Abstract
Human immunodeficiency virus (HIV) entry is mediated not only by the CD4 receptor, but also by interaction with closely related molecules that act as membrane coreceptors. We have analyzed mRNA expression and/or cell membrane exposition of the coreceptors most widely used by diverse HIV-1 strains (CXCR4, CCR5, and CCR3) on purified hematopoietic progenitor cells (HPCs) induced in liquid suspension culture to unilineage differentiation/maturation through the erythroid (E), granulocytic (G), megakaryocytic (Mk), and monocytic (Mo) lineages. Reverse transcriptase-polymerase chain reaction (RT-PCR) and cytofluorimetric analysis showed the presence of both CXCR4 and CCR5 in quiescent HPCs, but failed to detect CCR3-specific transcripts. Chemokine expression in HPC progenies showed that CXCR4 receptor is detected on the majority of MKs from early to late stages of maturation, whereas it is moderately decreased in the Mo lineage. In the G pathway, two distinct cell populations, CXCR4+ and CXCR4−, were observed: morphological analysis of the sorted populations showed that the CXCR4+ cells were largely eosinophils and the CXCR4− were granulocytes of the neutrophilic series. Furthermore, in the E pathway, CXCR4 was almost completely absent. CCR5 expression is restricted to Mo cultures, ie, ≈30% to 80% cells throughout all monocytopoietic differentiation/maturation stages. Finally, CCR3 mRNA is always absent in all the unilineage cultures. Evaluation of CD4 expression by flow cytometry on both quiescent HPCs and differentiating unilineage precursors showed that the CD4 receptor is present on ≈15% of the starting CD34+ HPC population, highly expressed in the Mo lineage up to 80% at terminal maturation, present on 20% to 30% of maturing Mks, and not detectable in either the E or G lineage. Expression of CD4 receptor together with CXCR4 and/or CCR5 coreceptor in the four lineages correlates with hematopoietic precursor susceptibility to T-lymphotropic and macrophage (M)-tropic HIV strains infection: (1) CD4− G and E cells were resistant to both M-tropic and T-lymphotropic strains; (2) HPC-derived Mks were susceptible to T-tropic, but resistant to M-tropic, infection; (3) Mo differentiating cells efficiently replicate both HIV strains. Furthermore, we showed that the CXCR4 and CCR5 ligands (stromal-derived factor 1 and macrophage-inflammatory protein-1 [MIP-1], MIP-1β and RANTES, respectively) inhibit HIV replication in both maturing Mo and Mk cells. Taken together, our data show a lineage-specific modulation of chemokine receptor/coreceptor during hematopoietic cell differentiation and extend previous observations on the relationship between the expression of HIV receptor/coreceptors, susceptibility, and chemokine-mediated resistance to HIV infection.
CHEMOKINE RECEPTORS belong to a family of seven transmembrane spanning G protein–coupled receptors whose activation leads to intracellular and extracellular signaling.1-3 Their role in the human immunodeficiency virus (HIV) entry process has been demonstrated by numerous studies.4-12 Particularly, chemokine receptor CXCR4 binds stromal-derived factor 1 (SDF-1) and has been shown to function as a coreceptor for T-cell tropic HIV-1 strains,4-6 while chemokine receptor CCR5 binds the β-chemokines macrophage-inflammatory protein-1α (MIP-1α), MIP-1β, and regulated-upon-activation, normal T-expressed and secreted (RANTES) and mediates binding and entry of the M-tropic HIV strain.7-12Therefore, HIV entry depends on the expression of receptor molecules that mediate physiological events of primary importance, such as chemotaxis, inflammation, and cell differentiation. In humans, HIV-1 strains that use CCR5 coreceptor predominate during the asymptomatic stages of infection; as disease proceeds, viruses emerge that use other receptors, most commonly the CXCR4.13
The pattern of expression of CXCR4 and CCR5 receptors has been investigated on CD34+ human progenitor cells. CXCR4 mRNA and protein were found on hematopoietic progenitor cells (HPCs) purified either from bone marrow (BM)14-18 or from peripheral blood (PB).19,20 Uncertain results were obtained for CCR5 expression: analysis of granulocyte colony-stimulating factor (G-CSF)–mobilized PB progenitors from single donors showed a low incidence of cell samples containing CCR5 mRNA20; negative18 or positive16 results were observed in BM-derived CD34+ cells. On the other hand, cytofluorimetric analysis revealed that CCR5 is undetectable on most BM hematopoietic progenitors,15,17 while its expression was found in PB-derived CD34+ cells.19 Little is known about HIV coreceptor regulation during the differentiation/maturation of HPCs. Recently, CXCR4 has been shown to be exposed on the cell surface of CD34+ cell-derived megakaryocytes, and its expression is increased during their maturation.14,17,21 22
We previously showed that both HPCs and maturing megakaryocytic (Mk) precursors are susceptible to T-tropic HIV infection.23,24These findings correlated with the membrane exposition of the CD4 molecule, the major HIV receptor. Taking advantage of a methodology for selective unilineage HPC differentiation cultures,25 we extended the studies to the expression of CD4 receptor and CXCR4, CCR5, and CCR3 coreceptors in HPCs and during their differentiation/maturation along the erythroid (E), granulocytic (G), monocytic (Mo), and Mk lineages. Furthermore, we correlated the receptor/coreceptor expression to the permissiveness of M- and T-tropic HIV strain replication and evaluated the degree of infection inhibition by the natural ligands of the HIV coreceptors.
MATERIALS AND METHODS
Hematopoietic Growth Factors (HGFs) and Culture Media
Recombinant human interleukin-3 (rhIL-3; 2 × 106U/mg), granulo-monocytic colony-stimulating factor (rhGM-CSF), and rhIL-6 were supplied by the Genetics Institute (Cambridge, MA); erythropoietin (rhEpo; 1.2 × 105 U/mg) by Amgen (Thousand Oaks, CA); and flt3-ligand (rhFL; 1.9 × 106U/mg) by Immunex (Seattle, WA). rhG-CSF (1 × 108U/mg) and rhM-CSF (6 × 107 U/mg) were purchased from R&D Systems Inc (Minneapolis, MN), and thrombopoietin (rhTpo) was from Pepro Tech, Inc (Rocky Hill, NJ). Iscove’s modified Dulbecco’s medium (IMDM; GIBCO, Grand Island, NY) was prepared weekly before each purification experiment.
HPC Purification
Adult PB was obtained from 20- to 40-year-old healthy male donors after informed consent. PB HPCs were purified from the buffy coat by a slight modification26,27 of the method described by Gabbianelli et al.28 Briefly, a negative selection of low-density cells was performed with an anti-T, -B, -natural killer (NK) lymphocyte, -monocyte, and -granulocyte monoclonal antibody (MoAb) cocktail supplemented with anti-CD45, -CD11a, and -CD71 MoAbs (Becton Dickinson, Mountain View, CA). The adherent fraction of PB mononuclear cells (PBMCs) obtained during HPC purification was used as the source of monocytic cells (92% to 96% CD14+). Neutrophils were isolated by sedimentation through a cushion of succinyl gelatine.
To obtain CD34+ cells, HPCs were incubated with a saturating amount of anti-CD34 directly conjugated with fluorescein isothiocyanate (FITC) (8G12 clone; Becton Dickinson), washed, and run on a FACSVantage (Becton Dickinson) cell sorter. The CD34+subpopulation was sorted on the basis of fluorescence emission.
HPC Characterization
Immunofluorescence assay.
Anti–CD34-FITC (8G12 clone) MoAb was used to characterize the membrane phenotype of purified HPCs. After the labeling procedure described by Testa et al,25 at least 4,000 cells for each determination were analyzed by fluorescence-activated cell sorting (FACScan; Becton Dickinson) for fluorescence intensity.
HPC clonogenetic assay.
In fetal calf serum (FCS)+ cultures, purified HPCs were seeded (1 × 102 cells/mL dish, 2 or 3 plates per point) and cultured in 0.9% methylcellulose, 40% FCS in IMDM supplemented with α-thioglycerol (10− 4mol/L) (Sigma, St Louis, MO), ferric ammonium citrate (10 μg/mL), pure human transferrin (0.7 mg/mL), and different HGFs as detailed below, at 37°C in a 5% CO2/5% O2/90% N2 humidified atmosphere. In FCS−cultures, FCS was substituted by bovine serum albumin (BSA), pure human transferrin, human low-density lipoproteins, insulin, sodium pyruvate, L-glutamine (2 × 10−3 mol/L), rare inorganic elements25 supplemented with iron sulfate (4 × 10−8 mol/L), and nucleosides. Both FCS+and FCS− cultures were supplemented with FL (100 ng/mL), kit ligand (KL; 100 ng), IL-3 (100 U), Tpo (100 ng), GM-CSF (10 ng), Epo (3 U), M-CSF (250 U), and G-CSF (500 U). CFU-GEMM, BFU-E, CFU-MK, and CFU-GM colonies were scored on days 14-15 and 16-17 in FCS+ and FCS− cultures, respectively.
Unilineage HPC Liquid Suspension Culture
HPCs were seeded at 5 × 104 cells/mL, grown in liquid suspension culture, and induced to specific differentiation by addition of appropriate HGF combinations: (1) in FCS−erythropoietic (E) culture, very low doses of IL-3 (0.01 U/mL) and GM-CSF (0.001 ng/mL), and a saturating level of Epo (3 U); (2) in FCS− granulopoietic (G) culture, low amounts of IL-3 (1 U) and GM-CSF (0.1 ng), and a saturating amount of G-CSF (500 U); (3) in FCS− megakaryocytopoietic (Mk) culture, saturating doses of Tpo (100 ng); and (4) in FCS+monocytopoietic (Mo) culture, IL-6 (1 ng) and saturating amounts of both FL (100 ng) and M-CSF (500 U). Cells were periodically counted and analyzed for morphology and membrane phenotype through the 12 to 14 days of culture.
For the morphology analysis, cells were cytocentrifuged onto glass slides and stained with May-Grünwald-Giemsa (Sigma).
Immunofluorescence Analysis
The following MoAbs (from Becton Dickinson, unless otherwise indicated) directly conjugated with FITC were used to characterize the membrane phenotype on cell samples collected at different days of culture: glycophorin A (Immunotech, Marseilles, France) for erythroid cells; CD11b or CD15 for granulocytic cells; CD14 for monocytic cells; and CD61 for megakaryocytic cells. CD4 expression was assessed using anti-CD4 MoAb Leu3A directly conjugated with phycoerythrin (PE). PE-labeled anti-CXCR4 (clone 12G5; Pharmigen, San Diego, CA) or anti-CCR5 (clone 2D7; Pharmigen) MoAbs were used for the study of chemokine receptors expression. Cells were washed twice in phosphate-buffered saline (PBS) and Fc-blocked by a 15-minute treatment at room temperature with 10 μg/mL human IgG. Cells were then incubated for 45 minutes at 4°C in the presence of appropriate amounts of specific MoAbs. After 3 washes with cold PBS containing 2 mg/mL BSA, cells were resuspended in 0.2 mL PBS/2.5% formaldehyde and analyzed by FACScan using the Lysis II program for fluorescence intensity analysis. CXCR4 and CCR5 expression on HPCs was investigated using anti-CXCR4 (clone 12G5) or anti-CCR5 (clone 2D7) MoAb along with CD34 MoAb, by the double-labeling procedure described above. HPCs were also labeled with a different CCR5 MoAb (clone 45502.111; R&D Systems) that requires a demasking step to enhance anti-CCR5 MoAb recognition: cells were first Fc-blocked by treatment with 10 μg/mL of human IgG, washed twice in PBS, and then incubated for 15 minutes at 4°C in PBS containing 4% formaldehyde. After 3 washings with PBS/BSA, the cells were incubated for 45 minutes with anti-CCR5-FITC and CD34-PE MoAbs, washed, and analyzed by flow cytometry.
Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR) Analysis
Total RNA from 1 to 3 × 104 cells derived from each lineage was extracted by the CsCl gradient technique, and Moloney murine leukemia virus (Mo-MuLV) reverse transcribed according to the manufacturer’s instructions (Boehringer, Mannheim, Germany). RT-PCR was performed in a final volume of 50 μL in the presence of 2.5 U of Taq polymerase (Perkin-Elmer Cetus, Norwalk, CT) as described.23 The cDNAs were normalized by the GAPDH gene as described in Carè et al.29
To evaluate coreceptor mRNA expression, aliquots of cDNA were amplified within the linear range by 40 PCR cycles with primers specific for CXCR4, CCR5, and CCR3. PCR products were then electrophoresed on agarose gel, transferred onto nylon membranes, and hybridized with the specific probes. The following primers and probes were used: CXCR4 forward 5′CCTCTATGCTTTCCTTGG3′, reverse 5′CCTGAAGACTCAGACTCA3′ and probe 5′AGCAGAGGGTCCAGCCTCAAGAT CCTCT3′; CCR5 forward 5′TCACTTGGGTGGTGGCTGTG3′, reverse 5′CCTGCATAGCTTGGTCCAAC3′ and probe 5′GAAATGAGAAGAAGAGGCACAGGGCTGTGA3′; CCR3 forward 5′CCAGCTCTGTCTCTCCATCCA 3′, reverse 5′GGGTTGATGATGAGTACGCTG3′ and probe 5′GGTCAGATGCAGAAAATTGCCTA3′.
Virus Preparations and Infections
HIV-1 BaL virus preparations were obtained as described by Gartner et al30 and titrated in terms of both picograms per milliliter of HIV p24 protein (by quantitative enzyme-linked immunosorbent assay [ELISA]; Abbott, North Chicago, IL) and 50% tissue culture infectious dose per milliliter (TCID50/mL) by the end dilution method31 performed on ex vivo monocyte-macrophage cultures. Four to 7 × 104 cells/mL were incubated overnight in the presence of lineage-specific HGFs described above at 0.01 to 0.1 multiplicity of infection (m.o.i.) of BaL.
NL4-3 lymphotropic HIV-1 strain was prepared as described by Chelucci et al,23 with viral titer ranging from 3 × 106 to 107 TCID50/mL. Cell pellets (4 to 7 × 104 cells) were challenged with 0.01 to 0.1 m.o.i. of NL4-3 for 2 hours at 37°C. Complete lineage-specific medium was then added and the cells were incubated overnight at 37°C.
Infected cells were extensively washed, diluted at 4 × 104/mL, and grown in liquid suspension culture for 12 to 14 days as described above. Aliquots of supernatants were removed every 2 to 3 days and stored at −80°C until used. Cell concentration was adjusted with fresh medium and HGFs to 1 × 105cells/mL.
A CD4+ UT-7 human megakaryoblastic cell line, resistant to HIV infection,32 and a CEMss cell line and ex vivo monocytic macrophagic cells were used as negative and positive controls, respectively.
Chemokine-Mediated HIV Inhibition
Cell pellets of Mk and Mo precursors (day 5) were incubated with 1 μg/mL of SDF-1 or a cocktail of 500 ng/mL each of MIP-1α, MIP-1β, and RANTES for 2 hours at 37°C in a final volume of 30 to 50 μL. The cells were then challenged with T-lymphotropic or M-tropic HIV strains as described above. After extensive washes, cells were resuspended in medium containing specific HGFs and supplemented with chemokines at the same concentration used in the preincubation step. Complete medium was collected every 2 days and the cells resuspended in fresh medium at 1 × 105/mL. Detection of p24 antigen was performed by ELISA on serially diluted culture supernatants. Positive and negative controls were those described for virus infection.
RESULTS
Unilineage HPC Differentiation/Maturation in Liquid Suspension Culture
The main characteristics of these culture systems have been extensively reported25-28,33-35 and reviewed.36 The purified HPC population is characterized by 92.3% ± 1.0% (mean ± SEM from 10 separate experiments) CD34+ cells as evaluated by flow cytometry. The mean frequency of HPCs (BFU-E + CFU-MK + CFU-GM + CFU-GEMM) was 91.0% ± 1.45% (mean ± SEM from 10 separate experiments) as evaluated by clonogenic assay, thus in line with previous reports.25,27,28 36
Highly purified HPCs grown in liquid suspension culture supplemented with the specific growth factors (see Materials and Methods) undergo a gradual and homogeneous wave of differentiation specifically along the E, G, Mk, and Mo pathway. Fluorescence studies showed that the gradual decrease of CD34+ membrane marker observed in the first days of culture is followed by the progressive increasing expression of glycophorin A, CD15, CD61, and CD14 markers in E, G, Mk, and Mo cultures, respectively. At the end of the cultures, cytofluorimetric and morphological analyses showed the following: 97% erythroid cells in E cultures, greater than 98% granulopoietic cells in G cultures, 97% to 99% megakaryocytes in Mk culture, and 95% to 100% monocytes/macrophages in Mo culture. These results indicate a unilineage HPC proliferation, ie, growth of E-, G-, Mk-, Mo-committed HPCs in the corresponding culture, as previously reported26 33-36 (also see Figs 3 and 4).
Chemokine Receptor mRNA and Protein Expression in Quiescent HPCs and During E, G, Mk, and Mo Differentiation/Maturation
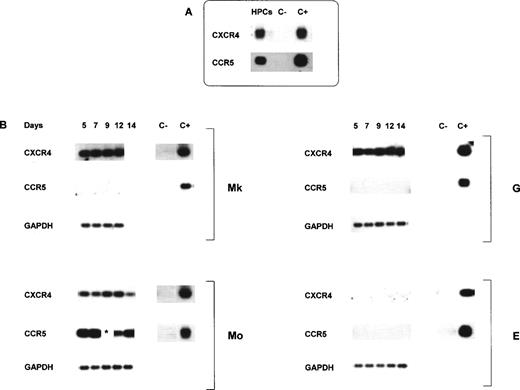
CXCR4, CCR5, and CCR3 mRNA expression was analyzed by RT-PCR on total cellular RNA on both quiescent HPCs and differentiating Mk, Mo, E, and G cells. Results showed that day 0 quiescent HPCs express significant levels of both CXCR4 and CCR5 (Fig 1A) and are negative for CCR3 mRNA (data not shown). Identical results were obtained on mRNA purified from CD34+ sorted HPCs (ie, >99.9% CD34+ cells), thus excluding positive hybridization signals from few contaminating cells present in purified HPCs (data not shown). Coreceptor expression was then examined in unilineage Mk, G, Mo, and E pathways: at different days of differentiation/maturation, total RNA was isolated and specifically amplified (Fig 1B). CXCR4 mRNA expression is sustained up to late stages of both Mk and G maturation, downmodulated at terminal days of Mo culture, and totally absent in E culture. CCR5 mRNA was constantly expressed from the early to late stages of Mo differentiation/maturation, and absent in Mk, G, and E lineages. CCR3 mRNA was not detected in any of the unilineage cultures (data not shown).
Representative results of CXCR4 and CCR5 mRNA expression analyzed by RT-PCR in quiescent HPCs after purification (A) and during in vitro megakaryocytic (Mk), monocytic (Mo), granulocytic (G), or erythroid (E) differentiation (B). In this experiment, HPCs were 97% CD34+ as evaluated by flow cytometry and 95% clonogenetic progenitors. Total RNA from CEM cell line (CXCR4) and monocytes (CCR5) and from UT-7 cell line was used as positive and negative controls, respectively. RT-PCR samples were normalized for GAPDH.
Representative results of CXCR4 and CCR5 mRNA expression analyzed by RT-PCR in quiescent HPCs after purification (A) and during in vitro megakaryocytic (Mk), monocytic (Mo), granulocytic (G), or erythroid (E) differentiation (B). In this experiment, HPCs were 97% CD34+ as evaluated by flow cytometry and 95% clonogenetic progenitors. Total RNA from CEM cell line (CXCR4) and monocytes (CCR5) and from UT-7 cell line was used as positive and negative controls, respectively. RT-PCR samples were normalized for GAPDH.
For CCR5 protein expression, 2 different MoAbs were used: 45502.111 clone that requires a demasking step to allow MoAb recognition, and 2D7 clone (see Materials and Methods). The latter MoAb was more suitable for double labeling of Mo precursor cells (see below). In fact, we observed that formaldehyde treatment used for 45502.111 clone labeling markedly reduced the CD14 MoAb recognition.
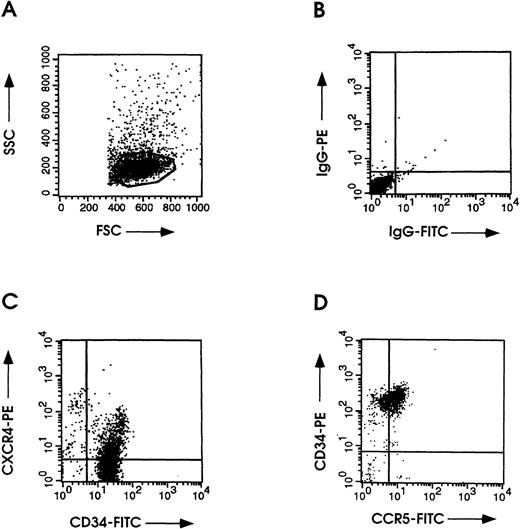
Double-fluorescence analyses using 12G5-PE CXCR4 or 45502.111-FITC CCR5 MoAb in combination with an MoAb specific for human CD34 showed surface expression of CXCR4 and CCR5 (Fig 2C and D). CXCR4 and CCR5 expression was found on ≈50% and ≈60%, respectively, of the CD34+ quiescent HPCs. Using the 2D7 CCR5 MoAb, less than 5% of CD34+/CCR5+ cells were found (not shown).
Flow cytometric analyses of CXCR4 and CCR5 on 97% CD34+ HPCs. (A) Unstained control for evaluation of HPC physical parameters. (B) HPCs stained with PE- or FITC-conjugated control antibodies. Cells were double-labeled with an FITC-conjugated anti-CD34 along with CXCR4-PE MoAb (C) or CD34-PE along with anti–CCR5-FITC MoAb (clone 45502.111) (D) as described in Materials and Methods.
Flow cytometric analyses of CXCR4 and CCR5 on 97% CD34+ HPCs. (A) Unstained control for evaluation of HPC physical parameters. (B) HPCs stained with PE- or FITC-conjugated control antibodies. Cells were double-labeled with an FITC-conjugated anti-CD34 along with CXCR4-PE MoAb (C) or CD34-PE along with anti–CCR5-FITC MoAb (clone 45502.111) (D) as described in Materials and Methods.
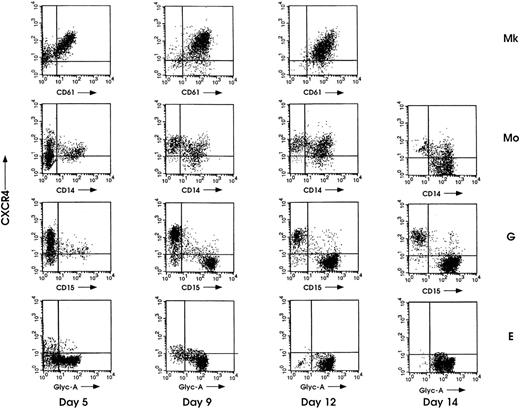
In Mk liquid differentiation culture, the fraction of CD61+/CXCR4+ cells rapidly increased starting from day 4-5, reaching more than 80% at terminal maturation (Fig 3). In the Mo lineage, a significant proportion (up to ≈80%) of CD14+/CXCR4+ cells was present until day 9-12 of maturation, and then decreased to ≈30% at day 14. Mo precursors showed a relatively broad doublet for CXCR4. We then investigated whether this phenomenon could be related to a different stage of maturation and/or to a heterogeneity in monocytic cell population. Thus, according to the intensity of CXCR4 single labeling, we sorted day 10 Mo precursors into CXCR4dim and CXCR4bright populations: morphological analysis showed that the former one was predominantly composed by mature monocytic cells (ie, 76% of mature monocytes, 24% of pro-monocytes), while the latter one mostly contained immature monocytic elements (ie, 35% mature monocytes and 65% pro-monocytes) (data not shown). This observation indicates that the different reactivity of anti-CXCR4 MoAb is related to monocytic maturation: immature Mo precursors exhibiting higher labeling with respect to mature Mo cells.
Flow cytometric analysis of CXCR4 along megakaryocytic (Mk), monocytic (Mo), granulocytic (G), and erythroid (E) HPC differentiation. Cells were collected for analysis at the indicated days of culture and double-labeled with PE-conjugated anti-CXCR4 MoAb along with the specific cell-lineage FITC-conjugated MoAbs (-CD61, -CD14, -CD15, -Glyc-A). Representative results from 1 of 3 independent experiments are shown.
Flow cytometric analysis of CXCR4 along megakaryocytic (Mk), monocytic (Mo), granulocytic (G), and erythroid (E) HPC differentiation. Cells were collected for analysis at the indicated days of culture and double-labeled with PE-conjugated anti-CXCR4 MoAb along with the specific cell-lineage FITC-conjugated MoAbs (-CD61, -CD14, -CD15, -Glyc-A). Representative results from 1 of 3 independent experiments are shown.
At day 5 of G differentiation, CXCR4+ cells represented ≈80% of the total population and decreased to 10% to 20% at the end of the culture (day 14). Double-immunofluorescence analysis with CXCR4 MoAb and CD15 MoAb that recognizes neutrophilic granulocytes showed 2 distinct cell populations, CXCR4+/CD15− and CXCR4−/CD15+, whose percentage decreased and increased, respectively, during maturation (Fig 3). These 2 cell populations were separated by sorting at day 12 of the culture, cytocentrifuged onto glass slides, and identified by morphology analysis. Most of the CXCR4−/CD15+ cells were neutrophilic granulocytes (60% mature band-neutrophils and 40% neutrophilic metamyelocytes), whereas the large majority (≈90%) of the CXCR4+/CD15− elements were eosinophils and the remaining cells immature myelocytes (≈10%) (data not shown). This result showed that the acquisition of the CD15 marker by maturing neutrophilic cells is coupled with the loss of the CXCR4 receptor, which is expressed on almost all mature eosinophils. Maturing E cells were negative for CXCR4 receptor at all differentiation/maturation stages (Fig 3).
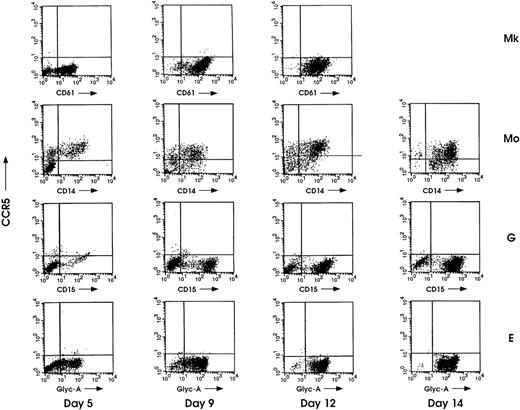
The same unilineage populations were analyzed for the expression of CCR5 receptor together with the lineage-specific membrane markers (Fig 4). Among the 4 lineages, only differentiating monocytes expressed the HIV CCR5 coreceptor; particularly, the CCR5+/CD14+ fraction progressively increased during the monocytic maturative process and reached ≈70% at the end of the culture. Moreover, labeling of maturing Mo cells by using the 45502.111 CCR5 MoAb revealed the same percentage of CCR5+ cells compared with those obtained in double-labeling experiments using 2D7 clone (data not shown).
Flow cytometric analysis of CCR5 during megakaryocytic (Mk), monocytic (Mo), granulocytic (G), and erythroid (E) differentiation of HPCs. Cells were collected for analysis at the indicated days of culture and double-labeled with PE-conjugated anti-CCR5 MoAb (clone 2D7) along with the specific FITC-conjugated cell lineage MoAbs. Representative results from 1 of 3 independent experiments are shown.
Flow cytometric analysis of CCR5 during megakaryocytic (Mk), monocytic (Mo), granulocytic (G), and erythroid (E) differentiation of HPCs. Cells were collected for analysis at the indicated days of culture and double-labeled with PE-conjugated anti-CCR5 MoAb (clone 2D7) along with the specific FITC-conjugated cell lineage MoAbs. Representative results from 1 of 3 independent experiments are shown.
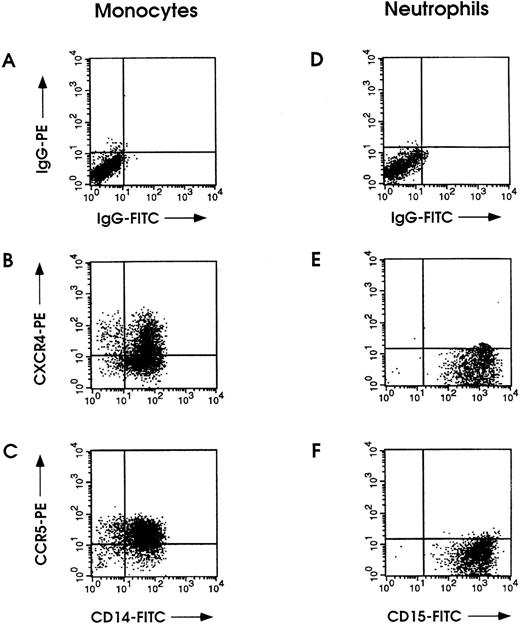
Control experiments performed on freshly isolated monocytes and neutrophils from PB showed a pattern of CXCR4 and CCR5 expression similar to that observed in Mo and neutrophilic cells grown in vitro (Fig 5).
Immunofluorescence labeling of CXCR4 and CCR5 in normal monocytes and neutrophilic granulocytes isolated from PB. (A and D) Staining using matched control antibodies. Monocytes were stained with either PE-conjugated CXCR4 (B) or CCR5 (C) MoAb along with CD14-FITC MoAb; neutrophils were labeled with either PE-conjugated CXCR4 (E) or CCR5 (F) MoAb along with CD15-FITC MoAb.
Immunofluorescence labeling of CXCR4 and CCR5 in normal monocytes and neutrophilic granulocytes isolated from PB. (A and D) Staining using matched control antibodies. Monocytes were stained with either PE-conjugated CXCR4 (B) or CCR5 (C) MoAb along with CD14-FITC MoAb; neutrophils were labeled with either PE-conjugated CXCR4 (E) or CCR5 (F) MoAb along with CD15-FITC MoAb.
CD4 Protein Expression
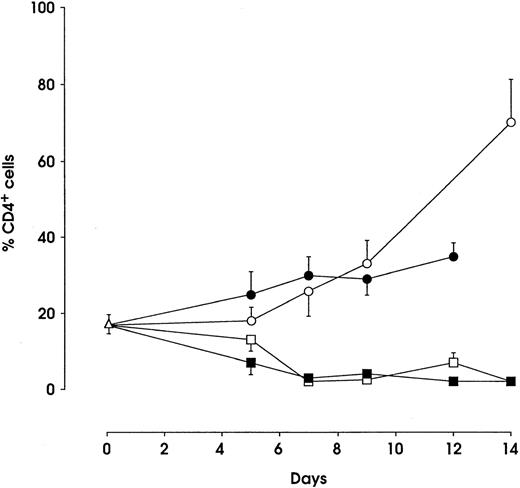
We have previously shown that a consistent fraction (≈15%) of HPCs are CD4+.23 In the Mo lineage, CD4+cells represent 15% of the total population during early differentiation (day 2-3) and reach greater than 80% at day 14-16 of culture when the cells are fully differentiated. In both E and G cultures, CD4 rapidly decreases, becoming barely detectable at the final days of culture. In the Mk lineage, CD4+ cells increased from ≈15% at day 0 to 30% at day 12 of culture as previously described24 (and Fig6).
Percentage of CD4+ cells during HPC (▵), monocytic (○), megakaryocytic (•), granulocytic (□), and erythroid (▪) differentiation (mean ± SEM values from 4 separate experiments). Cells from unilineage culture, collected at the indicated days, were labeled with PE-conjugated Leu3A MoAb and analyzed by FACS for fluorescence intensity.
Percentage of CD4+ cells during HPC (▵), monocytic (○), megakaryocytic (•), granulocytic (□), and erythroid (▪) differentiation (mean ± SEM values from 4 separate experiments). Cells from unilineage culture, collected at the indicated days, were labeled with PE-conjugated Leu3A MoAb and analyzed by FACS for fluorescence intensity.
T- and M-Tropic HIV Challenge of HPC and Mo, G, E, Mk Progenies: CXCR4 and CCR5 Natural Ligands Inhibit HIV Infection
HPCs were challenged at day 0 with NL4-3 (T-tropic) or BaL (M-tropic) viral strains as described in Materials and Methods. At day 1, after extensive washing the treated cells were resuspended at 1 × 105 cells/mL in Mk-, Mo-, G-, or E-specific media. Infection of HPCs was evaluated by measuring the p24 level in supernatants collected every 2 to 3 days from each unilineage differentiating culture starting from day 5 after infection. Both HIV strains were able to infect the CD34+ HPCs; in particular, challenge with T-tropic virus resulted in HIV replication in HPCs committed to Mk differentiation and with M-tropic HIV in Mo cultures. Finally, no viral replication was observed in HPCs committed to G or E differentiation (data not shown).
We then evaluated the infection susceptibility of hematopoietic precursors by challenging day 5 HPCs differentiating cells along the Mk, Mo, G, and E pathways with either BaL or NL4-3 HIV strains. Our results showed the following: (1) Mk precursors do not replicate M-tropic virus (data not shown) whereas, as already reported,24 they are fully permissive for T-tropic HIV replication (Fig 7A and B); (2) no p24 release was detected in supernatants from G and E cultures infected with either strain (data not shown); (3) Mo precursors were permissive to infection with M-tropic (BaL) and, to a lesser extent, with T-tropic (NL4-3) HIV strains (Fig 8A and B).
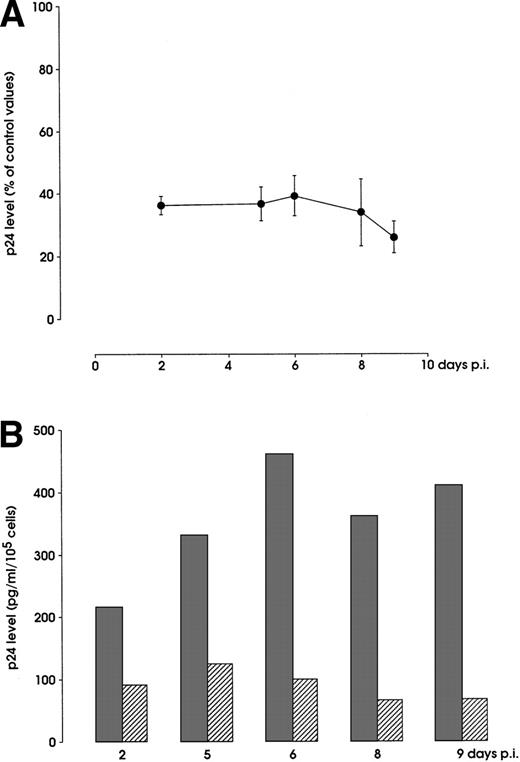
Inhibition of T-tropic HIV infection of Mk precursors by SDF-1. Day 5 Mk culture cells were treated or not with 1 μg/mL SDF-1 and then inoculated with 0.1 m.o.i of NL4-3. HIV p24 production was measured through 9 days postinfection (p.i.) as described in Materials and Methods. (A) Percentage of p24 level decrease in SDF-1 treated from untreated (control values) Mks (mean ± SEM values from 3 independent experiments). (B) Infection inhibition results from a representative experiment. (▩), NL4-3; (▨), NL4-3 + SDF-1.
Inhibition of T-tropic HIV infection of Mk precursors by SDF-1. Day 5 Mk culture cells were treated or not with 1 μg/mL SDF-1 and then inoculated with 0.1 m.o.i of NL4-3. HIV p24 production was measured through 9 days postinfection (p.i.) as described in Materials and Methods. (A) Percentage of p24 level decrease in SDF-1 treated from untreated (control values) Mks (mean ± SEM values from 3 independent experiments). (B) Infection inhibition results from a representative experiment. (▩), NL4-3; (▨), NL4-3 + SDF-1.
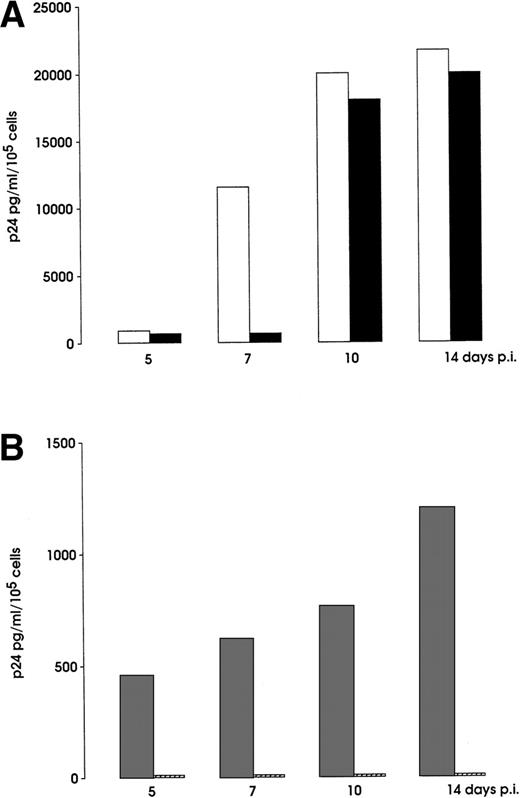
Inhibition of M-tropic (A) and T-tropic (B) HIV infection of day 5 Mo precursors. Cells were treated or not with a cocktail of MIP-1, MIP-1β, and RANTES or SDF-1, and then inoculated with either BaL or NL4-3 HIV strains, respectively (see Materials and Methods). HIV p24 production was measured for 14 days postinfection (p.i.). Representative results from 1 of 3 independent experiments are shown. (□), BaL; (▪), BaL + MIP-1, MIP-1β, RANTES; (▩), NL4-3; (▨), NL4-3 + SDF-1.
Inhibition of M-tropic (A) and T-tropic (B) HIV infection of day 5 Mo precursors. Cells were treated or not with a cocktail of MIP-1, MIP-1β, and RANTES or SDF-1, and then inoculated with either BaL or NL4-3 HIV strains, respectively (see Materials and Methods). HIV p24 production was measured for 14 days postinfection (p.i.). Representative results from 1 of 3 independent experiments are shown. (□), BaL; (▪), BaL + MIP-1, MIP-1β, RANTES; (▩), NL4-3; (▨), NL4-3 + SDF-1.
The role of CXCR4 and CCR5 in HIV infection was evaluated by blocking experiments on the hematopoietic precursors susceptible to infection with T-tropic (Mo and Mk) or M-tropic (Mo) HIV strains. At day 5 of HPC differentiation, precursor cells were incubated with the specific chemokines, SDF-1 or a cocktail of MIP-1α, MIP-1β, and RANTES, and then infected with NL4-3 and BaL HIV isolates, respectively. The results shown in Fig 7 demonstrate a significant level of inhibition of HIV spread on NL4-3–infected Mks similar to that previously obtained by pretreating Mk precursors with anti-CD4 antibodies.24 Furthermore, a total block of viral spread was observed in NL4-3–challenged Mo cells up to 14 days after infection (Fig 8B). Finally, treatment of Mo precursors with the MIP-1α, MIP-1β, RANTES cocktail inhibited the release of HIV at early culture times (day 5-9), while at later days viral release overcame the chemokine-induced antiviral effect (Fig 8A).
DISCUSSION
We previously showed in vitro infection of HPCs with T-tropic HIV strains by testing single HPC-derived colonies for HIV mRNA expression and/or p24 production.23 These results were supported by the finding that T-tropic strains also replicate in HPCs induced to Mk differentiation in liquid suspension culture.24 HPC and Mk susceptibility to HIV infection was attributed to detectable levels of the major HIV receptor, CD4, expressed on CD34+cells23 and Mk precursors.24 However, the recently demonstrated role of chemokine coreceptors in HIV entry and replication argues against an exclusive correlation between HIV susceptibility and CD4 receptor expression.
In the present study we evaluated the expression and role of the most commonly used HIV coreceptors, CXCR4, CCR5, and CCR3, in HPCs and precursor cells, particularly at discrete stages of HPC differentiation/maturation along the E, G, Mk, and Mo lineages.
Consistent with mRNA expression, cytofluorimetric analyses showed that ≈50% of CD34+ cells were CXCR4+. This finding is in line with our previous studies23,24 on the susceptibility to T-lymphotropic NL4-3 virus of day 0 HPCs. RT-PCR on purified CD34+ HPCs revealed the presence of CCR5 transcripts, in line with the cytofluorimetric results using the 45502.111 clone (≈60% of CCR5+ cells). A markedly lower CD34+/CCR5+ cell fraction (<5%) was found using the 2D7 clone. Because both anti-CCR5 MoAbs give a virtually identical labeling pattern on Mo precursors (Fig 4 and data not shown), the discrepancy found in CCR5 labeling of CD34+ cells may be related to a differential epitope conformation of CCR5 molecule expressed on HPCs. Along these lines, controversial data on CCR5 protein expression by CD34+ cells have been reported. Ruiz et al19 showed the presence of CCR5 protein on PB-derived CD34+ cells, whereas other investigators15 17did not detect CCR5 protein on most BM HPCs.
Productive infection by M-tropic BaL strain was observed only when challenged HPCs were induced to differentiate along the Mo lineage. This finding is not surprising considering that, differently than Mk, E, and G, Mo precursors exhibited a sustained CCR5 expression during the whole maturative process.
The study of the chemokine coreceptor pattern through Mo, Mk, G, or E, unilineage culture showed that CXCR4 mRNA and protein are significantly expressed in all the series, with the exception of E lineage. In the Mk culture, we observed a high and almost constant expression during all differentiation/maturation stages, while in the Mo pathway a downmodulation of CXCR4 is observed at both mRNA and protein level at the late stage of maturation. In the G lineage, progressive decrease of CXCR4 protein during maturation was shown coupled with the acquisition of CD15 marker by maturing neutrophilic granulocytes. This finding is in line with a recent study showing that circulating neutrophils are CXCR4−.37 Interestingly, in late G culture we observed that the minority of cells exhibiting the CXCR4+/CD15− phenotype were predominantly eosinophils, suggesting a possible role for SDF-1, the natural ligand of CXCR4, in the migration/chemotaxis of these cells. Finally, we showed that the expression of CCR5 is restricted to Mo lineage from early to late stages of differentiation-maturation, in agreement with the well-defined functions of CCR5 natural ligands (ie, MIP-1α, MIP-1β, and RANTES), involved mostly in the recruitment and activation of monocytic-macrophagic cells.1,3 38
The present data on HIV receptor and coreceptor expression correlate with the precursor infection susceptibility, in that (1) resistance of G and E cells to both HIV strains may be related to the virtual lack of CD4 expression; (2) Mk cells expressing CD4 and CXCR4 molecules were able to replicate T-tropic virus, but were resistant to M-tropic HIV replication; (3) Mo precursor cells expressing both CD4 and CCR5/CXCR4 receptors were permissive to replication of both HIV strains. Susceptibility to T-tropic HIV by Mo precursors is intriguing, considering the numerous initial reports on monocytic infectability restricted to M-tropic HIV stains. In this study, we infected an Mo precursor population which, at this stage of differentiation (day 5), is composed of 68% monoblasts, 27% promonocytes, 2% mature monocytes, and few contaminating (2%), but not infectable, granulocytic precursors (our unpublished results, 1994). Thus, our results are not comparable with those regarding fully mature PB-derived monocytic/macrophagic cells. Recent studies indicate that monocytes express CXCR4 and may be entered by T-lymphotropic HIV strains.39,40 However, as a consequence of an intracellular block, macrophage-derived monocytes lack the ability to productively replicate the virus.40 Thus, the macrophagic terminal differentiation may lead to the loss of intracellular factor(s) critical for the full replication of T-tropic HIV that, conversely, may be present in differentiating precursor cells. The reduced susceptibility of macrophages to infection by T-tropic HIV strains may also be related to the marked decline of CXCR4 expression observed during monocyte to macrophage maturation.41 Furthermore it has been reported that the T-tropic HIV-1MN strain can replicate in CD34+cell-derived monocytes, even at high m.o.i.,42 and a recent study showed that BM-derived Mo progenitors and precursors may be infected in vitro by both M- and T-tropic HIV strains.18Taken together, these observations suggest that cells pertaining to the monocytic lineage are infectable by both T- and M-tropic HIVs, from precursors up to the monocyte stage, while at terminal maturation (macrophages) they are only infectable by M-tropic viruses.
The role of chemokine receptors in HIV infection of Mo and Mk precursors was analyzed by inhibition experiments that consisted of treating cells with saturating amounts of either CXCR4 or CCR5 ligand, ie, SDF-1 (T-tropic HIV infection), or a cocktail of MIP-1α, MIP-1β, and RANTES (M-tropic infection), respectively. The significant reduction of viral spread observed in both cases strongly suggests that HIV entry in Mk and Mo precursors is largely mediated by the CD4/CXCR4 or CD4/CCR5 receptors, respectively. However, in the case of Mk precursors infected with T-tropic strain, the inhibition of replication did not appear complete, suggesting the existence of additional mechanisms of viral entry. This observation is in line with our previous study showing a similar partial inhibition in Mk precursors treated with anti-CD4 MoAb.24 Similarly, even large amounts of the MIP-1α, MIP-1β, and RANTES cocktail only delay the spread of M-tropic HIV in Mo cells. Consistent with this finding, an incomplete or ineffective block of HIV replication by the CCR5 ligands on monocytes-macrophages has been described.11 43Altogether, these data suggest the intrinsic ability of HIV to use alternative routes of entry (ie, endocytosis, phagocytic mechanisms) and/or different coreceptors to infect both Mk and Mo precursors.
In conclusion, our studies on the modulation of CXCR4 and CCR5 chemokine receptors along the 4 hematopoietic differentiation pathways shed light on the differential susceptibility of hematopoietic precursors to different HIV-1 strains and may represent a useful tool to investigate the role of these chemokines in the control of normal hematopoiesis.
ACKNOWLEDGMENT
The 12G5 MoAb was obtained through the AIDS Research and Reference Reagent Program, AIDS Program, National Institute of Allergy and Infective Diseases, National Institutes of Health, from Dr James Hoxie. We are grateful to Dr Ian Clark-Lewis (Biomedical Research Centre, University of British Columbia, Vancouver, Canada) for kindly providing SDF-1.
Supported by the AIDS Project (no. 10A/N) of the Ministry of Health, Rome, Italy.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to C. Peschle, MD, Thomas Jefferson University, Kimmel Cancer Center, Bluemle Life Sciences Bldg, Room 902, 233 S 10th St, Philadelphia, PA 19107-5541.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal