7 | +12 | + | +* | NT | NA | 0.286 | |
| 8 | +16 | + | NT | + | NA | 0.472 | |
Abbreviations: NA, not available; NT, not tested.
Repeated independent extractions from aliquots and amplifications.
Determination of HCV RNA by sequencing among subjects with prolonged antibody-negative periods in different blood compartments.
To verify all positive findings by RT-PCR in the different blood compartments from the 5 subjects, we performed sequence analyses of samples drawn before and after HCV seroconversion. HCV genotypes were determined using phylogenetic trees computed by the neighbour-joining algorithm, and genotype 1 was initially found in all subjects, although sequences were distinct at 1 or more nucleotide positions. As shown in Table 1, HCV RNA positivity was confirmed in independently collected plasma or PBMC samples of 3 subjects (1085, 3059, and 0146), but exclusively serum samples were available for testing in 2 subjects (0073 and 1083).
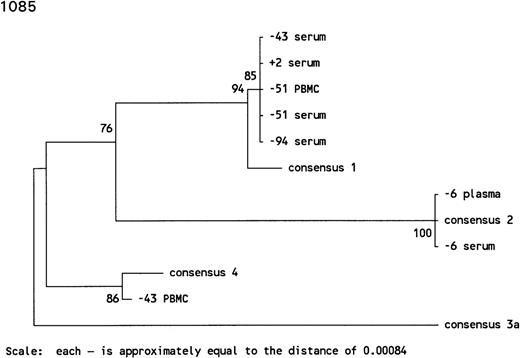
In subject 1085, there was an extremely long antibody-undetectable period of 94 months accompanied by intermittent detection of HCV RNA. The initial serum sample, drawn 94 months before HCV seroconversion, was positive for HCV RNA both by RT-PCR and bDNA testing and harbored an HCV genotype 1. The sample collected 51 months before HCV seroconversion was positive for HCV RNA by RT-PCR in serum and PBMCs and contained HCV genotype 1 in both compartments. The sample taken at 47 months before seroconversion was negative for HCV RNA, whereas the samples taken 4 months later were HCV RNA positive, and although HCV genotype 1 was found in serum, HCV genotype 4 was found in PBMCs sampled at the same time point. Thereafter, the HCV infection seemed to be cleared from blood for the next 37 months. Infection with HCV genotype 2 apparently occurred 6 months before HCV seroconversion, as detectable in plasma and serum. The sample collected 2 months before HCV seroconversion was HCV RNA negative, both in plasma and serum by RT-PCR, whereas HCV genotype 1 was found just after HCV seroconversion, indicating several reinfections in this individual (Figs 3A and 4).
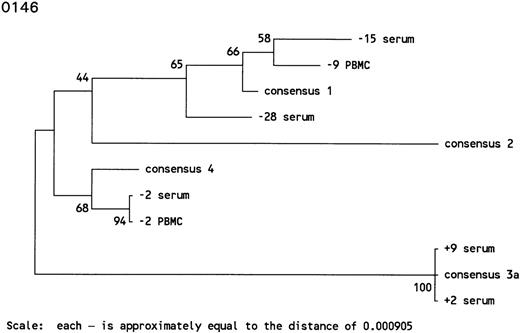
Phylogenetic trees of the 5′ noncoding region of HCV showing the genotypes identified in 3 individuals with prolonged antibody-undetectable periods. Patient specimens, consensus sequences, time in months before (−) and after (+) HCV seroconversion, and the 6 major consensus sequences are indicated. The trees were bootstrapped 500 times and the numbers represent the percentages of the 500 bootstraps where those branches were the same. The horizontal distances are represented by the scale bar in the lower part of the figure and the distance is indicated.
Phylogenetic trees of the 5′ noncoding region of HCV showing the genotypes identified in 3 individuals with prolonged antibody-undetectable periods. Patient specimens, consensus sequences, time in months before (−) and after (+) HCV seroconversion, and the 6 major consensus sequences are indicated. The trees were bootstrapped 500 times and the numbers represent the percentages of the 500 bootstraps where those branches were the same. The horizontal distances are represented by the scale bar in the lower part of the figure and the distance is indicated.
Alignment of all sequences found among the 3 individuals with prolonged seronegative periods. Patient specimens, consensus sequences, and time in months before (−) and after (+) HCV seroconversion are indicated.
Alignment of all sequences found among the 3 individuals with prolonged seronegative periods. Patient specimens, consensus sequences, and time in months before (−) and after (+) HCV seroconversion are indicated.
In subject 3059, the antibody-undetectable period of 38 months was accompanied by intermittent detection of HCV RNA. The sample taken at 38 months before HCV seroconversion was found to be positive by bDNA and RT-PCR in serum and PBMCs. Sequencing of virus isolated from serum and PBMC samples showed an infection with HCV genotype 1. HCV RNA remained detectable in both compartments by RT-PCR and bDNA in the sample taken at 34 months before HCV seroconversion; in PBMCs, genotype 1 was again detected by sequencing, but the virus in the serum sample was untypeable by sequencing and InnoLipa. The serum sample taken at 31 months before HCV seroconversion was repeatedly negative by RT-PCR, whereas the bDNA assay showed repeatedly positive results. HCV RNA was not detectable by RT-PCR or bDNA in the next 3 serum samples taken over a period of 17 months. Interestingly, in the sample drawn at 14 months before HCV seroconversion, genotyping showed HCV genotype 1 in PBMCs, whereas in serum of the same time point, HCV genotype 3a was found. Moreover, in samples taken 2 months before HCV seroconversion, genotype 3a was found in serum, plasma, and PBMCs. HCV genotype 3a remained present in the plasma samples tested after HCV seroconversion (Figs 3B and 4).
In subject 0146, the antibody-undetectable period of 28 months was accompanied by intermittent detection of HCV RNA. HCV genotype 1 was found in serum samples 28 and 15 months before HCV seroconversion. The serum sample collected 15 months before HCV seroconversion was positive by RT-PCR and bDNA. Interestingly, 9 months before HCV seroconversion, PBMCs were found to harbor HCV genotype 1, whereas the serum sample appeared to be HCV RNA negative. The sample taken 2 months before HCV seroconversion was HCV RNA-positive in serum and PBMCs, both of which were infected with HCV genotype 4. However, serum samples up to 9 months after HCV seroconversion appeared to harbor HCV genotype 3a, indicating several reinfections in this individual (Figs 3C and 4).
From subjects 0073 and 1083, only serum samples were available, and the antibody-undetectable period was accompanied by intermittent detection of HCV RNA. In subject 0073, HCV genotype 1 was detected in the serum samples drawn 31 and 25 months before HCV seroconversion. The next 2 samples were HCV RNA negative, but HCV genotype 1 was again found in consecutive serum samples drawn up to 25 months after HCV seroconversion. In subject 1083, the antibody-undetectable period of 13 months was accompanied by intermittent detection of HCV RNA. The initial serum sample, drawn 13 months before HCV seroconversion, contained HCV genotype 1, whereas samples drawn at 8 and 6 months before HCV seroconversion were HCV RNA-negative. Three months before HCV seroconversion, the serum sample was positive for both RT-PCR and bDNA, containing HCV genotype 1, and HCV seroconversion was observed in the next sample, accompanied by undetectable HCV RNA. However, HCV genotype 1 reappeared and was detectable both by RT-PCR and bDNA in the consecutive plasma and serum samples.
DISCUSSION
Detection of antibodies directed to HCV is still considered the gold standard for identification of HCV-infected individuals. A good correlation has been shown between antibodies to HCV and detectable HCV RNA, as well as between HCV seronegativity and undetectable RNA, among blood donors,13,14 hemodialysis patients,16hemophiliacs,17 and chronic HCV patients.19However, up to half of immunosuppressed patients, such as recipients of renal and liver transplants, fail to generate a humoral response if infected with HCV.25 Moreover, in studies of dialysis patients38 and on histologically verified patients with non-A, non-B hepatitis,19 commercially available antibody tests failed to identify a number of HCV-infected individuals. Thomas et al2 studied the sociodemographic and behavioral correlates of HCV infections among IDUs in Baltimore and found a high prevalence of HCV. They also found HCV RNA in 13 (30.2%) of 43 HCV-seronegative long-term drug users.
In the present study, we looked at the antibody-undetectable period for HCV RNA to gain insight into the proportion of undiscovered HCV carriers among IDUs. Because this population is at high risk for coinfections with other parenterally transmittable agents, ie, HIV, AIDS-associated immune-suppression could delay antibody response to HCV infection. However, we found a long HCV-seronegative period particularly in HIV-negative seroconverters (26.3%). The finding of long antibody-undetectable periods before HCV seroconversion may be explained in part by the transiently low detectable levels of HCV RNA present in most IDUs before seroconversion, which are apparently in some cases insufficient to elicit enough antibodies to be detected by diagnostic assays. Nevertheless, our 5 subjects with prolonged antibody-undetectable periods eventually seroconverted, apparently regardless of HCV genotype and viral load. Whether our data can be extrapolated to rarely exposed populations such as blood donors remains unanswered. However, in some antibody-negative samples, we found HCV RNA levels of more than 2 × 105 copies/mL that are in the range of 105 to 109 copies/mL as usually found in rejected blood donors during their preseroconversion periods of approximately 85 days.39 Although some of our IDUs may have comparable HCV RNA levels, results of quantification data obtained from different study groups by different assays may be hard to interpret. The question remains as to whether our observations warrant screening of blood donors or suspected HCV-positive patients only for antibodies to HCV by third-generation assays alone or in combination with HCV RNA. Previous studies of us showed that some patients have antibodies to HCV but low or no detectable HCV RNA in blood.12 Moreover, full and partial HCV seroreversions, with and without detectable HCV RNA, have been described in untreated immunocompetent humans.29,30 40-42 Such seroreversions may partially explain the antibody-undetectable period and may possibly indicate resolved HCV infections from blood. Loss of antibodies to HCV or intermittent seropositivity during follow-up may be interpreted as resolution of HCV from blood or latent infection with low HCV RNA levels and may be more common than generally suspected. Taken together, screening for HCV by PCR must be used in the acute preseroconversion phase, whereas screening for antibodies to HCV in combination with PCR may resolve all HCV-positive individuals. Thus, screening for antibodies to HCV in combination with PCR appears to be the safest way to identify all HCV-infected individuals.
Improper handling and storage of specimens may affect the detection of HCV RNA43 44 by yielding false-negative results, but such artefacts would not shorten the antibody-undetectable periods in our study. False RT-PCR–positive results can be introduced, either by improper handling patient specimen or by improper extraction and amplification of target RNA. To avoid such artefacts, the serum samples on one hand and plasma and PBMC samples on the other hand were sent to two different research institutes for final handling and storage. Moreover, the serum and EDTA-blood samples of the 19 HCV seroconverters were drawn at different Municipal Health stations in Amsterdam. To exclude false-positive serum results due to sampling, handling, or storage, plasma and PBMC samples were also tested when available. Moreover, all samples were randomly tested for the presence of HCV RNA, and all precautions were taken to avoid any possible contamination, using separated locations for the extraction of specimens, amplification of HCV RNA (using the GeneAmp PCR carry-over prevention kit), and analyses of PCR products.
In general, serum and PBMC samples drawn and analyzed at the same time-point contained the same HCV genotype. However, at some time points in subjects 1085 and 3059, the serum sample contained another HCV genotype than the genotype found in PBMCs. Especially remarkable findings were observed in samples taken before HCV seroconversion in subject 3059. In that patient, HCV genotype 1 was initially found in serum and PBMCs 38 months before HCV seroconversion. HCV genotype 1 remained detectable in the 2 following PBMC samples, whereas the serum samples were either untypeable or HCV-negative by RT-PCR. However, 14 months before HCV seroconversion, genotype 1 remained present in PBMCs, whereas the serum sample of that time contained genotype 3a. Strikingly, the samples drawn 2 months before HCV seroconversion contained genotype 3a in serum, plasma, and PBMCs, indicating a new infection that was found in all consecutive samples after HCV seroconversion. These findings could be explained by the fact that frequent drug injections lead to a mixture of HCV genotypes. Serotypes of developing antibodies may show how to interpret the final seroconversions observed. This is demonstrated by an earlier study by us in which a comparison was made between the obtained HCV genotypes and serotypes of the antibodies as they developed based on the antibody response to core and NS4 in the 19 HCV seroconverters.45The serotypes we found were in concordance with the HCV genotypes found after HCV seroconversion IDU 0146 (genotype 3a), 1085 (genotype 1), and 3059 (genotype 3a) and not with HCV genotypes found at several samples drawn during the prolonged antibody-undetectable periods. The serotypes did not change during follow-up, regardless of infections with heterologous HCV genotypes.45 Continuous reexposure and reinfections with HCV are often seen among IDUs, as exemplified in individual 3009 (Fig 1), in whom a new HCV seroconversion was detected by commercially available antibody assays. Superinfections and overtake phenomena in seropositive subjects have been reported in chimpanzees experimentally reinfected with HCV,24,46,47 in chronically HCV-positive humans reinfected with HCV through blood transfusion,48 and in chronically HCV-positive humans receiving HCV-positive liver grafts.49 According to these reports, neither humans nor chimpanzees chronically infected with HCV have adequate protective immunity against heterologous HCV genotypes or are protected against HCV strains of the same genotype or even against identical strains.
Another explanation, in addition to low levels of HCV RNA, might be a poor recognition or exposure of HCV antigens to helper T lymphocytes (TH cells), leading to diminished lymphokine secretion and impaired expansion, growth, and responsiveness of B cells. Among the 5 HIV-negative individuals, no differences were found in T-cell numbers during periods of at least 1 year before and after HCV seroconversion (results not shown). However, it has been described that exposure to opiates negatively influences T-cell responses, especially anti-CD3 responses. IDUs have reduced reactivity of T cells and higher levels of anergy than non-IDUs.50 This might be particularly relevant to our study, because long antibody-undetectable periods in our study were seen among HIV-negative individuals, excluding HIV as confounding factor. Extended antibody-undetectable periods were also found for HIV among IDUs compared with non-IDUs infected with HIV, although the delay between HIV detection and seroconversion is not as extreme as with HCV.51
In conclusion, we observed a prolonged period of HCV RNA-positivity before antibody seroconversion in a number of injecting drug users. Independent of HIV infection, the immune system appears to be sometimes incompetent to mount enough antibodies to HCV to be detected by commercially available assays, making antibody screening alone prone to false-negative results in the diagnosis of HCV infection. Our results indicate that HCV infection is not always characterized by persistent antibody responses and can be sustained at low levels without or delayed antibody responses to HCV. Therefore, we suggest screening for antibodies to HCV in combination with PCR to identify all HCV-positive individuals and potentially infectious blood donations. However, the percentage of silent carriers is hard to predict, and more studies are needed in subjects other than IDUs to extrapolate these findings to the general population.
ACKNOWLEDGMENT
The authors thank Lucy Phillips for editorial review, Martin McMorrow (Chiron Diagnostics) for bDNA assays, and Wim van Est for fine artwork.
Supported by the Health Research and Development Council (28-2370) and performed as part of the Amsterdam Cohort Studies on AIDS, a collaboration between the Academic Medical Centre, the Central Laboratory of the Netherlands Red Cross Blood Transfusion Service, and the Municipal Health Service, Amsterdam, The Netherlands. Approval was obtained from the Institutional Review Board for these studies. Informed consent was provided according to the Declaration of Helsinki.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Jaap Goudsmit, MD, PhD, Department of Human Retrovirology, Academic Medical Centre, University of Amsterdam Meibergdreef 15, 1105 AZ, Amsterdam, The Netherlands; e-mail: J.Goudsmit@AMC.UvA.NL.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal