Abstract
The high event-free survival rates of Down syndrome (DS) children with acute myeloid leukemia (AML) are due, in part, to increased in vitro sensitivity of DS myeloblasts to cytosine arabinoside (ara-C) and daunorubicin and the greater generation of ara-C triphosphate (ara-CTP) from ara-C compared with myeloblasts from non-DS patients (Taub et al,Blood 87:3395, 1996). This study further explores the molecular basis of chemotherapy sensitivity of DS AML patients by examining the expression of chromosome 21-localized genes in myeloblasts from newly diagnosed AML patients. Transcript levels of two chromosome 21-localized genes, cystathionine-β-synthase (CBS) and superoxide dismutase (SOD), measured by quantitative reverse transcriptase-polymerase chain reaction (RT-PCR), were 12.0- and 3.8-fold higher in DS compared with non-DS myeloblasts (P < .0001 and P < .0001, respectively). Conversely, there were no significant increases in transcripts for 2 other chromosome 21-localized genes, carbonyl reductase and the reduced folate carrier. CBS transcript levels correlated with both in vitro ara-C sensitivity measured by the 3-[4,5-dimethyl-thiazol-2-yl]-2,5-diphenyltetrazolium-bromide (MTT) assay (P = .003) and the generation of 3H-ara-C triphosphate (ara-CTP) after in vitro incubations with 5 μmol/L3H-ara-C (P = .0003). Transcripts of deoxycytidine kinase were 2.6-fold higher in DS compared with non-DS cells and may be a factor in the enhanced metabolism of ara-C in DS cells. There was no significant correlation of SOD transcripts with in vitro ara-C and daunorubicin sensitivities. Increased CBS transcripts could result in elevated CBS activity, which modulates ara-C metabolism by altering reduced folate pools, deoxycytidine triphosphate pools, S-adenosylmethionine levels, and/or methylation of the deoxycytidine kinase gene. The further identification of the molecular mechanisms of chemotherapy sensitivity of DS AML patients may lead to significant improvements in the treatment and cure of AML.
DESPITE SIGNIFICANT progress in the treatment of many childhood cancers, only modest progress has been made in the treatment of acute myeloid leukemia (AML), the second most common form of childhood leukemia. Childhood AML has the worst prognosis of all major childhood cancers, with 5-year relative survival rates of approximately 37%.1 This contrasts with the approximate 80% survival rate for acute lymphoblastic leukemia (ALL), the most common form of childhood cancer.
Although recent approaches have improved the overall outcome of treatment and prognosis for AML, they have not significantly impacted the outcome for the majority of patients.2,3 For instance, allogeneic bone marrow transplantation for AML in first remission has been more effective than chemotherapy but is limited in application due to the lack of suitable transplant donors in the majority of children. Autologous bone marrow transplantation in first remission does not appear to be clearly superior to intensive chemotherapy.4Salvage chemotherapy for relapsed/refractory AML has also not been very effective.5
Cytogenetic abnormalities in AML have been identified as one of the most important prognostic factors that predict treatment response in both children and adults. Favorable cytogenetic abnormalities include inv(16), t(8;21), and t(15;17), all of which are associated with higher remission induction rates and event-free survival (EFS) rates of greater than 40%.6 7
A recent unexpected finding is that Down syndrome (DS; trisomy 21) children with AML have the highest EFS rates of any subgroup of AML patients, with EFS rates of 68% to 100% when patients are treated with current intensive protocols.8-13 These studies reported that DS patients had leukemia relapse rates of less than 20%, indicating that significant leukemia cell kill occurs before the development of drug resistance. It was demonstrated by several groups (Pediatric Oncology Group, Children’s Cancer Group, Nordic Society of Pediatric Hematology Oncology, and Berlin-Frankfurt-Münster) that the improved response rate to therapy for DS children with AML coincided with the use of high-dose cytosine arabinoside (ara-C) for intensification,8,9,10,13 whereas Japanese and Canadian studies have used lower-dose ara-C successfully.11 12
In initial laboratory studies to investigate the basis for these compelling clinical findings, we reported that myeloblasts from newly diagnosed DS patients with AML were significantly more sensitive in vitro to both ara-C and daunorubicin compared with myeloblasts from non-DS patients.14 Furthermore, DS myeloblasts generated significantly higher levels of the active intracellular ara-C metabolite, ara-C triphosphate (ara-CTP), after in vitro incubations with ara-C.14
The enhanced chemotherapy sensitivity of DS myeloblasts may reflect the altered expression of chromosome 21-localized genes in DS cells. We previously hypothesized that alterations in expression of at least two such genes, cystathionine-β-synthase (CBS; 21q22.3)15 and zinc/copper superoxide dismutase (SOD; 21q22.1),16 may directly or indirectly contribute to the increased sensitivity of DS cells to ara-C and daunorubicin.5 14 In this study, we further explore the molecular basis of chemotherapy sensitivity of DS AML patients by examining the expression of these key chromosome 21-localized genes in DS and non-DS myeloblasts.
MATERIALS AND METHODS
Patient specimens.
Myeloblasts were obtained from (1) newly diagnosed pediatric AML patients registered for treatment on the Pediatric Oncology Group (POG) 9421 chemotherapy protocol (n = 53) before the initiation of therapy, (2) AML patients treated at Children’s Hospital of Michigan (CHM) from 1988 to 1998 (n = 7), and (3) newborn DS babies with the transient myeloproliferative disorder (TMD; n = 3). The mononuclear cells were isolated on a Ficoll-Hypaque gradient to obtain a highly purified mononuclear cell fraction consisting of mostly leukemic blasts. The majority of the specimens were obtained as bone marrow aspirates, although peripheral blood was used when the peripheral white blood cell (WBC) counts were sufficiently high (>50,000/μL). The AML samples received from member POG institutions were collected in sodium heparin diluted with equal amounts of RPMI, shipped overnight at room temperature, and purified at CHM upon receipt; the samples were then frozen in 20% fetal calf serum (FCS)/10% dimethyl sulfoxide (DMSO) at −195°C until further analysis. The leukemia samples were classified according to French-American-British (FAB) classification system as previously reported.8 The diagnosis of megakaryocytic leukemia (M7) was based on the expression of platelet-specific antigens recognized by the monoclonal antibodies CD61, CD41, and CD42. The protocol was approved by the hospital institutional review board.
Chemicals.
[5-3H]-Cytosine-β-D-arabinofuranoside (25 Ci/mmol) was obtained from Moravek Biochemicals (Brea, CA). Unlabeled cytosine arabinoside, daunorubicin, and 3-[4,5-dimethyl-thiazol-2-yl]-2,5-diphenyltetrazolium-bromide (MTT) were obtained from Sigma Chemical Co (St Louis, MO). Tissue culture reagents and supplies were purchased from assorted vendors. Polymerase chain reaction (PCR) primers were purchased from Genosys Biotechnologies, Inc (The Woodlands, TX).
Cell culture.
The CCRF-CEM and K562 leukemia cell lines were obtained from the American Type Culture Collection (Rockville, MD). Cells were maintained in RPMI 1640 containing 10% heat-inactivated calf serum, 100 U/mL penicillin, and 100 μg/mL streptomycin in a humidified atmosphere at 37°C in the presence of 5% CO2/95% air.
Reverse transcriptase-PCR (RT-PCR) analysis of transcripts of chromosome 21-localized genes.
Total RNA was isolated from 5 to 10 × 106patient myeloblasts or cultured cells by the phenol/chloroform extraction method of Chomczynski and Sacchi17 using Tri Reagent (Molecular Research Center, Cincinnati, OH). CBS, SOD, carbonyl reductase (CBR; localized to 21q22.1),18 reduced folate carrier (RFC; localized to 21q22.3),19 and deoxycytidine kinase (dCk; localized to chromosome 4)20 transcripts were assayed from the same cDNA mixtures using the CBS-1/CBS-2, SOD-1/SOD-2, CBR-1/CBR-2, RFC-1/RFC-2, and dCk-1/dCk-2 oligonucleotide primer pairs (Table 1).
Oligonucleotide Primers Used for PCR
| Gene . | Primers . | Primer Pairs . | Nucleotide Sequence (nt) . | Length of PCR Product (bp) . |
|---|---|---|---|---|
| CBS | CBS-1 | 5′-AGGACGGTGGTGGACAAGTGGTTCAAGAGCAA-3′ | 948-980 (exons 8/9) | 655 |
| CBS-2 | 5′-TGACCACCCCGAACACCATCTGCCGCTGACTG-3′ | 1571-1603 (exon 15) | ||
| SOD | SOD-1 | 5′-CCGTGTGCGTGCTGAAGGGCGAC-3′ | 14-37 (exon 1) | 436 |
| SOD-2 | 5′-ATTACACCACAAGCCAAACGACT-3′ | 427-450 (exon 5) | ||
| CBR | CBR-1 | 5′-GACTGGAGGCAACAAGGGCATCGG-3′ | 30-54 (exon 1) | 655 |
| CBR-2 | 5′-TGGGCAGCAGGCATTCAGGAGGAT-3′ | 660-684 (exon 3) | ||
| dCK | dCK-1 | 5′-GCCACCCCGCCCAAGAGAAGC-3′ | 4-24 (exon 1) | 762 |
| dCK-2 | 5′-GGATCCGACCTTTTCAACCAGACTTTC-3′ | 737-757 (exon 6/7) | ||
| RFC | RFC-1 | 5′-GATACGGCCAGGGGAGAGCTT-3′ | 215-235 (exon 3) | 452 |
| RFC-2 | 5′-GCCAGCGAGATGTAGTTGAGCGT-3′ | 645-667 (exon 4) | ||
| CBS (genomic) | CBS-3 | 5′-AGAATTCCTGAGCGACAGGTGGATGC-3′ | 1152-1174 (exon 11) | 422 |
| CBS-4 | 5′-TCGATCGATGGTGTGCCCACAGGTGA-3′ | 1285-1307 (exon 12) | ||
| β-actin (genomic) | Actin-1 | 5′-GGGAGAGCGGGAAATCGTGCGTGACATT-3′ | 1191-1215 (exon 3) | 211 |
| Actin-2 | 5′-AGACAGTCTCCACTCACCCAGGAAG-3′ | 1379-1404 (intron 3) |
| Gene . | Primers . | Primer Pairs . | Nucleotide Sequence (nt) . | Length of PCR Product (bp) . |
|---|---|---|---|---|
| CBS | CBS-1 | 5′-AGGACGGTGGTGGACAAGTGGTTCAAGAGCAA-3′ | 948-980 (exons 8/9) | 655 |
| CBS-2 | 5′-TGACCACCCCGAACACCATCTGCCGCTGACTG-3′ | 1571-1603 (exon 15) | ||
| SOD | SOD-1 | 5′-CCGTGTGCGTGCTGAAGGGCGAC-3′ | 14-37 (exon 1) | 436 |
| SOD-2 | 5′-ATTACACCACAAGCCAAACGACT-3′ | 427-450 (exon 5) | ||
| CBR | CBR-1 | 5′-GACTGGAGGCAACAAGGGCATCGG-3′ | 30-54 (exon 1) | 655 |
| CBR-2 | 5′-TGGGCAGCAGGCATTCAGGAGGAT-3′ | 660-684 (exon 3) | ||
| dCK | dCK-1 | 5′-GCCACCCCGCCCAAGAGAAGC-3′ | 4-24 (exon 1) | 762 |
| dCK-2 | 5′-GGATCCGACCTTTTCAACCAGACTTTC-3′ | 737-757 (exon 6/7) | ||
| RFC | RFC-1 | 5′-GATACGGCCAGGGGAGAGCTT-3′ | 215-235 (exon 3) | 452 |
| RFC-2 | 5′-GCCAGCGAGATGTAGTTGAGCGT-3′ | 645-667 (exon 4) | ||
| CBS (genomic) | CBS-3 | 5′-AGAATTCCTGAGCGACAGGTGGATGC-3′ | 1152-1174 (exon 11) | 422 |
| CBS-4 | 5′-TCGATCGATGGTGTGCCCACAGGTGA-3′ | 1285-1307 (exon 12) | ||
| β-actin (genomic) | Actin-1 | 5′-GGGAGAGCGGGAAATCGTGCGTGACATT-3′ | 1191-1215 (exon 3) | 211 |
| Actin-2 | 5′-AGACAGTCTCCACTCACCCAGGAAG-3′ | 1379-1404 (intron 3) |
The nucleotide sequence positions are relative to the ATG start.
For the analysis of CBS expression, first-strand cDNAs were synthesized from 2 μg of total RNA in 20 μL of a reaction mix containing 2.5 μmol/L of random hexadeoxynucleotide primers, 10 mmol/L Tris-HCl (pH 8.3), 50 mmol/L KCl, 5 mmol/L MgCl2, 0.5 mmol/L each dNTP, and 50 U of Moloney murine leukemia virus (MuLV) reverse transcriptase (Perkin Elmer, Foster City, CA). After 1 hour of incubation at 42°C, the reaction was stopped by heating at 99°C for 5 minutes. Aliquots of the cDNAs were amplified for PCR in a reaction mixture consisting of 1 μL of reverse transcription cDNA product, 10× PCR buffer (Perkin Elmer), 1.5 mmol/L MgCl2, 0.2 mmol/L of each dNTP, 0.2 μmol/L of each CBS primer, and 1.25 U Taq DNA polymerase (Promega, Madison, WI). PCR was performed for 28 cycles of 1 minute at 94°C (denaturation), 1 minute at 70°C (annealing), and 1 minute at 72°C (extension) in a Perkin Elmer GeneAmp PCR System 9600 using the CBS-1/CBS-2 primers (Table 1). Aliquots of the PCR reaction (10 μL) were analyzed on agarose gels (1.0%) that were stained with SYBR green I (FMC Bioproducts, Rockland, ME) for 1 hour. The fluorescent PCR products were quantitated with a Molecular Dynamics Storm 860 fluorescence and radioactivity imaging system and Image Quant software (Molecular Dynamics, Sunnyvale, CA). Levels of CBS transcripts were calculated from the intensities of the fluorescent PCR products and were normalized to the levels of 18S ribosomal RNA (localized to chromosome 13; Ambion Inc, Austin, TX).
PCR amplifications for the SOD, CBR, and RFC genes were for 30 cycles of 1 minute at 94°C, 1 minute at 60°C (SOD and RFC) or 66°C (CBR), and 1 minute at 72°C, and the PCR products were analyzed as for the CBS gene. PCR amplification of the dCk gene was for 28 cycles for 1 minute at 94°C, 1 minute at 60°C, and 1 minute at 72°C. For all primer sets, the linear range of exponential amplification was determined by preliminary reactions using dilutions of a series of cDNAs and a range of PCR cycles. Controls for the PCR reactions to detect CBS, SOD, and CBR transcripts included the K562 (positive) and CCRF-CEM (negative) leukemia cell lines.
PCR analysis of genomic DNA.
DNA was isolated from the AML samples using Tri Reagent according to the manufacturer’s guidelines. PCR amplification of the CBS gene was performed with 40 ng of genomic DNA in a 50 μL reaction volume containing 10 mmol/L Tris-HCl (pH 8.3), 1.5 mmol/L MgCl2, 50 mmol/L KCl, 5% DMSO, 200 μmol/L of each dNTP, 1.25 U Taq DNA polymerase, and 0.2 μmol/L of each CBS primer. β-Actin (localized to chromosome 7)21 was used as an internal control and amplified with gene-specific primers (0.2 μmol/L each; Table 1) in the same reaction. PCR amplification was performed for 29 cycles of 1 minute at 95°C, 1 minute at 68°C, and 1 minute at 72°C, followed by a final extension for 10 minutes using the CBS-3/CBS-4 primers and actin-1/actin-2 primers (Table 1). The PCR products were electrophoresed on a 2% agarose gel, stained with SYBR Green I, and the gel was analyzed as in the RT-PCR assays with the relative CBS gene copy number expressed as the ratio of CBS/β-actin level.
MTT colorimetric drug sensitivity assay.
The MTT assay was based on our previously reported method.14 Briefly, leukemia cells (2 × 104) were suspended in triplicate in 80 μL of RPMI 1640/20% dialyzed FCS in the presence of varying concentrations of ara-C, daunorubicin, or no drug (controls). After 72 hours in a humidified incubator at 37°C, MTT was added to a final concentration of 1 mmol/L. After 6 hours, the colored formazan crystals were dissolved with the addition of 100 μL acidified isopropranol and the optical densities read by a microplate reader at 540 nm. The absorbances were corrected for blank readings. Absorbance (A) at 540 nm directly correlates with cell number; the percentage of cell survival was calculated as follows: A (treated)/A (untreated control) × 100. The data were plotted as drug concentration versus percentage of cell survival, and the IC50s were determined by the MINSQ curve fitting program, (Dynacomp Inc, Livonia, NY), corresponding to the drug concentration that decreased cell survival by 50%.
Ara-C incubations and measurement of ara-CTP.
Incubation of leukemia cells with ara-C and the measurement of intracellular ara-CTP levels were performed as previously described.14 Briefly, cells suspended in Hank’s balanced salt solution were incubated with 5 μmol/L 3H-ara-C (250 μCi/μmol) at 37°C for 3 hours. The incubations were stopped with the addition of ice-cold phosphate-buffered saline (PBS; with 20 μmol/L dipyridamole to inhibit nucleoside transport). After additional PBS washes, the cells were aliquotted and processed as follows: (1) 1 sample was assayed for radioactivity and protein content (representing total intracellular accumulation of ara-C and metabolites) and (2) 1 sample was treated with 0.5 N ice-cold perchloric acid and the supernatant (acid-soluble fraction) was neutralized with 5 N KOH and 1 mol/L KH2PO4before high-performance liquid chromatography (HPLC) analysis.
The neutralized acid soluble fractions were analyzed for ara-C metabolites by an ion-pairing HPLC method on an Isco liquid chromatograph equipped with a 4-μm C-18 octadecylsilane column with a C-18 precolumn. The mobile phase consisted of 90 mmol/L potassium phosphate. The elution positions of the standard unlabeled ara-C metabolites (ara-C, ara-CMP, and ara-CTP) were monitored at 280 nm and correlated with those of the radioactive metabolites. The levels of intracellular metabolites (expressed in picomoles per milligram of protein) were calculated from the total intracellular radiolabel (in picomoles per milligram), the fraction comprising the PCA soluble pool, and the percentage of each compound from chromatographic analysis.
Statistical analysis.
Differences in age, WBC count, ara-C and daunorubicin IC50, ara-CTP, and transcript levels were compared between the DS and non-DS patient groups using the nonparametric Mann-Whitney 2-sample test. The nonparametric Spearman rank correlation coefficient was used to analyze transcript levels and their relationships to MTT and ara-CTP data.
RESULTS
Transcript levels of the chromosome 21-localized genes, CBS and SOD, in myeloblasts from pediatric patients (DS and TMD [n = 3]; DS and AML [n = 16]; and non-DS and AML [n = 44]) were assayed by quantitative RT-PCR (Fig 1). The goal was to determine whether differences in gene expression exist between DS and non-DS samples that might contribute to the dramatically increased EFS rates for DS over non-DS children with AML. Transcript levels were correlated with (1) ara-CTP levels generated during in vitro3H-ara-C incubations and (2) in vitro sensitivities to ara-C and daunorubicin determined by MTT assays. Patient characteristics and experimental results are summarized in Tables 2, 3, and 4.
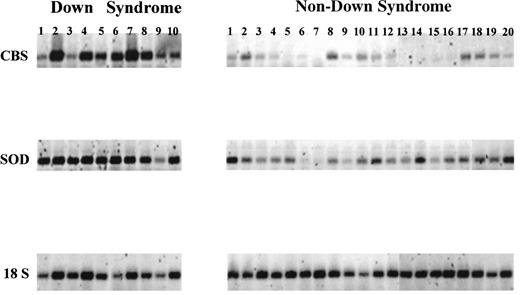
RT-PCR analysis of chromosome 21-localized transcript levels in myeloblasts obtained from newly diagnosed patients with AML or TMD. Lanes DS 1 through 10 and non-DS 1 through 20 correspond to the patients listed in Tables 1 and 2, respectively. CBS, cystathionine-β-synthase localized to 21q22.3 corresponding to a 655-bp product; SOD, superoxide dismutase localized to 21q22.1 corresponding to a 436-bp product. The transcript levels were normalized to 18S RNA transcript levels.
RT-PCR analysis of chromosome 21-localized transcript levels in myeloblasts obtained from newly diagnosed patients with AML or TMD. Lanes DS 1 through 10 and non-DS 1 through 20 correspond to the patients listed in Tables 1 and 2, respectively. CBS, cystathionine-β-synthase localized to 21q22.3 corresponding to a 655-bp product; SOD, superoxide dismutase localized to 21q22.1 corresponding to a 436-bp product. The transcript levels were normalized to 18S RNA transcript levels.
Patient Characteristics of DS AML Specimens
| Patient . | Age (yrs) . | WBC (×103/μL) . | FAB . |
|---|---|---|---|
| DS-1 | 1 | 15.2 | M7 |
| DS-2 | NB | 282.0 | M7 |
| DS-3 | 1.5 | 18.0 | M7 |
| DS-4 | 1.4 | 23.2 | M7 |
| DS-5 | 0.4 | 289.9 | M7 |
| DS-6 | 1.8 | 14.0 | M7 |
| DS-7 | 1.1 | 10.3 | M7 |
| DS-8 | 1.9 | 3.8 | M7 |
| DS-9 | NB | 26.1 | M7 |
| DS-10 | 3 | 17.9 | NA |
| DS-11 | 1 | 20.6 | M7 |
| DS-12 | 2.25 | 27.2 | M7 |
| DS-13 | NB | 47.1 | M7 |
| DS-14 | 2.1 | 10.3 | M7 |
| DS-15 | 2.5 | 29.8 | M7 |
| DS-16 | 2.75 | 32.2 | M7 |
| DS-17 | 0.63 | 137.3 | M7 |
| DS-18 | 0.83 | 21.0 | NA |
| DS-19 | 1 | 7.3 | M7 |
| Patient . | Age (yrs) . | WBC (×103/μL) . | FAB . |
|---|---|---|---|
| DS-1 | 1 | 15.2 | M7 |
| DS-2 | NB | 282.0 | M7 |
| DS-3 | 1.5 | 18.0 | M7 |
| DS-4 | 1.4 | 23.2 | M7 |
| DS-5 | 0.4 | 289.9 | M7 |
| DS-6 | 1.8 | 14.0 | M7 |
| DS-7 | 1.1 | 10.3 | M7 |
| DS-8 | 1.9 | 3.8 | M7 |
| DS-9 | NB | 26.1 | M7 |
| DS-10 | 3 | 17.9 | NA |
| DS-11 | 1 | 20.6 | M7 |
| DS-12 | 2.25 | 27.2 | M7 |
| DS-13 | NB | 47.1 | M7 |
| DS-14 | 2.1 | 10.3 | M7 |
| DS-15 | 2.5 | 29.8 | M7 |
| DS-16 | 2.75 | 32.2 | M7 |
| DS-17 | 0.63 | 137.3 | M7 |
| DS-18 | 0.83 | 21.0 | NA |
| DS-19 | 1 | 7.3 | M7 |
Abbreviations: NB, newborn infant with TMD; NA, not available.
Patient Characteristics of Non-DS AML Specimens
| Patient . | Age (yrs) . | WBC (×103/μL) . | FAB . |
|---|---|---|---|
| NDS-1 | 1.5 | 31.7 | M7 |
| NDS-2 | 14 | 74.0 | M3 |
| NDS-3 | 9 | 201.0 | M4 |
| NDS-4 | 14 | 147.1 | M1 |
| NDS-5 | 12 | 130.0 | M4 |
| NDS-6 | 0.67 | 15.4 | M5a |
| NDS-7 | 9 | 26.9 | M2 |
| NDS-8 | 15 | 2.0 | M4 |
| NDS-9 | 3 | 122.9 | M2 |
| NDS-10 | 8 | 6.8 | M6 |
| NDS-11 | 1.5 | 219.5 | M5b |
| NDS-12 | 6 | 167.2 | M5a |
| NDS-13 | 15 | 31.5 | M1 |
| NDS-14 | 14 | 110.8 | M1 |
| NDS-15 | 0.6 | 17.7 | M4 |
| NDS-16 | 0.8 | 6.3 | M2 |
| NDS-17 | 8 | 67.2 | M5a |
| NDS-18 | 11 | 8.5 | M2 |
| NDS-19 | 0.9 | 9.0 | M2 |
| NDS-20 | 9 | 1.7 | NA |
| NDS-21 | 15 | 21.6 | NA |
| NDS-22 | 0.67 | 21.6 | M5a |
| NDS-23 | 7 | 107.0 | M5a |
| NDS-24 | 17 | 7.0 | M4 |
| NDS-25 | 16 | 98.0 | M2 |
| NDS-26 | 1.5 | 19.0 | M4 |
| NDS-27 | 0.17 | 54.5 | M4 |
| NDS-28 | 19 | 13.3 | M4 |
| NDS-29 | 10 | 4.3 | NA |
| NDS-30 | 4 | 176.7 | M5 |
| NDS-31 | 4 | 43.7 | M1 |
| NDS-32 | 12 | 78.4 | M4 |
| NDS-33 | 15 | 27.1 | M1 |
| NDS-34 | 16 | 86.2 | M2 |
| NDS-35 | 17 | 207.1 | M4 |
| NDS-36 | 14 | 54.7 | M4 |
| NDS-37 | 1.75 | 86.6 | M5 |
| NDS-38 | 9 | 55.2 | M0 |
| NDS-39 | 2.5 | 15.1 | M2 |
| NDS-40 | 12 | 27.7 | M4 |
| NDS-41 | 7 | 57.2 | M2 |
| NDS-42 | 12 | 26.6 | M2 |
| NDS-43 | 11 | 241.8 | M1 |
| NDS-44 | 10 | 22.9 | M2 |
| Patient . | Age (yrs) . | WBC (×103/μL) . | FAB . |
|---|---|---|---|
| NDS-1 | 1.5 | 31.7 | M7 |
| NDS-2 | 14 | 74.0 | M3 |
| NDS-3 | 9 | 201.0 | M4 |
| NDS-4 | 14 | 147.1 | M1 |
| NDS-5 | 12 | 130.0 | M4 |
| NDS-6 | 0.67 | 15.4 | M5a |
| NDS-7 | 9 | 26.9 | M2 |
| NDS-8 | 15 | 2.0 | M4 |
| NDS-9 | 3 | 122.9 | M2 |
| NDS-10 | 8 | 6.8 | M6 |
| NDS-11 | 1.5 | 219.5 | M5b |
| NDS-12 | 6 | 167.2 | M5a |
| NDS-13 | 15 | 31.5 | M1 |
| NDS-14 | 14 | 110.8 | M1 |
| NDS-15 | 0.6 | 17.7 | M4 |
| NDS-16 | 0.8 | 6.3 | M2 |
| NDS-17 | 8 | 67.2 | M5a |
| NDS-18 | 11 | 8.5 | M2 |
| NDS-19 | 0.9 | 9.0 | M2 |
| NDS-20 | 9 | 1.7 | NA |
| NDS-21 | 15 | 21.6 | NA |
| NDS-22 | 0.67 | 21.6 | M5a |
| NDS-23 | 7 | 107.0 | M5a |
| NDS-24 | 17 | 7.0 | M4 |
| NDS-25 | 16 | 98.0 | M2 |
| NDS-26 | 1.5 | 19.0 | M4 |
| NDS-27 | 0.17 | 54.5 | M4 |
| NDS-28 | 19 | 13.3 | M4 |
| NDS-29 | 10 | 4.3 | NA |
| NDS-30 | 4 | 176.7 | M5 |
| NDS-31 | 4 | 43.7 | M1 |
| NDS-32 | 12 | 78.4 | M4 |
| NDS-33 | 15 | 27.1 | M1 |
| NDS-34 | 16 | 86.2 | M2 |
| NDS-35 | 17 | 207.1 | M4 |
| NDS-36 | 14 | 54.7 | M4 |
| NDS-37 | 1.75 | 86.6 | M5 |
| NDS-38 | 9 | 55.2 | M0 |
| NDS-39 | 2.5 | 15.1 | M2 |
| NDS-40 | 12 | 27.7 | M4 |
| NDS-41 | 7 | 57.2 | M2 |
| NDS-42 | 12 | 26.6 | M2 |
| NDS-43 | 11 | 241.8 | M1 |
| NDS-44 | 10 | 22.9 | M2 |
Abbreviation: NA, not available.
Summary of Transcript Levels and Drug Sensitivity Data in Patient Samples
| . | DS . | Non-DS . |
|---|---|---|
| CBS transcript | 0.660 (0.231-3.920) | 0.055 (0.001-0.673) |
| SOD transcript | 2.390 (1.328-12.044) | 0.634 (0.068-7.995) |
| CBR transcript | 0.567 (0.067-3.562) | 0.520 (0.016-9.100) |
| RFC transcript | 0.185 (0.042-0.246) | 0.139 (0.045-0.300) |
| dCK transcript | 2.477 (1.132-9.020) | 0.943 (0.059-11.283) |
| ara-C IC50 (nmol/L) | 100.5 (27.1-370.4) | 420.8 (47.2-3930.7) |
| Daunorubicin IC50 (nmol/L) | 6.75 (1.8-76.3) | 107.4 (4.0-885.0) |
| 3H-ara-CTP (pmol/mg prot) | 735.0 (416.0-1332.6) | 141.1 (18.4-480.0) |
| . | DS . | Non-DS . |
|---|---|---|
| CBS transcript | 0.660 (0.231-3.920) | 0.055 (0.001-0.673) |
| SOD transcript | 2.390 (1.328-12.044) | 0.634 (0.068-7.995) |
| CBR transcript | 0.567 (0.067-3.562) | 0.520 (0.016-9.100) |
| RFC transcript | 0.185 (0.042-0.246) | 0.139 (0.045-0.300) |
| dCK transcript | 2.477 (1.132-9.020) | 0.943 (0.059-11.283) |
| ara-C IC50 (nmol/L) | 100.5 (27.1-370.4) | 420.8 (47.2-3930.7) |
| Daunorubicin IC50 (nmol/L) | 6.75 (1.8-76.3) | 107.4 (4.0-885.0) |
| 3H-ara-CTP (pmol/mg prot) | 735.0 (416.0-1332.6) | 141.1 (18.4-480.0) |
Transcript levels determined by RT-PCR are normalized to 18S RNA transcripts. The in vitro sensitivity data were determined by the MTT assay and 3H-ara-CTP data were determined after in vitro incubations with 3H-ara-C. The values listed are median, with the range of values listed in parentheses.
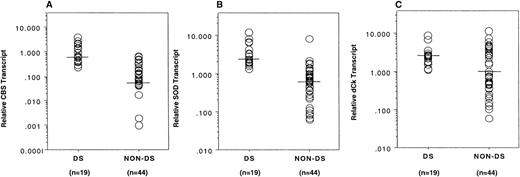
CBS mRNA transcripts were detectable in all 19 of the DS TMD/AML samples and in 29 of 44 (66%) of the non-DS blast cell samples; however, transcript levels in the DS samples were significantly higher compared with the non-DS samples (median, 0.660 v 0.055, respectively; P < .0001; the 15 non-DS samples in which CBS transcripts were not detectable were assigned a value of 0.001, which is the lower limit of sensitivity of our RT-PCR assay; Fig 2A). In contrast, when CBS gene copy numbers were measured for a subset of AML DNA samples (9 DS and 19 non-DS samples), gene copies differed by 1.3-fold (P = .0006), approximating the expected value (1.5-fold) for chromosome 21-localized genes between DS and non-DS cells.
Transcript levels of chromosome 21-localized genes in myeloblasts from newly diagnosed AML patients. Median CBS and SOD transcript levels were 12.0- and 3.8-fold higher, respectively, in DS myeloblasts compared with non-DS myeloblasts (A and B). Transcript levels of the dCk gene were 2.6-fold higher in DS compared with non-DS cells (C). Transcript levels were normalized to 18S RNA transcript levels. Horizontal bars represent the median values in each group.
Transcript levels of chromosome 21-localized genes in myeloblasts from newly diagnosed AML patients. Median CBS and SOD transcript levels were 12.0- and 3.8-fold higher, respectively, in DS myeloblasts compared with non-DS myeloblasts (A and B). Transcript levels of the dCk gene were 2.6-fold higher in DS compared with non-DS cells (C). Transcript levels were normalized to 18S RNA transcript levels. Horizontal bars represent the median values in each group.
SOD transcripts were detected in all 19 DS and 44 non-DS samples, and, again, marked differences were observed between the DS and non-DS samples (median, 2.390 v 0.634, respectively; P < .0001; Fig 2B). Conversely, there were no significant differences between the DS and non-DS samples in transcript levels for other chromosome 21-localized genes, including CBR (DS [n = 19] and non-DS [n = 44]; median, 0.567 v 0.520, respectively; P = .72) and RFC (DS [n = 10] and non-DS [n = 20]; median, 0.185v 0.139, respectively; P = .59). Deoxycytidine kinase transcripts were a median 2.6-fold higher in DS cells (n = 19) compared with non-DS cells (n = 44; median, 2.477 v 0.943, respectively;P = .03; Fig 2C).
There was no relationship between CBS transcripts and presenting WBC count for all of the patients (r = −.178, P = .16); there was a significant correlation between SOD transcripts and WBC count (r = −.271, P = .03). There was a significant correlation between age and transcript levels for the CBS (r = −.513; P < .0001) and SOD (r = −.454; P = .0003) genes, but the results may be skewed, because the DS patients were significantly younger in age compared with the non-DS patients (1.1 v 9.0 years, respectively; P< .0001).
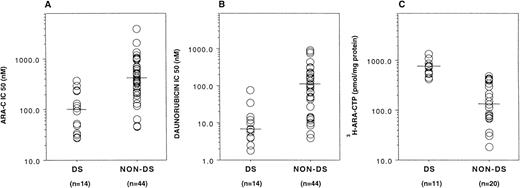
For a significant number of samples, there were sufficient blasts for assays of drug sensitivities and ara-C metabolism. DS myeloblasts (n = 14) were appreciably more sensitive than non-DS myeloblasts (n = 44) in vitro to both ara-C (median IC50, 100.5 v 420.8 nmol/L, respectively; P < .0001) and daunorubicin (median IC50, 6.75 v 107.4 nmol/L, respectively; P< .0001; Fig 3A and B). DS samples (n = 11) also generated significantly higher levels of3H-ara-CTP than non-DS cells (n = 20) during 3-hour in vitro incubations with 5 μmol/L 3H-ara-C (median, 735.0 v 141.1 pmol/mg protein, respectively; P< .0001; Fig 3C). This may result from enhanced dCk activity secondary to increased dCk transcripts, as previously predicted.5 14
In vitro sensitivity and metabolism of ara-C in myeloblasts from AML patients. DS myeloblasts were significantly more sensitive to both ara-C (A) and daunorubicin (B) compared with non-DS myeloblasts (P < .0001 and P < .0001, respectively). 3H-Ara-CTP generation in DS myeloblasts was significantly higher compared with non-DS myeloblasts after incubation with 3H-ara-C (P < .0001; C).
In vitro sensitivity and metabolism of ara-C in myeloblasts from AML patients. DS myeloblasts were significantly more sensitive to both ara-C (A) and daunorubicin (B) compared with non-DS myeloblasts (P < .0001 and P < .0001, respectively). 3H-Ara-CTP generation in DS myeloblasts was significantly higher compared with non-DS myeloblasts after incubation with 3H-ara-C (P < .0001; C).
Based on our hypothesis,5 14 increased CBS transcripts may lead to greater generation of ara-CTP and correlate with ara-C sensitivity; increased SOD transcripts may be associated with increased in vitro ara-C and daunorubicin sensitivities.
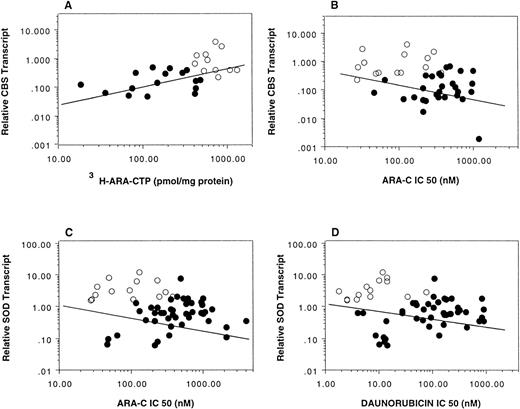
When the correlation between CBS transcript levels and ara-C sensitivity was evaluated for the DS and non-DS patients separately, the expected relationships were not seen. This may be due to sample size of the group and the fact that no CBS transcripts could be detected in up to one third of non-DS samples. As seen in Fig 2, there was a striking separation of the CBS transcript levels between DS and non-DS cases; only 8 of 44 non-DS cases exceeded the lowest transcript levels seen in the DS group and only 1 exceeded the median value for the DS cases. Interestingly, when the DS and non-DS groups were analyzed as a whole, CBS expression closely correlated with both in vitro ara-C sensitivity (14 DS and 44 non-DS samples; Spearman correlation coefficient r = −.397, P = .003) and in vitro 3H-ara-CTP generation (11 DS and 20 non-DS samples; r = .666, P = .0003; Fig 4A and B), suggesting a causal relationship. There was no significant correlation between SOD transcripts and sensitivities to ara-C and daunorubicin for the group analyzed together (14 DS and 44 non-DS samples; ara-C: r = −.236, P = .07; daunorubicin: r = −.217,P = .10; Fig 4C and D). Five DS samples were not included in the correlation analysis due to lack of MTT data secondary to insufficient cell numbers.
Correlation between CBS and SOD transcripts measured by RT-PCR in myeloblasts and in vitro drug activity and sensitivity. CBS transcripts correlated with both 3H-ara-CTP generation (Spearman correlation coefficient r = .666, P = .0003) and ara-C sensitivity (r = −.397, P = .003) (A and B, respectively). There were no significant correlations between SOD transcripts and ara-C (r = −.236, P = .07) or daunorubicin (r = −.217, P = .10) sensitivities (C and D, respectively). Transcript levels were normalized to 18S RNA transcript levels. (○) DS; (•) non-DS.
Correlation between CBS and SOD transcripts measured by RT-PCR in myeloblasts and in vitro drug activity and sensitivity. CBS transcripts correlated with both 3H-ara-CTP generation (Spearman correlation coefficient r = .666, P = .0003) and ara-C sensitivity (r = −.397, P = .003) (A and B, respectively). There were no significant correlations between SOD transcripts and ara-C (r = −.236, P = .07) or daunorubicin (r = −.217, P = .10) sensitivities (C and D, respectively). Transcript levels were normalized to 18S RNA transcript levels. (○) DS; (•) non-DS.
DISCUSSION
The significantly higher EFS rates of DS AML patients compared with non-DS patients are likely multifactorial and related to specific genetic alterations present in DS cells. Based on this premise, we previously speculated that key enzymes encoded by genes localized to chromosome 21 likely contribute to the sensitivity of myeloblasts to the chemotherapy drugs currently used in AML therapy, notably ara-C and daunorubicin.5,14 We previously reported that DS myeloblasts are more sensitive in vitro to both ara-C and daunorubicin compared with non-DS myeloblasts and hypothesized that at least 2 chromosome 21-localized genes, CBS and SOD, may be responsible.5 14
In this study, we found that the median CBS and SOD transcript levels in DS myeloblasts were 12.0- and 3.8-fold higher, respectively, than the levels in non-DS myeloblasts. For ara-C, increased drug sensitivity was associated with dramatically elevated synthesis of ara-CTP as well.
Increased CBS expression and enzyme activity could enhance the metabolism of ara-C to ara-CTP by 2 mechanisms5,14: (1) via effects on folate metabolism, decreased generation of deoxythymidine triphosphate, and, ultimately, decreased deoxycytidine triphosphate (dCTP), a feedback inhibitor of dCk required for activation of ara-C; and (2) via decreased generation of S-adenosylmethionine (AdoMet) and hypomethylation of the dCk gene.22,23 We previously reported that endogenous dCTP levels were lower in DS cells.14 In this study, we found that dCk transcript levels were also higher in DS cells than non-DS cells and likely contribute to differences in ara-CTP generation. It has been suggested that the in vitro cytostatic effects of ara-C were reversed by the administration of AdoMet, providing evidence that ara-C activity and AdoMet levels are linked,24 perhaps at the level of dCk. Most recently, we found that CCRF-CEM leukemia cells transfected with the CBS cDNA are more sensitive to ara-C due to higher levels of ara-CTP accumulation over wild-type cells.25
Although we did not detect a significant relationship between SOD expression and sensitivities to either ara-C or daunorubicin, a relationship may be more apparent with the analysis of a greater number of samples. Preliminary experiments from our laboratory have shown that transfection of CCRF-CEM cells with the SOD cDNA results in increased ara-C sensitivity, although there was no change in daunorubicin sensitivity compared with wild-type cells (unpublished data). Our results may not be entirely unexpected either in that SOD is not directly involved in the metabolism of either ara-C or daunorubicin. Rather, increased free radical generation may have an additive effect through lipid peroxidation and damage to cellular membranes and organelles.26,27 Imbalances of oxygen radicals secondary to increased SOD activity are known to be involved in the pathophysiology of DS.28 Increased SOD activity has been reported to be associated with increased apoptosis in thymic tissue of transgenic mice with elevated levels of SOD in vivo and DS fetal neurons in vitro that can be prevented by treatment with free-radical scavengers.29,30 This suggests that DS cells may have increased susceptibility to undergo apoptosis after exposure to apoptosis-inducing agents,31 thereby contributing to the enhanced sensitivity of DS cells to chemotherapy agents.
Acquired extra copies of chromosome 21 in pediatric B-precursor ALL are frequently associated with a favorable prognosis.32,33Although the basis for this effect in ALL is not well established, it partly reflects increased chromosome numbers, because expression of RFC (localized to 21q22.3) is elevated in hyperdiploid blasts containing acquired extra copies of chromosome 21.34 Acquired trisomy 21 was associated with an intermediate prognosis in a study of 1,612 pediatric and adult AML patients; however, in another study, this did not confer an improved treatment outcome due to the presence of additional cytogenetic abnormalities.7,35 In our study, 2 non-DS samples had acquired trisomy 21 (NDS 13 and 39, Table 3) and there was no pattern of increased transcript levels in these samples; CBS transcripts were undetectable in NDS 13 and approximately 4-fold higher in NDS 39 compared with the median non-DS value, whereas SOD transcripts in NDS 39 were below the median SOD value and only 1.1-fold higher for NDS 13. This suggests that the mere presence of extra chromosome 21 copies does not account for differences in CBS and SOD gene expression. Likewise, relative transcripts for CBS and SOD in DS myeloblasts far exceeded the minimum level in non-DS cells and the expected 1.5-fold increase from the 3 copies of chromosome 21 in DS cells. The lack of detectable CBS transcripts by RT-PCR in many of the non-DS AML samples was due to low levels of gene expression, rather than to gene deletions not apparent by karyotype analysis. By contrast, the median values for CBR and RFC more closely approximated the expected differences. Selective and disproportionate expression of chromosome 21-localized genes may reflect the presence of variable deletions in the 21q region that are not detectable by karyotype analysis36 and their effects on gene transcription. Similar findings of a disproportionate expression of chromosome 21-localized genes have been described in DS fetal tissue compared with non-DS tissue,37,38 and a 3.3-fold higher expression of the TIAM gene was reported in DS leukemia cells compared with a control group.39
In conclusion, this is the first study to demonstrate increased gene expression of chromosome 21-localized genes relevant to chemotherapy response in DS myeloblasts compared with non-DS myeloblasts from newly diagnosed AML patients. Our results imply that increased gene expression of chromosome 21-localized genes in DS myeloblasts contributes to the increased chemotherapy sensitivities of pediatric DS AML patients. The elevated CBS transcript levels are accompanied by increased sensitivities of DS myeloblasts to ara-C. The further identification of the molecular mechanisms of chemotherapy sensitivity of DS AML patients may lead to new approaches in the treatment and cure of AML.
ACKNOWLEDGMENT
The authors thank Rita Russo and Christine Menig for the patient data collection and Dr Long Zhang for providing the RFC primers.
Supported in part by the Leukemia Society of America Research Translational Grant (6203-98), Children’s Cancer Research Fund, Children’s Research Center of Michigan, The Litvak Foundation, BenePro Corp, and Leukemia Research, Life, Inc (Detroit, MI).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Jeffrey W. Taub, MD, Children’s Hospital of Michigan, 3901 Beaubien Blvd, Detroit, MI 48201.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal