Abstract
X-linked dyskeratosis congenita (DC) is a bone marrow failure syndrome caused by mutations in the DKC1 gene located at Xq28. By 20 years of age, most affected boys develop bone marrow failure, whereas female carriers show a skewed pattern of X-chromosome inactivation. The gene product, dyskerin, is homologous to a yeast protein involved in ribosomal RNA biogenesis, providing a unique insight into a cause of aplastic anemia. Whereas most causative mutations are single amino acid substitutions, and nonsense or frameshift mutations have not been observed, we present here a case of DC caused by a 2-kb deletion that removes the last exon of the gene. Normal levels of mRNA are produced from the deleted gene, with the transcripts using a cryptic polyadenylation site in the antisense strand of the adjacent MPP1 gene, normally located 1 kb downstream of DKC1 in a tail to tail orientation. The predicted truncated protein lacks a lysine-rich peptide that is less conserved than the rest of the dyskerin molecule and is dispensable in yeast, supporting the contention that it may retain some activity and that null mutations at this locus may be lethal. The affected boy had an unaffected brother with the same haplotype around the DKC1 gene and a sister who was heterozygous for the deletion. We conclude therefore that the mother must be a germline mosaic with respect to this deletion. Investigation of her blood cells and other somatic tissues showed that a small proportion of these cells also carried the deletion, making her a somatic mosaic and indicating that the deletion took place early in development.
A TRIAD OF CLINICAL features define the phenotype of dyskeratosis congenita (DC): nail dystrophy, leukoplakia, and skin pigmentation abnormalities.1-3 It has become clear that bone marrow failure is also commonly associated with the disease and is the principal cause of premature death.4 It seems that tissues that need continual renewal (skin, nails, and bone marrow) are most affected by the genetic lesion underlying the disorder.
Through a positional cloning strategy, the DKC1 gene responsible for the X-linked form of DC (MIM 305000) has recently been identified.5 This turns out to be a highly conserved gene, showing 61% protein sequence identity to the yeast centromere binding factor 5 (Cbf5p6) and 78% identity to the rat nucleolar-associated protein, NAP57.7 Functional studies indicate a role for these proteins in rRNA processing, specifically in the pseudouridylation of rRNA precursors.8 They may also be involved in centromere/microtubule binding6 or as ribosomal chaperones.7 It has been suggested, therefore, that DC represents a human ribosomopathy.9
Five of the original mutations described in the DKC1 gene give rise either to single amino acid substitutions or a single amino acid deletion.5 Subsequent mutation analysis has not shown any frameshift or nonsense mutations (Knight et al9a). The only gross change to the DKC1 gene observed so far is the 3′ end deletion, detected in a single case of DC, that facilitated the localization of the DKC1 gene in the candidate region.5 We report here on the characterization of this deletion and show that transcription of the DKC1 gene in this individual is rescued by a polyadenylation signal found on the noncoding strand of the closely neighboring MPP1 gene. We also demonstrate that the mother of the affected individual is a germline and somatic mosaic for the mutation.
MATERIALS AND METHODS
DNA and RNA extraction and Southern blot analysis were performed using standard techniques. Linkage analysis was performed by the analysis of radiolabeled polymerase chain reaction (PCR) products on 6% polyacrylamide gel electrophoresis (PAGE) gels for the Xq28 markers DXS8061, p39, DXS1073, and DXS1108 using the primer combinations described at the Genome Database (http://gdbwww.gdb.org).
The sequences of the primers (from GIBCO-BRL, Paisley, UK) used for PCR amplification of the DKC1/MPP1 genes from genomic DNA as well as cDNA are shown in Table 1, and their location and orientation with respect to each gene is shown in Fig 1. Standard PCR conditions were employed using a commercially supplied buffer (Advanced Biotechnologies, Epsom, UK), a magnesium concentration of 2.0 mmol/L, dNTPs at 0.2 mmol/L (Pharmacia, Uppsala, Sweden), and an annealing temperature of 58°C.
Oligonucleotide Sequences
| Name . | Sequence (5′ → 3′) . | Direction . | cDNA Position . |
|---|---|---|---|
| 931F | AAGGCCAAAGCTGGTCTG | Forward | 1524-1541 |
| XAP101F9 | GTTGACTACAGTGAGTCTGCC | Forward | 1341-1361 |
| XAP101R9 | ACCAGCTTTGGCCTTCTTGTCC | Reverse | 1538-1517 |
| XAP101F11 | GAGTAAGAAGGACAAGAAGGC | Forward | 1508-1528 |
| MPAF | CAAGCGTGCAGTTCTCCACA | Forward | 1457-1476 |
| MP3G | GTCCCGTGGTACAGTGGA | Forward | Genomic |
| MP3R2 | AGAGGGATGGGGTCCATTGCT | Reverse | 1814-1795 |
| MPBF | CAGGGCACTCAGACAGAAG | Forward | 1316-1334 |
| Name . | Sequence (5′ → 3′) . | Direction . | cDNA Position . |
|---|---|---|---|
| 931F | AAGGCCAAAGCTGGTCTG | Forward | 1524-1541 |
| XAP101F9 | GTTGACTACAGTGAGTCTGCC | Forward | 1341-1361 |
| XAP101R9 | ACCAGCTTTGGCCTTCTTGTCC | Reverse | 1538-1517 |
| XAP101F11 | GAGTAAGAAGGACAAGAAGGC | Forward | 1508-1528 |
| MPAF | CAAGCGTGCAGTTCTCCACA | Forward | 1457-1476 |
| MP3G | GTCCCGTGGTACAGTGGA | Forward | Genomic |
| MP3R2 | AGAGGGATGGGGTCCATTGCT | Reverse | 1814-1795 |
| MPBF | CAGGGCACTCAGACAGAAG | Forward | 1316-1334 |
The location of a deletion that removes the last exon of the DKC1 gene. The upper line shows the last exons of DKC1and MPP1 genes on the normal chromosome, with the direction of transcription indicated by the long arrows. The vertical bars show the position of the breakpoints of the deletion that gives rise to the deleted chromosome in patient HO, shown on the lower line. The DNA sequence at these breakpoints is shown. Solid boxes indicate coding sequences; shaded boxes indicate 3′UTR sequences. The location, direction, and names of the oligonucleotides used in various PCR experiments are shown as arrowheads.
The location of a deletion that removes the last exon of the DKC1 gene. The upper line shows the last exons of DKC1and MPP1 genes on the normal chromosome, with the direction of transcription indicated by the long arrows. The vertical bars show the position of the breakpoints of the deletion that gives rise to the deleted chromosome in patient HO, shown on the lower line. The DNA sequence at these breakpoints is shown. Solid boxes indicate coding sequences; shaded boxes indicate 3′UTR sequences. The location, direction, and names of the oligonucleotides used in various PCR experiments are shown as arrowheads.
3′ RACE PCR was performed as described elsewhere.10Briefly, first-strand DNA synthesis from total RNA was performed using the primer (dT)n-R1-R0(5′AAGGATCCGTCGACATCGATAATACGACTCACTATAGGGATTTTTTTTTTTTTTTTT3′). Two rounds of PCR amplification were performed using primers XAP101F1 and the outer adapter primer Ro(5′AAGGATCCGTCGACATC3′) and then the primer 931F and the inner adapter primer R1(5′GACATCGATAATACGAC3′). The product of this reaction was gel purified, cut with the restriction enzymes Pst I andCla I, and cloned into M13 before sequencing.
Bubble PCR, a one-sided PCR technique to amplify breakpoints, was performed as described.11 A duplex bubble oligo that contains 12 complementary bp at each end and 29 bp of noncomplementary sequence in the middle was obtained by annealing together the oligos BUB-T (5′AAGGATCCTAGTCTAGCTGTCTGTCGAAGGTAAGGAACGGACGAGCACTGAG3′) and BUB-B (5′CTCAGTGCTCGTAGTAATCGTTCGCACGAGAATCGCAAGATCTAGGATCCTT3′). A total of 1.5 μg of this oligo was ligated to 5 μg of genomic DNA that had been digested with Rsa I. After heating to 95°C for 5 minutes and removing excess oligonucleotide using a gene clean kit (Bio 101 Inc, La Jolla, CA), 2 rounds of PCR were performed with the primer XAP101F1 and the outer bubble oligo NVAMP-1 (5′TGCTCGTAGTAATCGTTCGCAC3′) and then the primer 931F and the inner bubble oligo NVAMP-2 (5′GTTCGCACGAGAATCGCAAGAT3′).
Analysis of the X-chromosome inactivation patterns (XCIP) was performed using the repeat polymorphism and methylation-sensitive Hpa II site at the androgen receptor gene locus (HUMARA) as described elsewhere.12 13 Sequencing was performed directly on gel purified PCR products (Qiagen, Crawley, UK) or on M13 DNA templates using an ABI automated sequencing machine (ABI, Foster City, CA).
RESULTS
A partial deletion of DKC1 in patient HO.
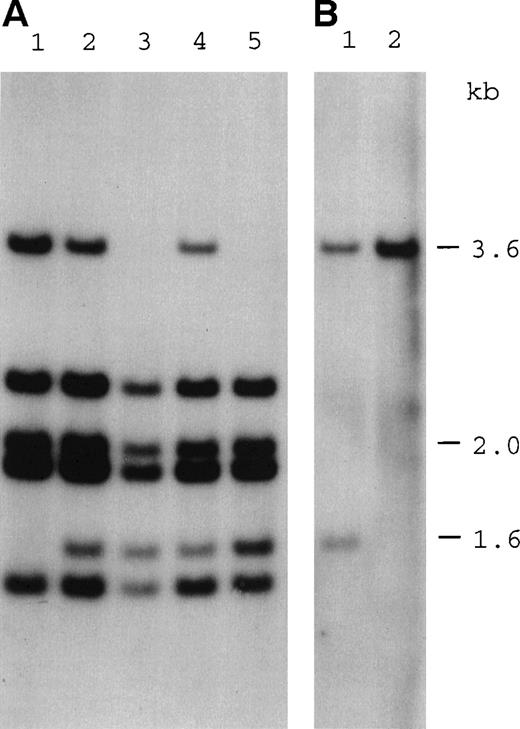
During the screening by Southern blot analysis of positional candidate genes for the DKC1 locus, a deletion affecting the 3′ end of cDNA XAP10114 was identified in 1 affected individual (patient HO) in pedigree DCR-015 (Fig 2A, lane 3). Through the identification of point mutations in this gene in other unrelated patients with DC, it was established that these mutations do indeed cause DC, and the gene was accordingly namedDKC1.5
Southern blot analysis showing the inheritance of a deletion at the 3′ end of the DKC1 gene. (A) Hybridization of a DKC1 probe 31g1K17 (cDNA nucleotides 744-1565) to Taq I digests of genomic DNA from the following family members: lane 1, mother of patient HO (I-1); lane 2, sister of patient HO (II-1); lane 3, patient HO (II-7); lane 4, niece of patient HO (III-1); and lane 5, nephew of patient HO (III-2). Numbers in brackets refer to the pedigree in Fig 5. Note that the 1.6-kb fragment produced by the deletion is not detected in the mother. (B) Hybridization of an MPP1 probe 39g1B21 (cDNA nucleotides 1117-1822) to Taq I digests of genomic DNA from the following: lane 1, patient HO; and lane 2, a normal control. Note that both probes hybridize to fragments of the same size. Two fragments of coincidental size are seen in the normal sample, 1 of which is deleted in patient HO.
Southern blot analysis showing the inheritance of a deletion at the 3′ end of the DKC1 gene. (A) Hybridization of a DKC1 probe 31g1K17 (cDNA nucleotides 744-1565) to Taq I digests of genomic DNA from the following family members: lane 1, mother of patient HO (I-1); lane 2, sister of patient HO (II-1); lane 3, patient HO (II-7); lane 4, niece of patient HO (III-1); and lane 5, nephew of patient HO (III-2). Numbers in brackets refer to the pedigree in Fig 5. Note that the 1.6-kb fragment produced by the deletion is not detected in the mother. (B) Hybridization of an MPP1 probe 39g1B21 (cDNA nucleotides 1117-1822) to Taq I digests of genomic DNA from the following: lane 1, patient HO; and lane 2, a normal control. Note that both probes hybridize to fragments of the same size. Two fragments of coincidental size are seen in the normal sample, 1 of which is deleted in patient HO.
To try and establish the extent of this deletion and because the orientation of the gene in Xq28 was unknown, both available flanking probes (DXYS64 and MPP1) were hybridized to Southern filters of the affected individual and a normal control. Although the DXYS64 probe showed only normal fragments, the cDNA probe 39g1B21, which covers the last 3 exons of the MPP1 gene (nt 1117-1822), not only showed an abnormal fragment of the same size as seen with the DKC1probe in the affected individual, but also normal fragments of the same size (Fig 2B). This suggested that the DKC1 gene and theMPP1 gene are located close to one another in a tail to tail configuration and that the deletion may also affect the 3′ end of the MPP1 gene. A probe to a more central region of theMPP1 cDNA (nt 565-1171) showed only normal fragments in the affected individual (not shown).
The deletion breakpoints.
The 5′ breakpoint of the deletion was established using a bubble PCR: the bubble oligonucleotide11 was ligated onto anRsa I digest of genomic DNA from patient HO. Nested PCR amplification was performed using oligonucleotides complementary to the bubble sequence and primers XAP101F11 and 931F from the DKC1gene. These 2 oligonucleotides were known to be present in the partially deleted DKC1 gene because a probe that is located toward the 3′ end of the gene (nt 1523-2143) had shown a very faint deleted restriction fragment in Southern blot analysis of the patient (not shown). This PCR gave rise to a discrete fragment of approximately 240 bp. The sequence of this fragment was found to diverge from the normal DKC1 genomic sequence (Knight et al9a) 50 bp into the last intron of the gene (Fig1).
Having seen the coincidental restriction fragments in Southern blots with probes to the 3′ end of the MPP1 and theDKC1 genes and predicting therefore that the genes lay tail to tail, a PCR was attempted between the 2 genes in patient HO. Using the primers 931F and MPAF (located on the sense strand of the MPP13′UTR), a fragment of approximately 1 kb was obtained, whereas no product was seen from a normal control. Sequencing of this fragment confirmed the 5′ breakpoint of the deletion established through the bubble PCR and showed that the other end of the deletion was 379 bp downstream of the 3′ end of the MPP1 gene (Fig 1). The sequence at each end of the deletion does not show any evidence of a repeat apart from sharing a TGG trinucleotide at the breakpoint.
Normal DKC1 and MPP1 are located tail to tail and separated by only 1 kb.
A new primer (MP3G) was derived from the sequence downstream of theMPP1 gene, determined from the inter DKC1-MPP1 PCR product obtained from patient HO. This was used in conjunction with theDKC1 primer 931F on normal genomic DNA in a PCR to show a clear band of approximately 1.8 kb. From this fragment, it was possible to complete the normal intergenic sequence between the 3′ ends of the DKC1 gene and the MPP1 gene. It is 988 bp in length and contains a stretch of 25 As. Knowing the normal sequence betweenMPP1 and DKC1, we can conclude that the deletion in patient HO has removed 1,931 bp (Fig 1).
The deleted DKC1 gene is transcribed and overlapsMPP1 at the 3′ end.
Insufficient RNA was obtained from the patient’s blood sample to perform Northern blot analysis. We therefore attempted semiquantitative reverse transcriptase-PCR (RT-PCR) of the DKC1gene from this RNA alongside a normal control sample that had been processed at the same time and in the same way, using a forward primer (XAP101F9) that spans an intron and a reverse primer XAP101R9. RT-PCR was performed for 21, 24, 27, and 30 cycles. Starting from equivalent amounts of cDNA, a fragment of the appropriate size was not seen at 24 cycles, was just visible after 27 cycles, and was clearly visible after 30 cycles in both the normal control and the patient HO (Fig 3). We conclude that patient HO does have a DKC1 transcript, which is detectable in peripheral blood at levels equivalent to normal. A similar experiment using primers for the MPP1 gene (MPBF and MP3R2) showed that patient HO also expresses this gene at levels equivalent to normal: a fragment of the appropriate size was just seen after 24 cycles and became clearly visible in both normal and patient after 27 cycles of amplification (Fig 3).
RT-PCR amplification of the DKC1 and MPP1genes from patient HO (p) and a control sample (c). The number of PCR cycles is shown. The molecular weight marker (m) is the plasmid pEMBL8 cut with Taq I and Pvu II.
RT-PCR amplification of the DKC1 and MPP1genes from patient HO (p) and a control sample (c). The number of PCR cycles is shown. The molecular weight marker (m) is the plasmid pEMBL8 cut with Taq I and Pvu II.
3′ RACE cloning established the nature of the 3′ end of theDKC1 transcript from the partially deleted gene. Nested amplification using primers XAP101F11 and 931F for the DKC1gene gave rise to a fragment of approximately 640 bp. As predicted from the size of this fragment, as well as from a search for a possible polyadenylation signal in the genomic sequence downstream of theDKC1 breakpoint in patient HO, sequence analysis showed that it overlaps the 3′ end of the MPP1 gene (Fig 4). A perfect polyadenylation signal, 5′AAUAAA3′, is found 11 bases from the polyA tail. Thus, as shown in Fig 4, the 2 genes overlap by 132 bp, which are transcribed from opposite strands to form the 3′ ends of the 2 genes.
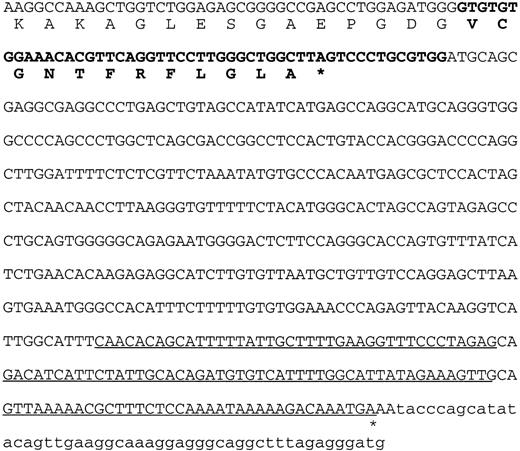
The sequence of the 3′ end of the DKC1 gene in patient HO. The last 45 nucleotides of the penultimate normal exon of the DKC1 gene, encoding amino acids 478-492, are shown in normal type. The nucleotides from the last intron of the DKC1gene and the amino acids they encode are shown in bold type. The underlined sequence shows the overlap with the MPP13′UTR. The asterisk indicates the position where the polyA tail is added to the mRNA.
The sequence of the 3′ end of the DKC1 gene in patient HO. The last 45 nucleotides of the penultimate normal exon of the DKC1 gene, encoding amino acids 478-492, are shown in normal type. The nucleotides from the last intron of the DKC1gene and the amino acids they encode are shown in bold type. The underlined sequence shows the overlap with the MPP13′UTR. The asterisk indicates the position where the polyA tail is added to the mRNA.
A truncated dyskerin.
The sequence derived from the deletion in patient HO predicts that the protein, dyskerin, encoded by his DKC1 gene will be truncated. Presuming that translation continues in frame beyond the penultimate exon and into the last intron, a stop codon is encountered after 39 bp (Fig 4). Twelve new amino acids (VCGNTFRFLGLA) will replace the 22 that are encoded in the last exon of the normal gene, which includes a remarkable stretch of eight consecutive lysine residues (DSDTTKKKKKKKKAKEVELVSE).
Family studies.
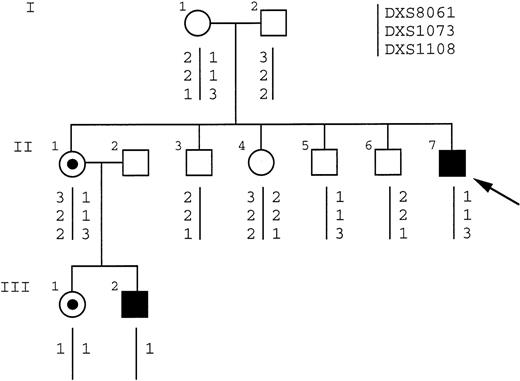
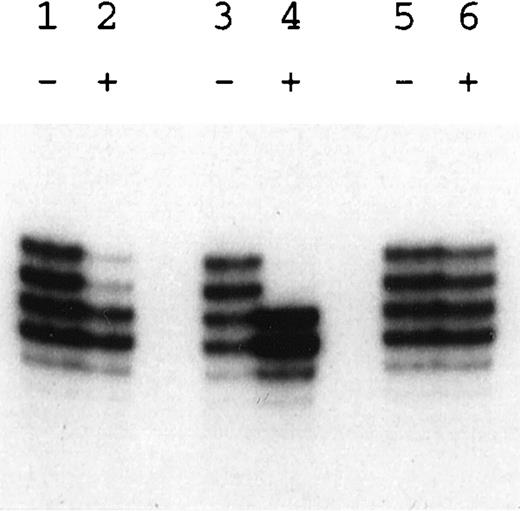
Patient HO is a sporadic case of DC. He has the classical mucocutaneous triad of skin pigmentation, nail dystrophy, and leukoplakia. He also has short stature, microcephaly, hypoplastic testes, and bone marrow failure (hemoglobin level, 8 g/dL; white blood cell count, 2.9 × 109/L; and platelet count, 20 × 109/L). Other members of this family, including the proband’s mother and sibs, had normal blood counts. Linkage analysis was performed on family members before the identification of the molecular lesion in theDKC1 gene in this family. Informative markers spanning Xq28 (Fig 5) showed that the haplotype of the affected boy (II:7) was shared by 1 of his sisters (II:1) but not the other (II:4). The analysis of XCIP, which are consistently skewed in carriers of X-linked DC,15 16 showed that II:1 did have the skewed pattern of a carrier (Fig 6, lanes 3 and 4), whereas II:4 did not (Fig 6, lanes 5 and 6), consistent with the haplotype data. This would imply that the mother, who showed an incompletely skewed XCIP (Fig 6, lanes 1 and 2), was a carrier of the disease. However, a sample from an unaffected brother of the patient (Fig 5, II:5) was found to share the affected haplotype.
Analysis of X-chromosome inactivation patterns. Amplification of the CAG repeat polymorphism at the HUMARA locus either without (−) or with (+) prior Hpa II digestion. Lanes 1 and 2, the mother of patient HO (I-1); lanes 3 through 6, the 2 sisters of patient HO (II-1 in lanes 3 and 4; II-4 in lanes 5 and 6). Note the completely skewed pattern in the carrier (lane 4), the random pattern in the normal sister (lane 6), and the incompletely skewed pattern in the mother (lane 2).
Analysis of X-chromosome inactivation patterns. Amplification of the CAG repeat polymorphism at the HUMARA locus either without (−) or with (+) prior Hpa II digestion. Lanes 1 and 2, the mother of patient HO (I-1); lanes 3 through 6, the 2 sisters of patient HO (II-1 in lanes 3 and 4; II-4 in lanes 5 and 6). Note the completely skewed pattern in the carrier (lane 4), the random pattern in the normal sister (lane 6), and the incompletely skewed pattern in the mother (lane 2).
Having identified the DKC1 deletion in this family, Southern blot analysis confirmed that the sister (II.1) who had been predicted to be a carrier indeed was (Fig 2A, lane 2). In addition, her daughter and son were both shown to have inherited the deletion (Fig 2A, lanes 4 and 5), as predicted by the linkage analysis (Fig 5, III:1 and III:2). However, only normal fragments were seen in the mother of the index case (Fig 2A, lane 1); she is therefore not a heterozygote for this mutation and yet has passed the affected allele to 2 of her children, whereas an affected and unaffected boy share the same haplotype. We conclude that she is a germline mosaic.
PCR amplification across the deletion breakpoint using primers 931F and MP3G gives rise to a deletion-specific fragment of approximately 200 bp. Testing all family members, we found that this fragment could be amplified from those shown to have the deletion but not from the unaffected sibs (Fig 7). However, a faint band was also seen from the mother of the index case (Fig 5, I:1) after 30 cycles of amplification, but not at 25 cycles, when it is clearly visible in the patient. This was true for DNA extracted from peripheral blood and mouthwash from the mother, and so we conclude that she is also a somatic mosaic for the mutation. The fact that we did not see a deletion-specific fragment in the Southern blot analysis of her DNA (Fig 2) indicates that less than 5% of the peripheral blood cells carry the deletion. Indeed, dilution experiments suggest that to get the level of signal seen in the PCR experiments (Fig 7), between 1:100 and 1:1,000 cells carry the deletion (data not shown). The proportion of cells bearing the deletion among the somatic tissues therefore appears to be smaller than the proportion present in the germline tissue, from which 2 of 3 chromosomes passed to her children (with the DXS8061:1, DXS1073:1, and DXS1108:3 haplotype, shown in Fig5) carry the deletion.
Amplification of a deletion-specific product from genomic DNA of the mother of patient HO. The number of PCR cycles is shown at the top. Family members are drawn above the appropriate lanes. C, a control sample. Note the faint product seen in the mother after 30 cycles of PCR, using primers that flank the deletion.
Amplification of a deletion-specific product from genomic DNA of the mother of patient HO. The number of PCR cycles is shown at the top. Family members are drawn above the appropriate lanes. C, a control sample. Note the faint product seen in the mother after 30 cycles of PCR, using primers that flank the deletion.
DISCUSSION
There are several interesting things about the deletion described in this report. First, its very existence led to the identification of theDKC1 gene that causes X-linked dyskeratosis congenita. Second, a transcript is still detected from the partially deleted gene, despite the fact that the last exon of the DKC1 gene has been lost. Third, the neighboring gene, MPP1, is normally located only approximately 1 kb away from the DKC1 gene and is arranged in a tail to tail configuration. Fourth, the mutant transcript overlaps theMPP1 gene at its 3′ end to terminate on the antisense strand of the MPP1 3′ UTR. Fifth, the mother of the affected boy is both a germline and somatic mosaic for this mutation.
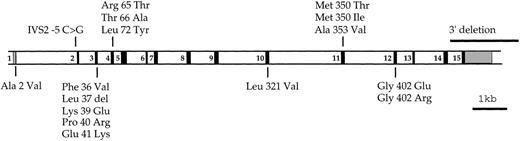
Having defined a region for the X-linked DKC1 gene of about 1.4 Mb in the distal part of chromosome Xq28,17 there remained some 28 known genes within the region.18-20 Whereas some could be discounted on the basis of a known disease association, the identification of the DKC1 gene resulted from the detection of a deletion by Southern blot analysis in a single patient with DC. In 5 additional patients for whom RNA was available, RT-PCR analysis of this gene led to the identification of 5 different mutations giving rise to 4 single amino acid substitutions and 1 single amino acid deletion.5 Subsequent mutation analysis using genomic DNA has led to the characterization of 11 additional mutations, which comprise 10 missense mutations (1 of which is found in 11 unrelated subjects) and 1 splice site mutation (Knight et al9a). None of them affects the 100 C-terminal amino acids of the protein; there is a small cluster of mutations around amino acids 36 to 41, but the functional significance of this is not yet clear (Fig 8).
The location of mutations identified in theDKC1 gene. Amino acid substitutions are drawn over and under a scale drawing of the DKC1 gene. Exons are shown as numbered solid boxes; untranslated regions are shown as shaded boxes. Data from Heiss et al,5 Knight et al,9a and this report.
It is therefore of significant interest that the deleted DKC1gene in the patient described here still gives rise to a presumably functional transcript. It seems reasonable to suggest that only relatively minor alterations to the gene can be tolerated and that a null phenotype may not be viable. By analogy with its highly conserved homologues in other species,7,8 the protein encoded by theDKC1 gene, called dyskerin, is likely to be involved in the nucleolus and possibly in the modification of rRNAs. We note that theCBF5 gene, encoding the yeast homologue, Cbf5p, is an essential gene in Saccharomyces cerevisiae, whereas the C-terminal KKD/E domain of this protein is dispensable in Kluyveromyces lactis.21 Conservation among the C-terminal amino acids is greatly reduced compared with the rest of this highly conserved protein.5 It may be that the loss of the last 22 amino acids in the predicted mutant dyskerin of patient HO, like the truncated K lactis protein, still retains sufficient function to be viable, whereas a complete null mutation would not.
The very close proximity of the DKC1 gene and the MPP1gene in the normal configuration has interesting implications for the termination of transcription and regulation of these ubiquitously expressed genes.6,22 Genes in this region of Xq28 do appear to be tightly packed,19 with, for example, the tail to tail Transketolase 2 and filamin genes separated by 4 kb, plexin and ITBA2 separated by 5 kb, and the QM protein and DNaseX genes separated by only 825 bp. Elsewhere, close tail to tail genes include the human histone H2A.X and hydroxymethylbilan genes, which are transcribed towards each other with their polyadenylation sites 330 bp apart,23 while the polyadenylation signals of human tuberous sclerosis 2 and polycystic kidney disease 1 genes lie just 60 bp apart24 and adjacent genes at the Surfeit locus are separated by very small distances, with 2 of the genes overlapping at their 3′ ends.25
Genes that overlap at their 3′ ends, as seen for the deletedDKC1 and the MPP1 genes in the patient described here, are uncommon in eukaryotes. Examples of normal genes that do this are the signal transducer and activator of transcription 6 (Stat6) and the immediate-early transcription factor NGF1-A binding protein (Nab2) genes that overlap by 58 bp in a region that is absolutely conserved between mouse and human.26 The mouse protein kinase gene PKN has a short transcript that overlaps the 3′UTR of the EP1 prostanoid receptor gene by 280 bp as well as a long alternatively spliced transcript that overlaps the whole gene.27Transcriptional interference is suggested here with a possible regulatory function of the antisense transcript. We presume that the overlap seen in our patient cannot unduly interfere with the transcription of either gene in that both appear to be transcribed at near normal levels. Whether both can be transcribed in the same cell has not been established, but if dyskerin is indispensable as suggested by the absence of null mutations, it would imply that they can.
A final point of interest is that the mother of this DC patient is a germline mosaic. In addition, a small number of her somatic cells also carry the mutation as detected by PCR but not by Southern blot analysis, implying that the mutation must have occurred early in development in a cell that contributed to both germline and somatic tissues. It may be that this is not such a rare event as previously suspected (reviewed in Zlotogora28). For example, 19% of all reported sporadic cases of the autosomal dominant disease facioscapulohumeral muscular dystrophy have a mosaic parent.29 Other X-linked disorders in which mosaicism has been demonstrated by molecular methods include X-linked severe combined immunodeficiency,30 the androgen insensitivity syndrome,31 Hunter syndrome,32 hemophilia A,33 and Duchenne muscular dystrophy.34 In one third of these cases, the mother was also found to be a somatic mosaic.
There are important implications for genetic counselling here: although the mother of the patient described here showed only an incompletely skewed pattern of X-inactivation, we had assumed that she was a carrier of DC, having an affected boy and a daughter with a completely skewed XCIP sharing the haplotype of the affected chromosome. Only when an unaffected brother sharing this haplotype became available did we suspect that the situation may be more complicated, as it indeed turned out to be.
ACKNOWLEDGMENT
The authors thank Lisa Lowery for running the automated sequencing facility and Dr Nick Cross for help with bubble and RACE PCR techniques.
Supported by The Wellcome Trust, Action Research, the Deutsche Forschungsgemeinschaft (DFG), and European Community (EU) Genome Analysis Program.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to T.J. Vulliamy, PhD, Department of Haematology, Imperial College School of Medicine, Hammersmith Hospital, Ducane Road, London W12 0NN, UK; e-mail:t.vulliamy@rpms.ac.uk.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal