Abstract
It is widely accepted that donor leukocytes survive within the recipient periphery after blood transfusion or solid organ transplantation. The significance of this microchimerism remains unclear, partially because of the insecurity of assays used to detect the donor-derived material. The techniques used to detect donor-derived DNA within recipient peripheral blood rely largely on major histocompatibility complex class II polymorphism. We and others have shown that the sensitivity of polymerase chain reaction with sequence-specific primers (PCR-SSP) typing for HLA class II alleles can be increased 100-fold by the addition of a primary amplification step (nested PCR-SSP). We have now extended this technique to encompass typing for HLA class I alleles, thereby adding flexibility to microchimerism testing by enabling testing of recipients HLA-DR matched with their donors. However, the high level of sensitivity achieved with the technique (1:100,000) leads to a concomitant decrease in the specificity that results in the amplification of unexpected products, a phenomenon we encountered in the development of our nested PCR-SSP typing system for HLA class II alleles. We describe here how it is possible to compensate for these anomalies by including multiple testing of a pretransfusion sample that acts as a specificity control, establishing a rigorous baseline for subsequent analysis.
IT IS WIDELY ACCEPTED that donor leukocytes can survive within a recipient after transplantation1-4 or blood transfusion,5-8 yet the relevance of this microchimerism remains a contentious issue.4 9-13 Further controlled studies are required to achieve an improved understanding of the functional importance of microchimerism.
The majority of assays used to detect donor-derived material in recipient blood exploit sex mismatching between donor and recipient1,14-16 or the polymorphism of the HLA-DR region of the major histocompatibility complex (MHC).2,4,9,10,17 The most commonly used techniques based on HLA-DR polymorphism are the polymerase chain reaction with either sequence-specific primers (PCR-SSP) or sequence-specific oligonucleotides (PCR-SSO). These methods are commonly used to HLA type donors and recipients before transplantation,18,19 but may be applied to the detection of microchimerism. The sensitivity of detection of such techniques is variable and can range from 0.5% to 0.01%,3,20,21 but the sensitivity of PCR-SSP for HLA-DR alleles has been increased by the introduction of a preliminary PCR (nested PCR-SSP), amplifying exon 2 of HLA-DRB1.4,17,22,23 It has been demonstrated that the sensitivity of nested PCR-SSP typing for HLA-DR alleles is at least 100-fold higher than that of standard PCR-SSP typing.17,22Nevertheless, the use of nested PCR-SSP typing for HLA-DR alleles is not applicable when the donor and recipient are HLA-DR matched. In such cases, it would be useful to be able to increase the power of detection of the technique by applying the recently developed molecular typing methods for HLA class I alleles.19 24-26
We describe here the development of a nested PCR-SSP typing method for the detection of HLA class I alleles. During the development of this method, we detected bands on the gel that were not indicative of donor or recipient HLA type, entirely consistent with our previous findings when developing a nested PCR-SSP typing system for HLA-DR alleles.22 These nonspecific bands do not arise from contamination, but result from the nonspecific amplification of recipient DNA with certain primer sets. The application of the technique to clinical samples is described, emphasizing the importance of rigorous controls for this type of assay system.
MATERIALS AND METHODS
Clinical material.
Anticoagulated peripheral blood was obtained from selected HLA-typed healthy volunteers for the sensitivity experiments. To validate the technique, peripheral blood samples were obtained from patients at the Oxford Transplant Centre (Oxford, UK) awaiting a renal transplant who had received planned transfusions of HLA-typed blood. Each patient received 50 mL of freshly isolated blood (<36 hours from donation) from 2 healthy donors. Blood samples were obtained at 2 time points before the transfusion and at defined time points after transfusion.
DNA isolation.
Genomic DNA was isolated from leukocytes obtained from anticoagulated blood using a salting out procedure,27 was precipitated with ethanol, and was resuspended in sterile water. The genomic DNA was quantitated and purity was assessed by spectroscopic absorbance at 260 and 280 nm.
Nested PCR-SSP typing.
Two hundred nanograms of DNA was initially amplified in a buffer containing 67 mmol/L Tris, pH 8.8; 16.6 mmol/L NH4SO4; 200 μmol/L of each dATP, dCTP, dGTP, and dTTP; 1.0 mmol/L MgCl2; 0.5 μmol/L forward and reverse primer (Table 1); and 0.25 U BioTaq polymerase (Bioline, London, UK) for 30 cycles according to Cereb et al28 in a PTC200-96v thermal cycler (Genetic Research Instrumentation, Essex, UK). The resultant PCR product was diluted 1:500 in water before PCR-SSP typing. Extreme care was taken to avoid contamination at any stage.29 Filter tips were used when pipetting (BioExpress UK Ltd, London, UK), and the work was performed in a clean air cabinet (Heto-Holten UK Ltd, Camberley, UK).
Nested PCR-SSP First-Round Primers
| Primer . | Binding Site . | Sequence . |
|---|---|---|
| HLA-A forward | Intron 1: 21-46 | 5′ GAA ACS GCC TCT GYG GGG AGA AGC AA |
| HLA-A reverse | Intron 3: 66-89 | 5′ TGT TGG TCC CAA TTG TCT CCC CTC |
| HLA-B forward | Intron 1: 36-57 | 5′ GGG AGG AGC GAG GGG ACC SCA G |
| HLA-B reverse | Intron 3: 37-59 | 5′ GGA GGC CAT CCC CGG CGA CCT AT |
| HLA-C forward | Intron 1: 42-61 | 5′ AGC GAG GKG CCC GCC CGG CGA |
| HLA-C reverse | Intron 3: 12-35 | 5′ GGA GAT GGG GAA GGC TCC CCA CT |
| Primer . | Binding Site . | Sequence . |
|---|---|---|
| HLA-A forward | Intron 1: 21-46 | 5′ GAA ACS GCC TCT GYG GGG AGA AGC AA |
| HLA-A reverse | Intron 3: 66-89 | 5′ TGT TGG TCC CAA TTG TCT CCC CTC |
| HLA-B forward | Intron 1: 36-57 | 5′ GGG AGG AGC GAG GGG ACC SCA G |
| HLA-B reverse | Intron 3: 37-59 | 5′ GGA GGC CAT CCC CGG CGA CCT AT |
| HLA-C forward | Intron 1: 42-61 | 5′ AGC GAG GKG CCC GCC CGG CGA |
| HLA-C reverse | Intron 3: 12-35 | 5′ GGA GAT GGG GAA GGC TCC CCA CT |
Sequences of HLA-A, -B, and -C specific primers used in first-round PCR amplification (purchased from Genosys Europe, Cambridge, UK). These primers amplify a 979-bp region of HLA-A, a 940-bp region of HLA-B, and a 909-bp region of HLA-C.28Redundancies (bolded) are identified using the International Union of Biochemistry Group Codes, where K = G/T, S = C/G, and Y = C/T.
PCR-SSP typing.
Allele-specific primers (0.5 μmol/L) designed on the basis of published sequences19,30 (Table2) were used in multiple amplification reactions consisting of diluted first-round PCR product (1/500); 67 mmol/L Tris, pH 8.8; 16.6 mmol/L NH4SO4; 200 μmol/L of each dATP, dCTP, dGTP, and dTTP; 2.0 mmol/L MgCl2; and 0.25 U BioTaq polymerase. PCR amplifications were performed in a PTC200-96v thermal cycler, and the products were subsequently analyzed by agarose gel electrophoresis using the conditions exactly as previously described.19
Primer Mix Composition for HLA Class I PCR-SSP Typing
| Lane . | PM . | Primer . | Size (bp) . | Alleles amplified . |
|---|---|---|---|---|
| 1 | 151 | 286 + 431 | 629 | A*0101-4N |
| 2 | 3 | 296 + 302 | 489 | A*0201-27 |
| 3 | 4 | 291 + 299 | 628 | A*0301-4 |
| 4 | 5 | 284 + 302 | 464 | A*2301,A*2402-14 |
| 5 | 6 | 292 + 249 | 557 | A*2301,A*2413 |
| 6 | 9 | 193 + 298 | 440 | A*2501-2,A*2601-11N,A*3401-2,A*6601-3 |
| 7 | 10 | 294 + 298 | 400 | A*2501-2 |
| 8 | 12 | 174 + 298 | 442 | A*4301 |
| 9 | 13 | 239 + 451 + 168 | 170 | A*2609,A*3401-2 |
| 10 | 14 | 290 + 475 + 298 | 419 | A*2502,A*3401-2,A*6601-2 |
| 11 | 225 | 290 + 303 + 167 | 520/552 | A*1101-4,A*2502,A*6601 |
| 12 | 152 | 174 + 300 | 465 | A*2901-3 |
| 13 | 18 | 295 + 301 | 561 | A*3001-6 |
| 14 | 184 | 434 + 486 | 198 | A*31012 |
| 15 | 153 | 173 + 234 | 259 | A*3201-2 |
| 16 | 154 | 434 + 429 | 200 | A*3301/3 |
| 17 | 226 | 193 + 302 | 468 | A*6801-8,A*6901 |
| 18 | 24 | 290 + 171 | 383 | A*6901 |
| 19 | 34 | 193 + 221 | 619 | B*0702-9,B*8101 |
| 20 | 155 | 195 + 220 | 606 | B*0801-3 |
| 21 | 160 | 202 + 272 + 285 + 393 | 480/546 | B*1804,B*3519,B*4003/9/18,B*4402-7/9 |
| 22 | 42 | 192 + 220 | 605 | B*4101-3 |
| 23 | 44 | 192 + 247 | 465 | B*1533,B*4001-6/9-12/14-18,B*4101-3,B*4801/3/4 |
| 24 | 45 | 272 + 218 | 627 | B*4001-4/6-11/14/16-18,B*4701-3 |
| 25 | 46 | 280 + 229 | 607 | B*4001/7 |
| 26 | 47 | 243 + 215 | 486 | B*1301-3 |
| 27 | 48 | 197 + 127 | 389 | B*1401-4 |
| 28 | 49 | 205 + 232 | 182 | B*1402-3/5,B*3904 |
| 29 | 50 | 207 + 217 | 507 | B*3901-12,B*6701 |
| 30 | 172 | 208 + 435 + 217 | 498/508 | B*3801-2 |
| 31 | 56 | 194 + 213 | 374 | B*5801-2 |
| 32 | 55 | 194 + 224 | 351 | B*5701-4 |
| 33 | 57 | 187 + 214 | 458 | B*1801-5 |
| 34 | 58 | 209 + 236 | 422 | B*3906,B*5501-3/5,B*5601,B*5901,B*7301 |
| 35 | 59 | 242 + 215 | 383 | B*4501,B*5001-2,B*5401,B*5501-3/5,B*5601-2/4,B*8201 |
| 36 | 60 | 203 + 238 | 551 | B*5601-4 |
| 37 | 61 | 395 + 236 | 421 | B*5401 |
| 38 | 62 | 280 + 281 + 282 | 149/150 | B*2701-11 |
| 39 | 64 | 192 + 392 | 422 | B*3701,B*3902/8 |
| 40 | 65 | 192 + 228 | 414 | B*3702,B*4701-3 |
| 41 | 67 | 203 + 220 | 594 | B*4021-2 |
| 42 | 69 | 194 + 225 | 516 | B*1516-17 |
| 43 | 70 | 240 + 241 | 460 | B*4601 |
| 44 | 72 | 243 + 250 | 124 | B*1501-2/4-8/11-12/14-15/19-21/25-28/30-35/38-40 |
| 45 | 73 | 192 + 214 | 421 | B*1304,B*1501/3-7/12/14/19-20/24-27/32-35/38-40,B*4802 |
| 46 | 79 | 193 + 223 | 369 | B*1502/13/21,B*3501-4/6-9/11-12/15/18-21/23-24,B*4406,B*5104,B*5301-2 |
| 47 | 80 | 188 + 237 | 128 | B*1522,B*1801-5,B*3501-13/15-18/20-24,B*7801-2 |
| 48 | 81 | 195 + 213 + 277 | 389/390 | B*3501-9/11/15/17-19/21/23-24,B*5301-2 |
| 49 | 82 | 207 + 216 | 400 | B*1509,B*7801-2 |
| 50 | 84 | 193 + 216 | 451 | B*1509,B*5101-9/11N,B*7801-2 |
| 51 | 85 | 192 + 216 | 440 | B*5201 |
| 52 | 86 | 368 + 315 | 340 | Cw*0102-3 |
| 53 | 87 | 366 + 145 | 522 | Cw*0202-3,Cw*1701-2 |
| 54 | 88 | 368 + 389 | 564 | Cw*0302-6/8-9 |
| 55 | 89 | 366 + 143 | 331 | Cw*0401-5,Cw*1801-2 |
| 56 | 90 | 366 + 379 | 564 | Cw*0501 |
| 57 | 91 | 367 + 127 | 297 | Cw*0602-4 |
| 58 | 93 | 313 + 184 | 516 | Cw*0701/6/7 |
| 59 | 94 | 367 + 183 | 302 | Cw*0702/3/10 |
| 60 | 275 | 271 + 143 | 500 | B*8201,Cw*0708,Cw*1801-2 |
| 61 | 96 | 367 + 379 | 536 | Cw*0704 |
| 62 | 182 | 165 + 166 + 317 | 160/625 | Cw*0801-4 |
| 63 | 99 | 159 + 389 | 523 | Cw*0303 |
| 64 | 201 | 130 + 160 + 389 | 206/522 | Cw*0302/4-6/8-9 |
| 65 | 245 | 507 + 215 | 407 | Cw*1502 |
| 66 | 102 | 369 + 126 | 538 | Cw*1202 |
| 67 | 103 | 368 + 157 | 446 | Cw*1203/6 |
| 68 | 104 | 371 + 388 | 541 | Cw*1402-3 |
| 69 | 106 | 366 + 223 | 318 | Cw*1502-6 |
| 70 | 107 | 366 + 382 | 502 | Cw*0203,Cw*0404,Cw*0604,Cw*0707,Cw*1502-6,Cw*1701-2 |
| 71 | 109 | 368 + 146 | 513 | Cw*1601 |
| 72 | 110 | 366 + 146 | 503 | Cw*1602 |
| 73 | 111 | 366 + 377 | 502 | Cw*0202,Cw*0602-3,Cw*1204-5 |
| Lane . | PM . | Primer . | Size (bp) . | Alleles amplified . |
|---|---|---|---|---|
| 1 | 151 | 286 + 431 | 629 | A*0101-4N |
| 2 | 3 | 296 + 302 | 489 | A*0201-27 |
| 3 | 4 | 291 + 299 | 628 | A*0301-4 |
| 4 | 5 | 284 + 302 | 464 | A*2301,A*2402-14 |
| 5 | 6 | 292 + 249 | 557 | A*2301,A*2413 |
| 6 | 9 | 193 + 298 | 440 | A*2501-2,A*2601-11N,A*3401-2,A*6601-3 |
| 7 | 10 | 294 + 298 | 400 | A*2501-2 |
| 8 | 12 | 174 + 298 | 442 | A*4301 |
| 9 | 13 | 239 + 451 + 168 | 170 | A*2609,A*3401-2 |
| 10 | 14 | 290 + 475 + 298 | 419 | A*2502,A*3401-2,A*6601-2 |
| 11 | 225 | 290 + 303 + 167 | 520/552 | A*1101-4,A*2502,A*6601 |
| 12 | 152 | 174 + 300 | 465 | A*2901-3 |
| 13 | 18 | 295 + 301 | 561 | A*3001-6 |
| 14 | 184 | 434 + 486 | 198 | A*31012 |
| 15 | 153 | 173 + 234 | 259 | A*3201-2 |
| 16 | 154 | 434 + 429 | 200 | A*3301/3 |
| 17 | 226 | 193 + 302 | 468 | A*6801-8,A*6901 |
| 18 | 24 | 290 + 171 | 383 | A*6901 |
| 19 | 34 | 193 + 221 | 619 | B*0702-9,B*8101 |
| 20 | 155 | 195 + 220 | 606 | B*0801-3 |
| 21 | 160 | 202 + 272 + 285 + 393 | 480/546 | B*1804,B*3519,B*4003/9/18,B*4402-7/9 |
| 22 | 42 | 192 + 220 | 605 | B*4101-3 |
| 23 | 44 | 192 + 247 | 465 | B*1533,B*4001-6/9-12/14-18,B*4101-3,B*4801/3/4 |
| 24 | 45 | 272 + 218 | 627 | B*4001-4/6-11/14/16-18,B*4701-3 |
| 25 | 46 | 280 + 229 | 607 | B*4001/7 |
| 26 | 47 | 243 + 215 | 486 | B*1301-3 |
| 27 | 48 | 197 + 127 | 389 | B*1401-4 |
| 28 | 49 | 205 + 232 | 182 | B*1402-3/5,B*3904 |
| 29 | 50 | 207 + 217 | 507 | B*3901-12,B*6701 |
| 30 | 172 | 208 + 435 + 217 | 498/508 | B*3801-2 |
| 31 | 56 | 194 + 213 | 374 | B*5801-2 |
| 32 | 55 | 194 + 224 | 351 | B*5701-4 |
| 33 | 57 | 187 + 214 | 458 | B*1801-5 |
| 34 | 58 | 209 + 236 | 422 | B*3906,B*5501-3/5,B*5601,B*5901,B*7301 |
| 35 | 59 | 242 + 215 | 383 | B*4501,B*5001-2,B*5401,B*5501-3/5,B*5601-2/4,B*8201 |
| 36 | 60 | 203 + 238 | 551 | B*5601-4 |
| 37 | 61 | 395 + 236 | 421 | B*5401 |
| 38 | 62 | 280 + 281 + 282 | 149/150 | B*2701-11 |
| 39 | 64 | 192 + 392 | 422 | B*3701,B*3902/8 |
| 40 | 65 | 192 + 228 | 414 | B*3702,B*4701-3 |
| 41 | 67 | 203 + 220 | 594 | B*4021-2 |
| 42 | 69 | 194 + 225 | 516 | B*1516-17 |
| 43 | 70 | 240 + 241 | 460 | B*4601 |
| 44 | 72 | 243 + 250 | 124 | B*1501-2/4-8/11-12/14-15/19-21/25-28/30-35/38-40 |
| 45 | 73 | 192 + 214 | 421 | B*1304,B*1501/3-7/12/14/19-20/24-27/32-35/38-40,B*4802 |
| 46 | 79 | 193 + 223 | 369 | B*1502/13/21,B*3501-4/6-9/11-12/15/18-21/23-24,B*4406,B*5104,B*5301-2 |
| 47 | 80 | 188 + 237 | 128 | B*1522,B*1801-5,B*3501-13/15-18/20-24,B*7801-2 |
| 48 | 81 | 195 + 213 + 277 | 389/390 | B*3501-9/11/15/17-19/21/23-24,B*5301-2 |
| 49 | 82 | 207 + 216 | 400 | B*1509,B*7801-2 |
| 50 | 84 | 193 + 216 | 451 | B*1509,B*5101-9/11N,B*7801-2 |
| 51 | 85 | 192 + 216 | 440 | B*5201 |
| 52 | 86 | 368 + 315 | 340 | Cw*0102-3 |
| 53 | 87 | 366 + 145 | 522 | Cw*0202-3,Cw*1701-2 |
| 54 | 88 | 368 + 389 | 564 | Cw*0302-6/8-9 |
| 55 | 89 | 366 + 143 | 331 | Cw*0401-5,Cw*1801-2 |
| 56 | 90 | 366 + 379 | 564 | Cw*0501 |
| 57 | 91 | 367 + 127 | 297 | Cw*0602-4 |
| 58 | 93 | 313 + 184 | 516 | Cw*0701/6/7 |
| 59 | 94 | 367 + 183 | 302 | Cw*0702/3/10 |
| 60 | 275 | 271 + 143 | 500 | B*8201,Cw*0708,Cw*1801-2 |
| 61 | 96 | 367 + 379 | 536 | Cw*0704 |
| 62 | 182 | 165 + 166 + 317 | 160/625 | Cw*0801-4 |
| 63 | 99 | 159 + 389 | 523 | Cw*0303 |
| 64 | 201 | 130 + 160 + 389 | 206/522 | Cw*0302/4-6/8-9 |
| 65 | 245 | 507 + 215 | 407 | Cw*1502 |
| 66 | 102 | 369 + 126 | 538 | Cw*1202 |
| 67 | 103 | 368 + 157 | 446 | Cw*1203/6 |
| 68 | 104 | 371 + 388 | 541 | Cw*1402-3 |
| 69 | 106 | 366 + 223 | 318 | Cw*1502-6 |
| 70 | 107 | 366 + 382 | 502 | Cw*0203,Cw*0404,Cw*0604,Cw*0707,Cw*1502-6,Cw*1701-2 |
| 71 | 109 | 368 + 146 | 513 | Cw*1601 |
| 72 | 110 | 366 + 146 | 503 | Cw*1602 |
| 73 | 111 | 366 + 377 | 502 | Cw*0202,Cw*0602-3,Cw*1204-5 |
All primer mixes have previously been described in Bunce et al.19
Abbreviations: PM, primer mix; Size, size of PCR product.
Sensitivity of nested PCR-SSP typing.
Primer mixes 151 (A*0101-4N), 9 (A*2501-2, A*2601-11N, A*3401-2, A*6601-3), 155 (B*0801-3), 47 (B*1301-3), 86 (Cw*0102-3), and 106 (Cw*1502-6) (Table 2) were randomly selected to determine the sensitivity of nested PCR typing. The experiments were performed as previously described and repeated on three occasions.22Briefly, decreasing amounts of DNA from selected HLA-typed healthy volunteers were mixed with DNA of known HLA type, both at a starting concentration of 200 ng/μL, to give final concentrations of 1%, 0.1%, 0.01%, and 0.001% (vol/vol). The mixtures were then subjected to nested PCR-SSP typing. All products were subsequently analyzed by agarose gel electrophoresis.
RESULTS
Sensitivity of nested PCR-SSP typing for HLA class I alleles.
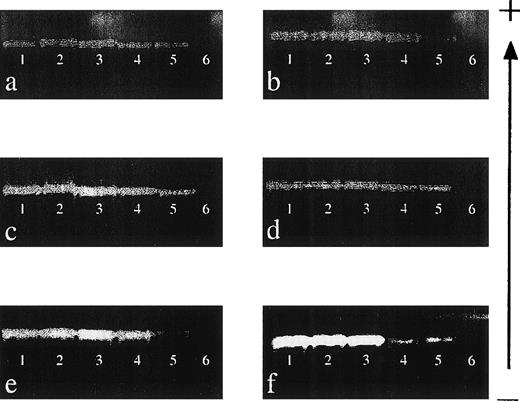
The sensitivity of detection of 6 of the HLA class I primer mixes (151, 9, 155, 47, 86, and 106; see Table 2) was determined by performing mixing experiments, and the results are shown in Fig 1. Each primer mix was capable of reproducibly detecting DNA to a level of 0.001% (equivalent to a dilution of DNA of 1:100,000), demonstrating that the sensitivity of this technique is comparable to that of other nested PCR-SSP techniques.17 22
Sensitivity of nested PCR-SSP typing for HLA class I alleles. (a) Primer mix (PM) 151 (A*0101-4N); (b) PM 9 (A*2501-2, A*2601-11N, A*3401-2, A*6601-3); (c) PM 155 (B*0801-3); (d) PM 47 (B*1301-3); (e) PM 86 (Cw*0102-3); and (f) PM 106 (Cw*1502-6). DNA of known HLA type (200 ng/μL) was mixed with an irrelevant DNA (200 ng/μL) to give relative final concentrations of 10% (lane 1), 1% (lane 2), 0.1% (lane 3), 0.01% (lane 4), and 0.001% (lane 5). Lane 6 is a specificity control in which the irrelevant DNA was amplified on its own. Each mix was then subject to a primary amplification with the appropriate set of first-round primers.28 The resultant product was diluted 1:500 in water before PCR-SSP typing using the method of Bunce et al.19 The gel is run from negative (−) to positive (+). Sensitivity experiments were performed on 3 occasions, and each primer mix was reproducibly capable of detecting DNA at a final concentration of 0.001%.
Sensitivity of nested PCR-SSP typing for HLA class I alleles. (a) Primer mix (PM) 151 (A*0101-4N); (b) PM 9 (A*2501-2, A*2601-11N, A*3401-2, A*6601-3); (c) PM 155 (B*0801-3); (d) PM 47 (B*1301-3); (e) PM 86 (Cw*0102-3); and (f) PM 106 (Cw*1502-6). DNA of known HLA type (200 ng/μL) was mixed with an irrelevant DNA (200 ng/μL) to give relative final concentrations of 10% (lane 1), 1% (lane 2), 0.1% (lane 3), 0.01% (lane 4), and 0.001% (lane 5). Lane 6 is a specificity control in which the irrelevant DNA was amplified on its own. Each mix was then subject to a primary amplification with the appropriate set of first-round primers.28 The resultant product was diluted 1:500 in water before PCR-SSP typing using the method of Bunce et al.19 The gel is run from negative (−) to positive (+). Sensitivity experiments were performed on 3 occasions, and each primer mix was reproducibly capable of detecting DNA at a final concentration of 0.001%.
Application of nested PCR-SSP typing to the detection of microchimerism.
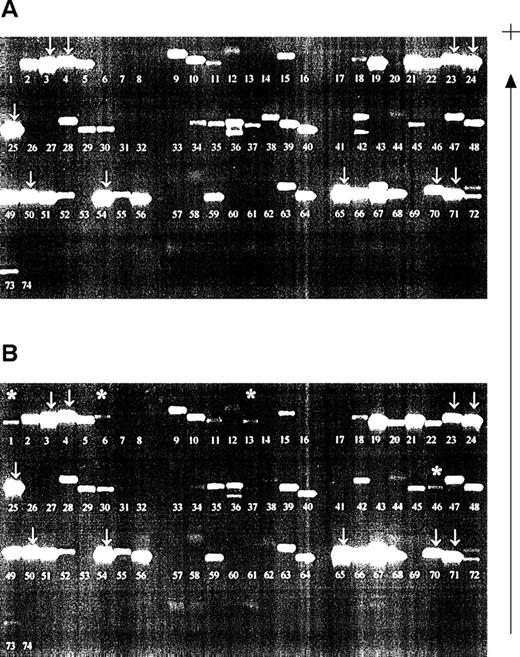
Samples were investigated from a patient who had received planned HLA-typed blood transfusions mismatched for HLA class I alleles. Samples obtained before and 2 days after blood transfusion were compared (Fig 2). All recipient alleles were detected in both samples (arrowed) and, in the posttransfusion samples, additional bands corresponding to the HLA alleles of both blood transfusion donors were evident (asterisked). However, nonspecific products also appeared in certain lanes on the gels, a finding consistent with our results of nested PCR-SSP typing for HLA-DR.22 The bands were not compatible with the known HLA type of the donors or the recipient. The pattern of bands generated by the nested PCR-SSP typing of the pretransfusion sample remains consistent in the posttransfusion samples and, thus, by inference, is recipient HLA-type dependent.
Detection of microchimerism using nested PCR-SSP typing. (A) Pretransfusion DNA sample (recipient HLA type: A*0301, A*2402; B*4001, B*51011; Cw*0304, Cw*1502, all recipient bands arrowed; other bands are nonspecific). (B) Posttransfusion DNA sample (blood donor 1 HLA type: A*0101; B*0801, B*4402; Cw*0701, Cw*0704; blood donor 2 HLA type: A*2501, A*3002; B*3501, B*5501; Cw*0303, Cw*0401). Asterisked bands are those donor alleles detected; the pattern of recipient alleles and nonspecific bands is the same as that of a pretransfusion sample. Patient DNA was isolated from peripheral blood leukocytes and used in a primary amplification using HLA-A, -B, or -C primers.28 The resultant products were diluted 1:500 and used in a PCR-SSP typing system.19 The gel is run from negative (−) to positive (+).
Detection of microchimerism using nested PCR-SSP typing. (A) Pretransfusion DNA sample (recipient HLA type: A*0301, A*2402; B*4001, B*51011; Cw*0304, Cw*1502, all recipient bands arrowed; other bands are nonspecific). (B) Posttransfusion DNA sample (blood donor 1 HLA type: A*0101; B*0801, B*4402; Cw*0701, Cw*0704; blood donor 2 HLA type: A*2501, A*3002; B*3501, B*5501; Cw*0303, Cw*0401). Asterisked bands are those donor alleles detected; the pattern of recipient alleles and nonspecific bands is the same as that of a pretransfusion sample. Patient DNA was isolated from peripheral blood leukocytes and used in a primary amplification using HLA-A, -B, or -C primers.28 The resultant products were diluted 1:500 and used in a PCR-SSP typing system.19 The gel is run from negative (−) to positive (+).
To control for the presence of the nonspecific products resulting from mispriming, a specificity control in the form of a pretransfusion sample is required. Rigorous analysis of this sample establishes a baseline for the analysis of posttransfusion samples. Therefore, nested PCR-SSP typing is routinely performed 5 times on 2 samples obtained before transfusion and on samples after transfusion. The results from one such analysis of the HLA-A locus are shown in Fig 3.
Validation of nested PCR-SSP typing. Recipient 01 (HLA-A type: A*0301, A*2402) was transfused with fresh blood from 2 healthy, HLA-typed volunteers (donor 1: A*0101; donor 2: A*2501, A*3002). The bar graph shows the results of amplification of 2 pretransfusion samples and a posttransfusion sample by primer mixes for the HLA-A locus alleles used in this system (Table 2). Analysis of both pretransfusion samples produces a similar pattern of amplification in which primer mixes 4 and 5 represent recipient alleles (A*0301) and (A*2402), respectively (dark shading). Potentially informative primer mixes are those that do not give rise to nonspecific products in any of the multiple tests of the pretransfusion samples. In this case, these are primer mixes 151, 9, 12, 152, 18, 184, and 154. Analysis of the posttransfusion samples shows additional amplification with primer mix 151, which is indicative of donor 1 (A*0101), and primer mixes 9 and 18, which are both indicative of donor 2 (A*2501 and A*3002, respectively; hatched shading). Primer mixes 3, 6, 10, 13, 14, 15, 153, 23, and 24 detect nonspecific products in the pretransfusion samples and thus are noninformative in the analysis of posttransfusion samples (light shading). The requirement for testing pretransfusion samples is demonstrated by the results with primer mix 10, which potentially should amplify A*2501 from donor 2. In this example, although there was amplification in the posttransfusion sample, the reaction has to be excluded from the analysis because of the nonspecific amplification present in the pretransfusion samples.
Validation of nested PCR-SSP typing. Recipient 01 (HLA-A type: A*0301, A*2402) was transfused with fresh blood from 2 healthy, HLA-typed volunteers (donor 1: A*0101; donor 2: A*2501, A*3002). The bar graph shows the results of amplification of 2 pretransfusion samples and a posttransfusion sample by primer mixes for the HLA-A locus alleles used in this system (Table 2). Analysis of both pretransfusion samples produces a similar pattern of amplification in which primer mixes 4 and 5 represent recipient alleles (A*0301) and (A*2402), respectively (dark shading). Potentially informative primer mixes are those that do not give rise to nonspecific products in any of the multiple tests of the pretransfusion samples. In this case, these are primer mixes 151, 9, 12, 152, 18, 184, and 154. Analysis of the posttransfusion samples shows additional amplification with primer mix 151, which is indicative of donor 1 (A*0101), and primer mixes 9 and 18, which are both indicative of donor 2 (A*2501 and A*3002, respectively; hatched shading). Primer mixes 3, 6, 10, 13, 14, 15, 153, 23, and 24 detect nonspecific products in the pretransfusion samples and thus are noninformative in the analysis of posttransfusion samples (light shading). The requirement for testing pretransfusion samples is demonstrated by the results with primer mix 10, which potentially should amplify A*2501 from donor 2. In this example, although there was amplification in the posttransfusion sample, the reaction has to be excluded from the analysis because of the nonspecific amplification present in the pretransfusion samples.
The recipient HLA type (A*0301, A*2402) is present in all repeats of the pretransfusion and posttransfusion samples (PM 4 and 5; dark shading). Potentially informative reactions are those reactions that do not yield a band in the pretransfusion samples (in this case, primer mixes 151, 9, 12, 152, 18, 184, and 154). The blood donors were typed as HLA-A*0101 (donor 1) and HLA-A*2501, A*3002 (donor 2), and these alleles are recognized by primer mixes 151 (A*0101), 9 (A*2501), 10 (A*2501), and 18 (A*3002). All of these primer mixes gave rise to a band when the posttransfusion sample was analyzed, but in the case of primer mix 10, a band was also evident in both pretransfusion samples. The reaction with primer mix 10 is therefore termed a noninformative reaction. Furthermore, there were other primer mixes that gave rise to bands in the pretransfusion samples that were also present in posttransfusion samples (PM 3, 6, 13, 14, 15, 153, and 24). The presence of these nonspecific products is dependent on the recipient HLA type.
DISCUSSION
In this report, we have described a method for the detection of HLA class I donor alleles after blood transfusion using the nested PCR-SSP technology previously developed for HLA-DR alleles.17,22,31The technique is finely tuned to achieve maximum sensitivity while retaining specificity; for example, increased sensitivity can be achieved by increasing the amount of DNA in the first-round amplification, but such a modification will result in a decrease in specificity. The sensitivity of the technique has been demonstrated using mixing experiments and DNA can be detected to a level of at least 0.001% (equivalent to a dilution of 1:100,000), comparable to the techniques developed for the detection of HLA-DR alleles.17 22
The high level of sensitivity achieved with the technique leads to a concomitant decrease in specificity. When the nested PCR-SSP typing was applied to the detection of donor-type microchimerism in a patient receiving an HLA-mismatched blood transfusion, nonspecific products appeared that did not correspond to the donor or recipient HLA type. This finding is entirely consistent with our previous results obtained while developing a similar system for the detection of HLA-DR alleles. In the previous study, we sequenced several of these nonspecific products and determined that they did not result from contamination but rather from mispriming events that led to amplification of recipient-derived HLA alleles or associated pseudogenes.22It is therefore possible that some of the nonspecific products seen in the HLA class I nested PCR-SSP typing system result from the amplification of HLA class I-associated pseudogenes, of which 6 have been currently identified.32 33 Therefore, to use nested PCR-SSP typing to analyze clinical samples, a means of carefully controlling the system is required.
In the system we have devised, a pretransfusion blood sample is analyzed to act as a baseline and specificity control. Multiple analysis of this sample with nested PCR-SSP typing allows a rigorous baseline to be established for all subsequent analyses (Figs 2 and 3). If such a sample is not included in the analysis, then amplification appearing in posttransfusion samples may be wrongly assumed to be indicative of donor alleles. For example, in Fig 2, primer mix 10, which amplifies the donor allele HLA-A*2501, yields a band in the posttransfusion sample suggesting the presence of microchimerism. However, this band is also detected in the pretransfusion sample; therefore, the amplification in the posttransfusion samples is noninformative. The requirement for a pretransfusion/transplant sample has also been shown by Elwood et al,4 who, by using nested PCR-SSP typing for HLA-DR alleles, demonstrated that false-positive results for microchimerism would have been obtained in 17% of patients had a recipient pretransplant DNA sample not been included in the analysis.
The inclusion of HLA-A, -B, and -C alleles in the nested PCR-SSP system significantly increases the power of this type of analysis where donors and recipients are frequently matched for HLA-DR alleles. The technique is flexible and widely applicable to the detection of microchimerism after blood transfusion or solid organ transplantation and may provide the means for understanding the true relevance of microchimerism.
Supported by a grant from the National Kidney Research Fund.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Susan V. Fuggle, D. Phil., Nuffield Department of Surgery, University of Oxford, John Radcliffe Hospital, Oxford, OX3 9DU, UK; e-mail: susan.fuggle@nds.ox.ac.uk.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal