Abstract
Human herpesvirus 6 (HHV-6) genome has been detected in several human lymphoproliferative disorders with no signs of active viral infection, and found to be integrated into chromosomes in some cases. We previously reported a woman with HHV-6–infected Burkitt’s lymphoma. Fluorescence in situ hybridization showed that the viral genome was integrated into the long arm of chromosome 22 (22q13). The patient’s asymptomatic husband also carried HHV-6 DNA integrated at chromosome locus 1q44. To assess the possibility of chromosomal transmission of HHV-6 DNA, we looked for HHV-6 DNA in the peripheral blood of their daughter. She had HHV-6 DNA on both chromosomes 22q13 and 1q44, identical to the site of viral integration of her mother and father, respectively. The findings suggested that her viral genomes were inherited chromosomally from both parents. The 3 family members were all seropositive for HHV-6, but showed no serological signs of active infection. To confirm the presence of HHV-6 DNA sequences, we performed polymerase chain reaction (PCR) with 7 distinct primer pairs that target different regions of HHV-6. The viral sequences were consistently detected by single-step PCR in all 3 family members. We propose a novel latent form for HHV-6, in which integrated viral genome can be chromosomally transmitted. The possible role of the chromosomally integrated HHV-6 in the pathogenesis of lymphoproliferative diseases remains to be explained.
SINCE THE FIRST isolation of human herpesvirus 6 (HHV-6),1 it has become apparent that infection by this virus is widespread. As with other herpesviruses, HHV-6 remains latent in the host after primary infection.2Transmission of HHV-6 via saliva from mother to infant is thought to be the most common route.2 Besides being an infectious agent, HHV-6 has been cited as a possible etiological factor or as a modulating element of certain human neoplastic diseases, particularly lymphoproliferative disorders. HHV-6 genome was detected in Hodgkin’s disease, various types of non-Hodgkin’s lymphomas, and acute lymphoblastic leukemia (ALL),3-10 but the question of how HHV-6 exists in these neoplastic cells has not yet been fully answered. We recently showed the integration of HHV-6 in the leukemic blast cells of a patient with ALL4 and proposed that the chromosomally integrated HHV-6 could be transmitted from generation to generation.11 Supportive evidence for this novel mode of viral transmission has been desired.
We previously reported a case of HHV-6 genome-positive Burkitt’s lymphoma,3 in which the viral genome was found to be integrated into the long arm of chromosome 22 (22q13) of the lymphoma cells.12 This case provided a unique opportunity to investigate chromosomal transmission of HHV-6 in the patient’s family.
MATERIALS AND METHODS
Fluorescence in situ hybridization (FISH) on metaphase chromosomes.
Metaphase chromosome preparations were obtained from leukemic peripheral blood from the patient with HHV-6+ Burkitt’s lymphoma3 and phytohemagglutinin (PHA)-stimulated peripheral blood mononuclear cells (PBMCs) from the patient’s family members and 3 HHV-6–seropositive healthy donors. The cells were synchronized by treatment with 5-bromodeoxyuridine for 16 hours, and thereafter the cells were released from the block by incubation in fresh medium containing thymidine for 6 hours as previously described.13 Metaphase cells and chromosome spreads were obtained according to standard procedures. Probe containing aBamHI fragment (6.9 kb) of HHV-6, which was cloned DNA inserted into a plasmid (pH6Z-101) (supplied by Dr P.E. Pellett, Centers for Disease Control and Prevention, Atlanta, GA),14 was labeled with biotinylated 14-deoxyadenosine triphosphate (14-dATP) by nick translation. This probe proved to be specific for HHV-6. Forty microliters of hybridization solution (hybrisol VII; Oncor, Gaithersberg, MD) containing 50 ng of the biotinylated probe was denatured at 70°C for 2 minutes and applied to RNase-treated chromosome preparations, which were then incubated at 37°C overnight in a humidified chamber. The slides were washed twice for 10 minutes in 50% formamide in 2× standard saline citrate (SSC) (0.3 mol/L sodium chloride, 30 mmol/L sodium citrate, pH 7.0) at 43°C, followed by two rinses in 2 × SSC at 37°C. The hybridized probe was detected by incubation with fluorescein isothiocyanate-conjugated avidin. The chromosomes were counterstained with propidium iodide and diamidino-2-phenylindole (DAPI). The slides were observed with a BX50 epifluorescence microscope (Olympus, Tokyo, Japan) and microphotographs were taken on Provia Fujichrome 100 film (Fuji Film, Tokyo, Japan).
Primers and polymerase chain reaction (PCR).
For HHV-6 DNA amplification by PCR, 7 sets of primers from different regions of the HHV-6 genome were used (Table1). DNA was obtained using the phenol chloroform-extraction technique after proteinase K digestion. A total of 0.1 μg of genomic DNA was amplified in 50 μL of PCR buffer (10 mmol/L Tris-HCl, pH 8.3, 50 mmol/L KCl, 1.5 mmol/L MgCl2) with 0.2 mmol/L of each deoxynucleoside triphosphate, 1.5 U Taq polymerase enzyme (Takara Shuzo, Shiga, Japan), and 0.2 μmol/L of each pair of primers. Reaction mixtures were incubated at 94°C for 3 minutes for denaturation followed by 25 cycles at 94°C for 1 minute, 57°C for 1 minute, and 72°C for 1 minute. A terminal extension at 72°C for 5 minutes was performed after completion of the 25 cycles. A total of 20% of the amplification products (10 μL) was electrophoresed on a 2% agarose gel followed by ethidium bromide staining and visualization under ultraviolet light for the presence of DNA bands of appropriate sizes.
Oligonucleotide Primers Used for HHV-6 DNA Amplification
| Region of HHV-6 Genome . | Sequences (5′_3′) . | Size of PCR Product (bp) . |
|---|---|---|
| U4 | GATCCATGGTCGTCTTTCCAC | |
| ATTGTGATGTACGTGGCCGTC | 392 | |
| U31 | CCCATTTACGATTTCCTGCAC | |
| TTCAGGGACCGTTATGTCATT | 186 | |
| U57 | CTTTGTGTAGGTGGTCGAATGCGAC | |
| ACAGCGCAGCAACATGTTTCAGAGC | 494 | |
| U67 | GCGTTTTCAGTGTGTAGTTCGGCAG | |
| TGGCCGCATTCGTACAGATACGGAGG | 520 | |
| U89 | CATCATTGTTATCGCTTTCACTCTC | |
| TTCTCCAGAAGTCCCAGGGAAATCC | 553* | |
| U94 | CGCTGGGAATTCCATCTAAAGC | |
| ATCCGAGTGATGATTTCTGGAC | 330 | |
| Sequence between U91 and U92 | CTGAACTGGCTGTAACTTCTGC GCAAACGAGAACTCTAAAATCT | 550 |
| Region of HHV-6 Genome . | Sequences (5′_3′) . | Size of PCR Product (bp) . |
|---|---|---|
| U4 | GATCCATGGTCGTCTTTCCAC | |
| ATTGTGATGTACGTGGCCGTC | 392 | |
| U31 | CCCATTTACGATTTCCTGCAC | |
| TTCAGGGACCGTTATGTCATT | 186 | |
| U57 | CTTTGTGTAGGTGGTCGAATGCGAC | |
| ACAGCGCAGCAACATGTTTCAGAGC | 494 | |
| U67 | GCGTTTTCAGTGTGTAGTTCGGCAG | |
| TGGCCGCATTCGTACAGATACGGAGG | 520 | |
| U89 | CATCATTGTTATCGCTTTCACTCTC | |
| TTCTCCAGAAGTCCCAGGGAAATCC | 553* | |
| U94 | CGCTGGGAATTCCATCTAAAGC | |
| ATCCGAGTGATGATTTCTGGAC | 330 | |
| Sequence between U91 and U92 | CTGAACTGGCTGTAACTTCTGC GCAAACGAGAACTCTAAAATCT | 550 |
For HHV-6 variant B.
RESULTS
The patient was a 58-year-old Japanese woman with Burkitt’s lymphoma. Her blood samples were positive for HHV-6 DNA by PCR.3 By using the HHV-6+ lymphoma cell line established from the patient, we showed integration of HHV-6 at chromosome locus 22q13 in the lymphoma cells.12 The presence of HHV-6 DNA was also shown by Southern blot hybridization using the BamHI fragment of HHV-6 as probe.3,12 In this study, FISH with the HHV-6–specific pH6Z-101 probe was also performed on metaphase chromosomes from the peripheral blood in a leukemic phase. FISH allows the identification of integrated viral genome as well as episomal viral genome, because episomes should be associated with chromosomes randomly, whereas integrated copies should give rise to symmetrical hybridization signals at distinct chromosomal sites.15 16FISH enabled us to directly visualize the integrated HHV-6 DNA at the single-cell level. Specific symmetrical doublet signals on both chromatids of a single homolog of chromosome 22 were observed in approximately 70% of the peripheral blood metaphase cells examined (Fig 1A). Q-banding–like DAPI staining indicated that the signals on chromosome 22 were located at q13. The finding confirmed that the patient carried integrated HHV-6 DNA on chromosome 22q13.
FISH on metaphase chromosomes from the lymphoma patient (A), patient’s husband (B), and their daughter (C). Hybridization with an HHV-6–specific probe showed HHV-6 integration with symmetrical doublet signals at homologous sites of both chromatids. Arrow and arrow head indicate the hybridization signals on chromosome loci 1q44 and 22q13, respectively. Chromosomes were counterstained with propidium iodide.
FISH on metaphase chromosomes from the lymphoma patient (A), patient’s husband (B), and their daughter (C). Hybridization with an HHV-6–specific probe showed HHV-6 integration with symmetrical doublet signals at homologous sites of both chromatids. Arrow and arrow head indicate the hybridization signals on chromosome loci 1q44 and 22q13, respectively. Chromosomes were counterstained with propidium iodide.
We next examined the patient’s family members for the presence of HHV-6 genome. After obtaining informed consent, we obtained peripheral blood from her asymptomatic 68-year-old husband. PBMCs were incubated with PHA for 3 days and subjected to FISH analysis. Unexpectedly, symmetrical doublet hybridization signals were seen on chromosome 1q44 in about 60% of metaphase cells (Fig 2A) and, therefore, he was also thought to carry the HHV-6 genome integrated on chromosome 1q44. To assess chromosomal transmission of HHV-6 DNA, we looked for the viral genome in their healthy 35-year-old daughter. HHV-6 DNA was detected on both chromosome loci 22q13 and 1q44 in about 90% of PHA-stimulated PBMCs (Fig 1C), identical to the site of viral integration of her mother and father, respectively. The results strongly suggested that her viral genomes were inherited chromosomally from both parents. The 3 family members were all seropositive for HHV-6, but showed no serological signs of active infection (anti–HHV-6 immunoglobin G (IgG), 1: 160; anti–HHV-6 IgM, <1:10). In parallel experiments, PHA-stimulated PBMCs from 3 control HHV-6–seropositive healthy adults did not show any signals after hybridization.
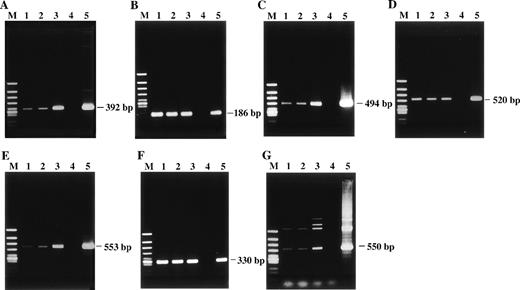
Detection of HHV-6 DNA by PCR with 7 distinct primer pairs that target different regions of the HHV-6 genome. The PCR products were subjected to electrophoresis and stained with ethidium bromide. The products were all of the predicted sizes. (A) HHV-6 U4 gene. (B) U31 gene. (C) U57 gene. (D) U67 gene. (E) U89 gene. (F) U94 gene. (G) HHV-6 sequence located between U91 and U92 genes. Lane 1, patient; lane 2, patient’s husband; lane 3, their daughter; lane 4, healthy adult donor as a negative control; lane 5, HHV-6B–infected cord blood cells as a positive control; lane M, ◊X174/HincII-cut DNA size marker.
Detection of HHV-6 DNA by PCR with 7 distinct primer pairs that target different regions of the HHV-6 genome. The PCR products were subjected to electrophoresis and stained with ethidium bromide. The products were all of the predicted sizes. (A) HHV-6 U4 gene. (B) U31 gene. (C) U57 gene. (D) U67 gene. (E) U89 gene. (F) U94 gene. (G) HHV-6 sequence located between U91 and U92 genes. Lane 1, patient; lane 2, patient’s husband; lane 3, their daughter; lane 4, healthy adult donor as a negative control; lane 5, HHV-6B–infected cord blood cells as a positive control; lane M, ◊X174/HincII-cut DNA size marker.
To confirm the presence of HHV-6 DNA sequences, we performed extensive PCR analysis with 7 distinct primer pairs that target different regions of HHV-6, including U4, U31, U57, U67, U89, U94 genes, and a sequence located between U91 and U92 genes. These HHV-6 DNA sequences are located away from the sequences that hybridize with the pH6Z-101 probe.17 The viral sequences were consistently detected by single-step PCR in all 3 family members (Fig 2), suggesting high copy numbers of HHV-6 genome. Under the same PCR conditions, no HHV-6 DNA sequences were detected in PBMCs of our control HHV-6–seropositive adults.
Furthermore, attempts were made to establish lymphoblastoid cell lines from the PBMCs of the patient’s husband and daughter. Epstein-Barr virus (EBV) and Herpesvirus saimiri (HVS) selectively infect B and T cells, respectively, and immortalize them to give rise to continuously growing cell lines.18-21 By exploiting this viral transforming capacity, we have successfully established 2 HHV-6–carrying B- and T-cell lines from each of them. The presence of HHV-6 DNA in these cell lines was confirmed by PCR with 7 sets of primers as described above as well as Southern blot hybridization with the pH6Z-101 probe (data not shown). The EBV-immortalized cell lines possessed the activated B-cell phenotype, whereas the HVS-immortalized cell lines were CD8+ T-cell lines. Both B- and T-cell lines from the patient’s husband carried integrated HHV-6 DNA at chromosome 1q44, while both B- and T-cell lines derived from the daughter had the viral genome integrated at both chromosomes 22q13 and 1q44 (data not shown).
DISCUSSION
In this study, we detected the integrated HHV-6 genome at chromosome locus 22q13 in a patient with Burkitt’s lymphoma and at 1q44 in the patient’s husband. Moreover, we showed that their daughter carried the HHV-6 genome at both 22q13 and 1q44, which are the identical sites of HHV-6 integration of her mother and father, respectively. We also reported a latent form of HHV-6 in an ALL family, in which chromosomally integrated viral genome at 1q44 was shown to be transmitted serially in 3 generations.11 These observations provide convincing evidence for chromosomal transmission of the HHV-6 genome, which is a phenomenon to be known for the first time for the Herpesviridae. HHV-6 is the causative agent of exanthem subitum in early childhood,22 and most adults are thought to be latently infected with this virus. However, such HHV-6 latency in adults is usually characterized by a very low copy number of HHV-6 genome in peripheral blood, detectable only by highly sensitive nested-PCR.23 In this respect, chromosomally integrated HHV-6 with a high copy number of HHV-6 genome should be distinct from the latency after primary infection. Indeed, HHV-6 DNA sequences in our 3 family members were detectable by single-step PCR with only 25-cycle amplification using 0.1 μg genomic DNA as template, whereas in our parallel experiments neither FISH nor PCR detected HHV-6 DNA in peripheral blood of 3 control HHV-6–seropositive adults.
It may be argued that we are detecting a cellular gene with homology to a viral gene in the family members. However, this would be unlikely, because HHV-6 DNA sequences were consistently detected by Southern blot hybridization as well as PCR with 7 primer pairs from different regions of HHV-6 including U4, U31, U57, U67, U89, U94 genes, and a sequence located between U91 and U92 genes. It also excludes the possibility that only a small fragment of HHV-6 is integrated, but further analysis is needed to clarify whether the integrated viral genome is complete. Our FISH analysis showed that not all metaphase cells were positive for hybridization signals. This may appear to be contradictory to our proposal of chromosomal transmission of HHV-6 DNA because HHV-6 should be present in all cells if it is constitutionally integrated. However, signal-negative cells may have been false-negative for HHV-6 DNA because of technical difficulty of FISH, and the hybridization efficiency would be increased if longer HHV-6 DNA fragments are used as probes.
Although the most common mode of infection with HHV-6 is by salivary transmission, several studies suggested that intrauterine or perinatal transmission may occur. Aubin et al24 reported that 1 of 52 aborted fetuses was positive for HHV-6 DNA by PCR in PBMCs, thymus, liver, spleen, brain, and cerebrospinal fluid. HHV-6 DNA sequences were also found in PBMCs of the mother, but no HHV-6–specific IgM antibody was detected in maternal serum and fetal plasma. Adams et al25 showed that HHV-6 DNA was detected in 5 of 305 cord blood samples, but HHV-6 IgM antibody could not be found in the fetal sera of the HHV-6 DNA+ cases. Although these findings were interpreted as indicative of intrauterine infection, the possibility of chromosomal transmission of HHV-6 from parents to offspring should be explored. To survey the prevalence of carriers of chromosomally integrated HHV-6 in a population from our geographic area, we also investigated the presence of HHV-6 DNA in cord blood samples. HHV-6 DNA could not be found in any of our 58 cord blood specimens.26The incidence of chromosomal transmission of HHV-6 genome appears to be rare, but the present findings should alert investigators to the presence of HHV-6 as an inheritable chromosomal element. Of 3 family members carrying the integrated HHV-6 DNA in this study, 2 remain healthy. Further investigations are needed to clarify its role in the pathogenesis of lymphoproliferative diseases.
ACKNOWLEDGMENT
We thank P.E. Pellett for the gift of HHV-6–specific DNA probe, pH6Z-101. We are also grateful to M. Yasukawa for providing HVS.
Supported by a grant-in-aid for scientific research from the Japanese Ministry of Education, Science and Culture (to M.D.).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Masanori Daibata, MD, Department of Medicine, Kochi Medical School, Kochi 783-8505, Japan.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal