Abstract
Tumor necrosis factor (TNF) is a pluripotent activator of inflammation by inducing a proinflammatory cytokine cascade. This phenomenon is mediated, in part, through inducible expression of the CXC chemokine, interleukin-8 (IL-8). In this study, we investigate the role of TNF-inducible reactive oxygen species (ROS) in IL-8 expression by “monocyte-like” U937 histiocytic lymphoma cells. TNF is a rapid activator of IL-8 gene expression by U937, producing a 50-fold induction of mRNA within 1 hour of treatment. In gene transfection assays, the effect of TNF requires the presence of an inducible nuclear factor-κB (NF-κB) (Rel A) binding site in the IL-8 promoter. TNF treatment induces a rapid translocation of the 65 kD transcriptional activator NF-κB subunit, Rel A, whose binding in the nucleus occurs before changes in intracellular ROS. Pretreatment (or up to 15 minutes posttreatment) relative to TNF with the antioxidant dimethyl sulfoxide (DMSO) (2% [vol/vol]) blocks 80% of NF-κB–dependent transcription. Surprisingly, however, DMSO has no effect on inducible Rel A binding. Similar selective effects on NF-κB transcription are seen with the unrelated antioxidants, N-acetylcysteine (NAC) and vitamin C. These data indicate that TNF induces a delayed ROS-dependent signalling pathway that is required for NF-κB transcriptional activation and is separable from that required for its nuclear translocation. Further definition of this pathway will yield new insights into inflammation initiated by TNF signalling.
THE PLURIPOTENT CYTOKINE tumor necrosis factor α (TNFα) has been shown to be an endogenous mediator of inflammation and cellular immune responses.1 Binding to ubiquitously expressed receptors, TNFα is capable of eliciting a wide spectrum of physiologic and cellular responses to acute endotoxemia, acute respiratory distress syndrome, and infection with protozoal, bacterial, and viral pathogens. In circulating blood, monocytes are important target cells of TNFα action where it induces parallel cellular (enhanced cytotoxic capacity) and genetic programs (enhanced secretion of additional inflammatory mediators, including interleukin-1 [IL-1], IL-8, and platelet activating factor) that allow acquisition of a phagocytic phenotype. In fact, monocytic cells represent the major source of TNFα-inducible IL-8 secretion in human blood.2
The actions of TNFα are mediated by single transmembrane spanning receptors lacking intrinsic kinase activity (reviewed in Smith et al3). Circulating and binding as a trimeric peptide, TNFα induces receptor trimerization. Trimerization, an event required for receptor activation, results in protein recruitment to the intracellular (cytoplasmic) domain of the receptor.4 These signal transducing proteins, termed TNF receptor death domain protein (TRADD), and TRADD-associated factors (TRAFs) apparently control the activity of intracellular serine-threonine kinase cascades and protease activation.4 In addition, it has been appreciated that TNF receptor activation also results in the generation of putative second messenger molecules including ceramide, 1,2-diacylglycerol, arachidonic acid, and reactive oxygen species (ROS).
IL-8 is an important paracrine mediator of inflammation of the CXC chemokine family that amplifies inflammatory signals by demargination, activation, and chemotaxis of polymorphonuclear leukocytes.5,6 Encoded by a highly inducible gene, TNFα is a potent inducer of IL-8 secretion in a variety of cell types through a transcriptional mechanism primarily regulated by nuclear factor-κB (NF-κB).2,7-11 NF-κB is a heterodimeric protein composed of the transactivating subunit (Rel A) associated with the DNA binding subunit (NF-κB1) sequestered in a latent cytoplasmic form by association with IκBα inhibitor. In response to cytokine stimulation, Rel A-IκBα dissociates and IκBα is proteolyzed, allowing the liberated cytoplasmic NF-κB to be translocated into the nucleus, where it binds to genomic targets and initiates transcription.12
ROS are ubiquitous highly diffusable and reactive molecules produced as a result of reduction of molecular oxygen, and include species such as hydrogen peroxide, superoxide anion, and hydroxyl radical.13 Recently, a role of ROS in cellular responses to growth factor signalling has been described for platelet-derived growth factor14 and basic fibroblast-derived growth factor.15 In these examples, hormone receptor-activated ROS were involved in proliferative and programmed cell death. The contribution of ROS as second messenger molecules in TNFα signalling is controversial. Although others have shown that extracellular oxidants (H2O2) and enzymatic oxidative stress-inducing systems are capable of activating IL-8 secretion,16 these systems may be artifactual because they may not reproduce hormone receptor-induced ROS production in magnitude, kinetics, or ROS concentrations in proper subcellular compartments.17
Here we investigate the role of ROS in mediating TNFα-inducible expression of IL-8 in the histiocytic lymphoma cell line U937. U937 cells share phenotypic (IgG receptor expression and inducible differentiation) and functional (inducible cytokine expression) features with normal monocytes.18,19 We show that TNFα is a potent and rapid inducer of IL-8 protein secretion and gene expression. This effect is, in part, through enhanced transcription mediated through a single inducible cis regulatory element that binds to the inducible NF-κB transcription factor. In parallel, TNFα induces ROS generation as measured by the specific fluorescent 2’,7’dichlorofluescein oxidation assay.20 Pretreatment of U937 cells with the antioxidant dimethyl sulfoxide (DMSO) blocks inducible ROS generation and NF-κB transcriptional activity. Surprisingly, the effect of DMSO occurs without altering either NF-κB nuclear abundance or DNA-binding activity. The unrelated antioxidants, vitamin C and N-acetylcysteine (NAC), also selectively inhibit NF-κB transcriptional activity without detectable effects on NF-κB binding. These data indicate that an antioxidant pathway is required for NF-κB transcriptional activity that is separate and independent of signals coupled to NF-κB nuclear translocation.
MATERIALS AND METHODS
Materials.
Recombinant human TNFα was obtained from Calbiochem (San Diego, CA). DMSO, NAC, vitamin C, and vitamin E were purchased from Sigma (St Louis, MO). N-[2-hydroxyethyl]piperazine-N’-[2-ethanesulfonic acid] (HEPES), sodium lauryl sulfate (SDS), sodium chloride, sodium citrate, and ethylenediaminetetraacetic Acid (EDTA) were from Fischer Scientific (Pittsburgh, PA). Dichlorofluorescein diacetate (DCF-DA) was obtained from Molecular Probes (Eugene, OR).
Cell culture and treatment.
U937 human histiocytoma lymphoma cells were obtained from the American Type Culture Collection (ATCC; Rockville, MD) and grown as a suspension in RPMI medium containing 10% (vol/vol) fetal bovine serum, 10 mmol/L glutamine, 100 IU/mL penicillin, and 100 μg/mL streptomycin (GIBCO, Gaithersburg, MD) in an atmosphere of 5% CO2, at 37°C. TNFα was added to the medium at a final concentration of 20 ng/mL, unless otherwise stated. For antioxidant treatments, cells were pretreated either with indicated concentrations of DMSO (1 hour before TNFα, unless otherwise indicated), or NAC (7.5 mmol/L, 2 hours before TNFα), or with vitamin C (4 mmol/L, 2 hours before TNFα).
Assesment of intracellular ROS generation.
U937 cells were preloaded with 5 μmol/L DCF-DA in fully supplied culture medium containing 20 mmol/L HEPES, pH 7.4 for 20 minutes at 37°C. The cells were washed by centrifugation at 200g and resuspended in serum-containing fresh medium buffered in 20 mmol/L HEPES pH 7.4. After stimulation with TNFα, the cells were placed in a Becton Dickinson FACScan flow cytometer (excitation 485 nm; emission 530 nm; Becton Dickinson, Franklin Lakes, NJ) to quantitate oxidation into fluorescent DCF (an indicator of the intracellular ROS production20). A minimum of 10,000 cells was analyzed and the results expressed as fluorescence mean ± standard deviation (SD) of n = 3 independent experiments.
IL-8 enzyme-linked immunosorbent assay (ELISA).
Immunoreactive IL-8 was quantitated in cell culture supernatants by a double-antibody ELISA kit using recombinant IL-8 as a standard (R&D Systems, Minneapolis, MN) following the manufacturer’s protocol. This assay has a sensitivity of detection of 200 pg/mL.
Northern blot analysis.
Total RNA was extracted from control, TNFα-treated, or antioxidant plus TNFα-treated cells by the RNAzol kit (Teltest, Friendswood, TX) and RNA abundance quantitated spectrophotometrically. Twenty micrograms of RNA was fractionated on a 1.2% agarose-formaldehyde gel and transferred to nylon-reinforced nitrocellulose membrane (MSI, Westboro, MA). The RNA was then hybridized using polymerase chain reaction (PCR)-generated body-labeled cDNA probe for IL-8,7 followed by an 18S rRNA-cDNA probe using previously reported conditions.21Blots were washed in 5% SDS, 1 × sodium chloride sodium citrate (SSC) buffer at 50°C 3 times, 15 minutes each, and quantitated by exposure to a Molecular Dynamics (Sunnyvale, CA) Phosphorimager cassette. After quantitation, the blots were exposed to a Kodak XAR5 film (Rochester, NY).
Plasmid construction and transient transfections.
5′ deletion constructs of the human IL-8 (hIL-8) promoter2‘ were produced using the PCR with -1498/+44 hIL-8/Luc reporter plasmid8 as a template and a downstream oligonucleotide hybridizing +86 to +557 of the luciferase (LUC) cDNA.22 Upstream primers were used to produce 5′ deletions at nucleotide –162, -132, -99, and -54 by incorporating a unique Bam H1 restriction site immediately upstream. The PCR products were restricted with Bam H1 and Hind III, gel purified, and subcloned into the poLUC reporter vector.22Site-directed mutagenesis of the NF-κB site in the context of -162/+44 hIL-8 were introduced using the technique of PCR “SOEING”8 with the mutagenic primers (mutations underlined): 5′-TTCATTATGTCAGATTAAATTAAACGATTT-3′ and 5′-TTGCAAATCGTTTAATTTAATCTGACAATA-3′. For the NF-IL–6 binding site-mutation, the primers 5′-GCCATCAGCTACGAGTCGTGGAATTTCCTCTGA-3′ and 5′-GAAATTCCACGACTCGTAGCTGATGGCCCATCC-3′ were used. For the activator protein (AP)-1 binding site mutation, the primers 5′-GAGTGTGATATCTCAGGTTTGCCCTGA-3′ and 5′-CAAACCTGAGATATCACACTTCCTA-3′ were used. Multimeric binding sites were constructed by ligation of 3 copies of the NF-κB 5′-GATCCATCAGCTACGAGTCGTGGAATTTCCTCTA-3′, AP-1 5′-GATCCGAGTGTGATGACTCAGGTTTGCCCTTTA-3′ and NF-IL–6 5′-GATCCATCAGTTGCAAATCGTTTAATTTCCTCTA-3′ DNA sequences (having previously annealed them to complementary overlapping oligonucleotides) upstream of the –54 hIL-8/LUC promoter. Plasmids for use in transfection were purified by ion exchange (Qiagen, Chatsworth, CA) and sequenced to verify authenticity.
Transient transfections were performed in 107logarithmically growing U937 cells using a mixture of 60 μg diethylaminoethyl-dextran with 45 μg of hIL-8/LUC reporter and 9 μg of SV40/alkaline phosphatase-internal control plasmid. After 20 minutes at room temperature, the cells were centrifuged (at 300xg), resuspended in fresh culture medium, and distributed into 9 60-mm plates, and returned to the incubator. Cells were treated 16 hours after transfection. Six hours after treatment, cells were harvested, cytoplasmic lysates prepared, and luciferase activity measured.8 As an internal control for transfection efficiency, alkaline phosphatase activity was measured in 50 μg cell lysate by the dephosphorylation of alkaline phosphatase substrate (Sigma) in DEA buffer (1 mol/L Diethanolamine, pH 9.85, 0.28 mol/L NaCl, 0.5 mol/L MgCl2). Fold induction of reporter activity (by treatment with TNFα) was calculated by division of the mean normalized luciferase activity from 3 treated cultures, by the mean normalized luciferase activity from 3 untreated cultures.
Electrophoretic mobility shift assays (EMSAs) and microaffinity purification.
Sucrose-cushion purified nuclear extracts (NE) of U937 cells were prepared using hypotonic/nonionic detergent lysis as described previously.7 23 After extraction, nuclear protein was normalized by protein assay and used to bind to duplex oligonucleotides corresponding to –96 to –69 bp of hIL-8 promoter shown below (underlines indicate site mutations that disrupt NF-κB binding).NF-κB: GATCCATCAGTTGCAAATCGTGGAATTTCCTCTA GTAGTCAACGTTTAGCACCTTAAAGGAGATCTAG NF-κB mut: GATCCATCAGTTGCAAATCGTTTAATTTAATCTA GTAGTCAACGTTTAGCAAATTAAATTAGATCTAG
EMSAs included 10 μg of nuclear protein, 1.5 μg of polydeoxyadenylic-thymidylic acid (dA/dT), and 30,000 cpm α32P-labeled double-stranded IL-8 probe. For competition 50-fold molar excess of unlabeled competitor was included in the initial binding reaction. Antibody interference assays were as described for the supershift.8
Microaffinity purification of proteins binding to NF-κB wild-type (WT) was performed using a 2-step biotinylated DNA-streptavidin capture assay.7 In this assay, duplex NF-κB WT oligonucleotides were chemically synthesized containing 5′ biotin (Bt) on a flexible linker (Genosys, The Woodlands, TX). Identical amounts of nuclear protein from control and hormone-stimulated extracts were incubated with 50 pmoles Bt-NF-κB WT DNA in the presence of 8 μg poly dA/dT (as nonspecific competitor) in 800 μL (final volume) of binding buffer (8% [vol/vol] glycerol, 5 mmol/L MgCl2, 1 mmol/L dithiothreitol [DTT], 60 mmol/L KCl, 1 mmol/L EDTA, 12 mmol/L HEPES, pH 7.8) at 4°C for 1 hour. A total of 100 μL of a 50% slurry of prewashed streptavidin-agarose beads was then added to the sample and incubated at 4°C for an additional 20 minutes with shaking. Pellets were washed twice with 500 μL binding buffer and then resuspended in 100 μL 1X sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) buffer for analysis by Western immunoblot.
Western immunoblot.
Proteins were fractionated by SDS-PAGE and transferred onto polyvinylene difluoride membranes as described.23,24Affinity-purified rabbit polyclonal antibodies to Rel A and IκBα were obtained commercially (Santa Cruz Biotechnology, Santa Cruz, CA). Mouse monoclonal antibody to β-Actin was from Sigma. Secondary detection was using horseradish peroxidase-coupled donkey antirabbit or goat antimouse antibody in the ECL enhanced chemiluminescence assay (Amersham Life Sciences Arlington Heights, IL) as described.8 23
Statistical analysis.
Data from experiments involving multiple samples subject to each treatment were analyzed by the Student Newman Keuls t-test for multiple pairwise comparisons.
RESULTS
Induction of the IL-8 gene expression by TNFα.
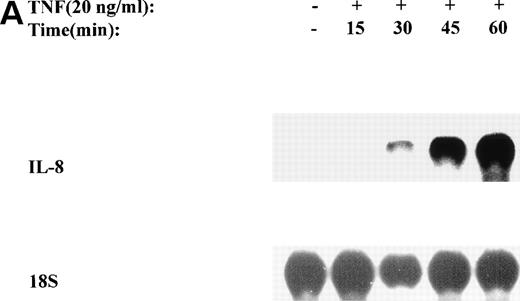
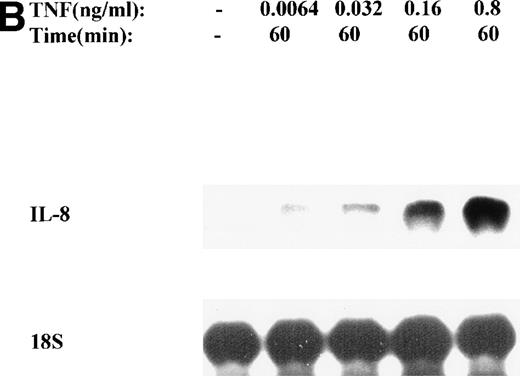
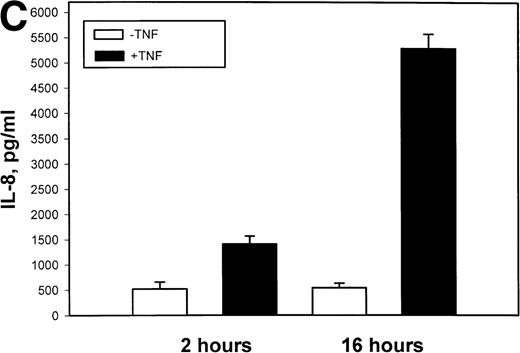
The mechanism for TNFα-induced IL-8 expression was studied in U937 histiocytic lymphoma cells. First, U937 cells were stimulated by maximal doses of TNFα and changes in IL-8 mRNA detected by Northern blot assay at various times of stimulation (Fig 1A). TNFα induced the rapid appearance of a 1.8-kb transcript, detectable at 10-fold (relative to control) with 30 minutes and peaked at a ≈50-fold induction between 60 to 180 minutes. At later times, IL-8 expression began to fall, indicating the effect of TNFα was rapid, but transient (not shown). The dose-response relationship of IL-8 mRNA was next investigated. At TNFα concentrations between 0.0064 ng/mL to 0.8 ng/mL, IL-8 gene expression was strongly induced in a dose-dependent manner (Fig 1B). Further increasing the dose of TNFα produced a flat dose-response curve (not shown), indicating receptor saturation. To determine whether IL-8 was secreted in parallel to the changes in IL-8 gene expression, IL-8 protein was assayed in U937 cell culture supernatants (Fig 1C). Although a 3-fold increase in IL-8 could be detected after 2 hours of TNFα, significant secretion of the protein was maximal at 16 hours. The delay relative to mRNA accumulation presumably reflects the time required for mRNA translation and protein secretion.
TNF inducible IL-8 expression in the U937 monocytic cell line. (A) TNF rapidly induces IL-8 mRNA abundance. U937 cells were untreated (control) or stimulated with 20 ng/mL TNF for the indicated times (in minutes, at top). Total RNA was extracted and analyzed by Northern analysis by hybridization with a hIL-8 cDNA probe (top) and an 18S cDNA probe (bottom) as an internal control. Shown is a representative Northern blot. Relative to control, IL-8 increases 10-fold (30 minutes), 37-fold (45 minutes), and 56-fold (60 minutes). This experiment was reproduced 2 times with similar results. (B) TNF induces a dose-dependent increase in IL-8 mRNA abundance. Cells unstimulated/stimulated with the indicated doses (in ng/mL) TNF for 1 hour were lysed and the total RNA subject to Northern analysis by hybridization with a hIL-8 cDNA probe (top) and an 18S cDNA probe (bottom) as an internal control. Relative to control, 0.0064 ng/mL TNF increased normalized IL-8 signal by 2-fold, 0.032 ng/mL TNF increased IL-8 signal by 7-fold, 0.16 ng/mL TNF increased IL-8 signal by 21-fold; 0.8 ng/mL increased IL-8 signal by 38-fold. This experiment was reproduced 2 times with similar results. (C) TNF induces a time-dependent increase in IL-8 protein secretion. Duplicate cultures were untreated (control) or stimulated with 20 ng/mL TNF. At indicated times, cell culture supernatants were harvested and immunoreactive IL-8 determined by ELISA. Shown is the mean ± SD of n = 2 independent experiments. TNF increases IL-8 secretion by 2.4-fold at 2 hours and 10-fold by 16 hours. Basal secretion is 500 pg/mL and unchanged any time of this experiment (P< .0001 for unstimulated v stimulated).
TNF inducible IL-8 expression in the U937 monocytic cell line. (A) TNF rapidly induces IL-8 mRNA abundance. U937 cells were untreated (control) or stimulated with 20 ng/mL TNF for the indicated times (in minutes, at top). Total RNA was extracted and analyzed by Northern analysis by hybridization with a hIL-8 cDNA probe (top) and an 18S cDNA probe (bottom) as an internal control. Shown is a representative Northern blot. Relative to control, IL-8 increases 10-fold (30 minutes), 37-fold (45 minutes), and 56-fold (60 minutes). This experiment was reproduced 2 times with similar results. (B) TNF induces a dose-dependent increase in IL-8 mRNA abundance. Cells unstimulated/stimulated with the indicated doses (in ng/mL) TNF for 1 hour were lysed and the total RNA subject to Northern analysis by hybridization with a hIL-8 cDNA probe (top) and an 18S cDNA probe (bottom) as an internal control. Relative to control, 0.0064 ng/mL TNF increased normalized IL-8 signal by 2-fold, 0.032 ng/mL TNF increased IL-8 signal by 7-fold, 0.16 ng/mL TNF increased IL-8 signal by 21-fold; 0.8 ng/mL increased IL-8 signal by 38-fold. This experiment was reproduced 2 times with similar results. (C) TNF induces a time-dependent increase in IL-8 protein secretion. Duplicate cultures were untreated (control) or stimulated with 20 ng/mL TNF. At indicated times, cell culture supernatants were harvested and immunoreactive IL-8 determined by ELISA. Shown is the mean ± SD of n = 2 independent experiments. TNF increases IL-8 secretion by 2.4-fold at 2 hours and 10-fold by 16 hours. Basal secretion is 500 pg/mL and unchanged any time of this experiment (P< .0001 for unstimulated v stimulated).
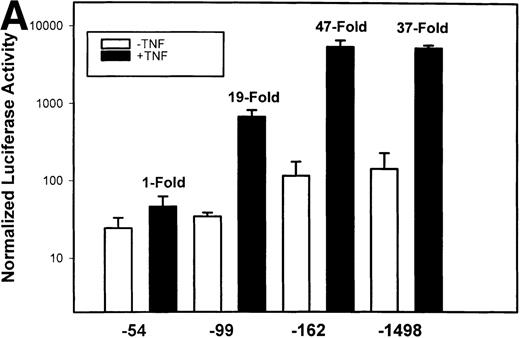
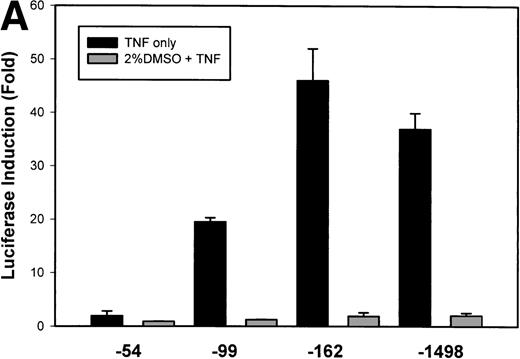
To test whether transcription was a component of IL-8 gene induction by TNFα, the reporter gene luciferase under the control of various lengths of IL-8 promoter7 was transfected into U937 cells. TNFα stimulation resulted in a strong induction of the native –1498 nucleotide (nt) IL-8 promoter-mediated luciferase activity (Fig 2A). 5′ deletion from –1498 to –162 nt resulted in no significant change in either basal or inducible luciferase activity. However, deletion from–162 to –99 nt reduced the basal and fold induction by approximately 2-fold (47-fold became 19-fold). Stimulation of –54 hIL-8/LUC showed it to be completely inert to TNFα. These data indicate that TNFα induction of the IL-8 promoter requires 2 domains, the first between –162 to –99 and the second between –99 to –54 for full inducibility.
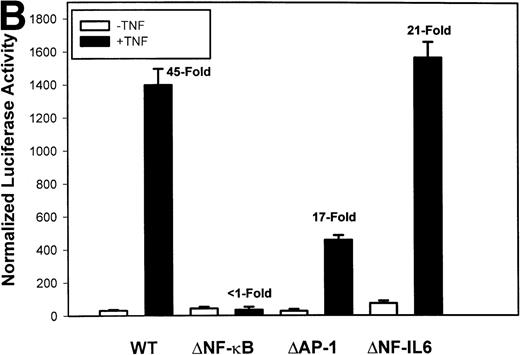
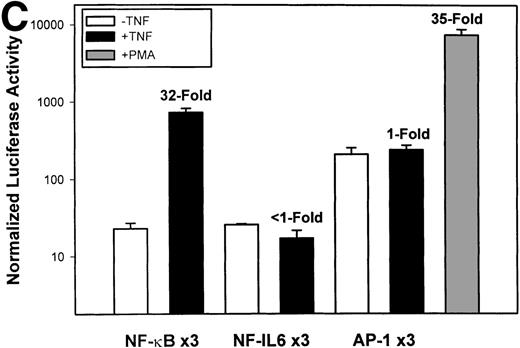
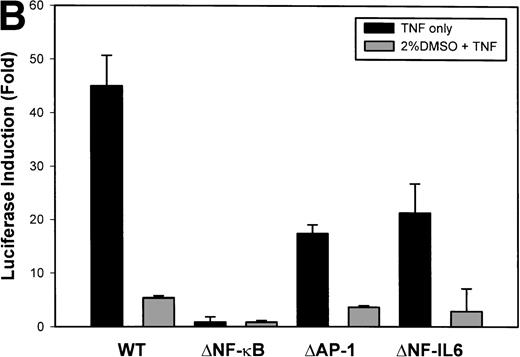
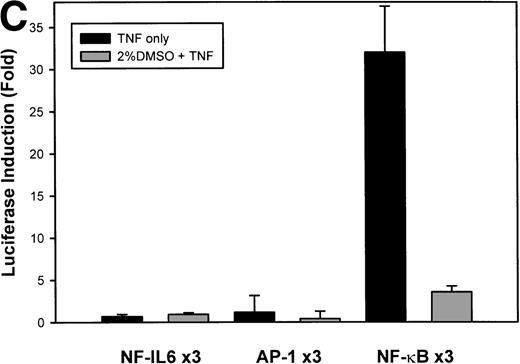
Identification of TNF-inducible IL-8 cis elements in U937 cells. (A) 5′-deletions of the human IL-8 promoter/Luciferase reporter (hIL-8/LUC) were transfected into U937 cells. Sixteen hours later, cells were stimulated with 20 ng/mL TNF for 6 hours before luciferase assay. Shown is the normalized Luciferase activity from a representative transfection plotted on a semilogarithmic graph. Above the TNF-stimulated bar is the fold activation of the Luciferase activity by TNF (fold: activity of stimulated divided by activity of unstimulated). (B) Site mutations of NF-κB, NF-IL6, and AP-1 sites in the context of the –162hIL-8/LUC were analyzed for their inducibility by TNF. Shown is the normalized Luciferase activity in a representative transfection (mean ± SD,P < .0001 for all comparisons of unstimulated vstimulated, except ▵NF-κB). (C) Multimers of NF-κB, NF-IL6, and AP-1 sites ligated upstream of an inert hIL-8 TATA box were analyzed for their inducibility by TNF. As a positive control, the AP-1 multimer was treated for 6 hours with 1 μmol/L phorbol myristyl acetate (PMA). Shown is the normalized Luciferase activity from a representative transfection. TNF stimulated the NF-κB multimer, and PMA stimulated the AP-1 multimer (P < .0001 for comparisons of unstimulated v stimulated).
Identification of TNF-inducible IL-8 cis elements in U937 cells. (A) 5′-deletions of the human IL-8 promoter/Luciferase reporter (hIL-8/LUC) were transfected into U937 cells. Sixteen hours later, cells were stimulated with 20 ng/mL TNF for 6 hours before luciferase assay. Shown is the normalized Luciferase activity from a representative transfection plotted on a semilogarithmic graph. Above the TNF-stimulated bar is the fold activation of the Luciferase activity by TNF (fold: activity of stimulated divided by activity of unstimulated). (B) Site mutations of NF-κB, NF-IL6, and AP-1 sites in the context of the –162hIL-8/LUC were analyzed for their inducibility by TNF. Shown is the normalized Luciferase activity in a representative transfection (mean ± SD,P < .0001 for all comparisons of unstimulated vstimulated, except ▵NF-κB). (C) Multimers of NF-κB, NF-IL6, and AP-1 sites ligated upstream of an inert hIL-8 TATA box were analyzed for their inducibility by TNF. As a positive control, the AP-1 multimer was treated for 6 hours with 1 μmol/L phorbol myristyl acetate (PMA). Shown is the normalized Luciferase activity from a representative transfection. TNF stimulated the NF-κB multimer, and PMA stimulated the AP-1 multimer (P < .0001 for comparisons of unstimulated v stimulated).
The IL-8 gene promoter contains 3 binding sites for known transcription factors: (1) AP-1, a protein binding between –127 and –119 nt; (2) NF-IL6, a protein that binds between –94 and –81 nt; and (3) NF-κB, a protein that binds between –80 and –70 nt.10 We next tested the individual role for each of these sites by producing site-directed mutations in the context of the –162 hIL-8 promoter. As shown in Fig 2B, mutation of the NF-κB site completely abolished TNFα-inducible transcription. Mutation of the AP-1 site reduced induction by ≈2-fold (from 45-fold to 20-fold), whereas mutation of the NF-IL6 site was silent. These data indicate that although the AP-1 site participates in basal and TNFα-inducible activity of the IL-8 promoter, only the NF-κB site is absolutely required. To determine whether these sites are independently TNFα-inducible, reporter genes containing multimers of either the AP-1, NF-κB, or NF-IL6 elements ligated upstream of an inert TATA box were transfected into U937 and stimulated with TNFα (Fig 2C). The NF-κB multimer was 32-fold inducible by TNF, whereas other sites were not significantly TNF-inducible. Although the AP-1 site was not TNFα-inducible, it was strongly induced by the diacylglycerol agonist, phorbol 12-myristate 13-acetate. We conclude that the NF-κB site is the only TNFα-inducible promoter element in the IL-8 promoter, whose presence is both necessary and sufficient for the TNFα transcription.
TNFα induces NF-κB binding and IκBα proteolysis.
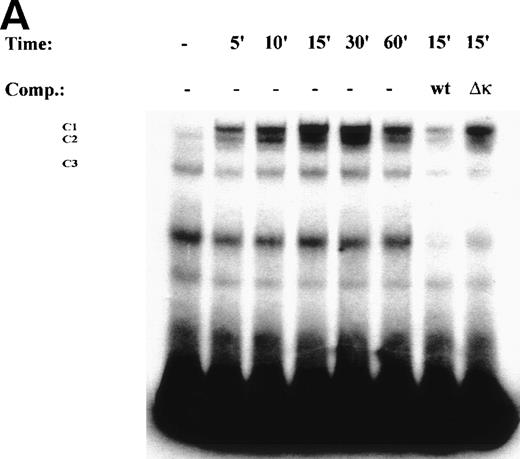
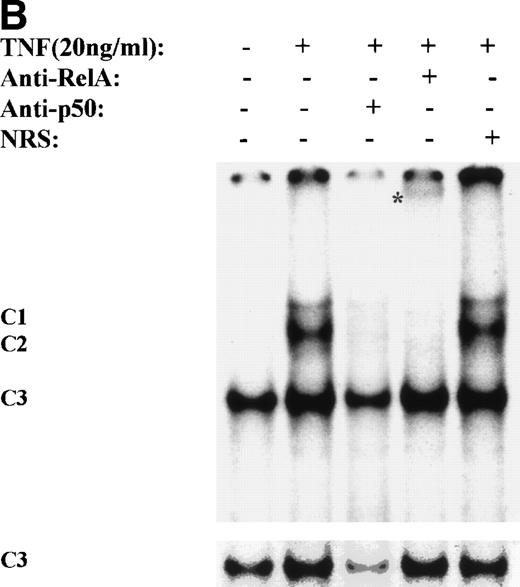
One mechanism for NF-κB activation is enhanced nuclear DNA binding of the Rel A transactivator subunit. To verify this mechanism, sucrose cushion-purified nuclear extracts from TNFα-stimulated U937 cells were assayed for NF-κB binding in EMSA (Fig 3A). In control extracts, a constitutive binding activity (C3) was observed. Within 5 minutes stimulation, 2 additional closely comigrating complexes (C1 and C2) were induced to bind that apparently peaked at 30 minutes. Sequence specificity of C1, 2, and 3 complexes is seen by ability of 50-fold molar excess of unlabeled wild-type, but not site mutation, of the NF-κB contact points23 to compete for their binding (Fig3A). Inducible complexes C1 and C2 contain the Rel A transactivating subunit as indicated by ability of Rel A antibody, but not preimmune sera, to selectively attenuate their binding (Fig 3B).
TNF induces NF-κB binding and IκB proteolysis. (A) TNF rapidly induces nuclear NF-κB Rel A DNA binding. U937 cells unstimulated or stimulated with 20 ng/mL TNF for the indicated times (at top). Cells were lysed, the nuclei were isolated, and subjected to EMSA analysis with a radiolabeled IL-8 NF-κB site. The bound complexes (C1-C3) are indicated. Unlabeled duplex wild-type (wt) or mutant NF-κB (▵κ) competitors were included where indicated. (B) Antibody interference. EMSA of TNF-stimulated nuclear extract was prepared. Either normal rabbit serum (NRS), anti-p50, or anti-Rel A antibodies were preincubated for 1 hour before the assay as indicated. Asterix is a faint supershifted band. C1 and C2 are completely attenuated by the Rel A antibody. Bottom: lighter exposure. C3, C2, and C1 are attenuated by the p50 antibody. (C) TNF induces time-dependent proteolysis of the IκB protein. Cells unstimulated/stimulated with 20 ng/mL TNF for the indicated times (top) were lysed and the cytosols were prepared. Top panel, Western immunoblot with anti-IκB antibody; bottom panel, Western immunoblot with an anti-β–actin antibody (internal control).
TNF induces NF-κB binding and IκB proteolysis. (A) TNF rapidly induces nuclear NF-κB Rel A DNA binding. U937 cells unstimulated or stimulated with 20 ng/mL TNF for the indicated times (at top). Cells were lysed, the nuclei were isolated, and subjected to EMSA analysis with a radiolabeled IL-8 NF-κB site. The bound complexes (C1-C3) are indicated. Unlabeled duplex wild-type (wt) or mutant NF-κB (▵κ) competitors were included where indicated. (B) Antibody interference. EMSA of TNF-stimulated nuclear extract was prepared. Either normal rabbit serum (NRS), anti-p50, or anti-Rel A antibodies were preincubated for 1 hour before the assay as indicated. Asterix is a faint supershifted band. C1 and C2 are completely attenuated by the Rel A antibody. Bottom: lighter exposure. C3, C2, and C1 are attenuated by the p50 antibody. (C) TNF induces time-dependent proteolysis of the IκB protein. Cells unstimulated/stimulated with 20 ng/mL TNF for the indicated times (top) were lysed and the cytosols were prepared. Top panel, Western immunoblot with anti-IκB antibody; bottom panel, Western immunoblot with an anti-β–actin antibody (internal control).
We and others have shown that Rel A is tethered in the cytoplasm by association with IκBα inhibitor, which must be degraded to release Rel A into the nucleus.23,25 26 Western immunoblot of cytoplasmic extracts from control and treated U937 cells was performed to determine changes in steady state IκBα protein (Fig 3C). TNFα induced a rapid proteolysis of IκBα at 5 minutes, followed by its reappearance (due to resynthesis) at 60 minutes. This data indicates NF-κB is activated by TNFα in a conventional pathway requiring IκBα proteolysis.
TNFα induces ROS formation.
The precise role of ROS in TNFα signaling is controversial.17 Because extracellular oxidants can activate NF-κB binding in some cell lines and high concentrations of antioxidants block TNFα–induced NF-κB activation, NF-κB is considered to be an ROS-responsive transcription factor.17,25,27,28 It is, however, unclear whether these pharmacological studies are relevant to hormone-induced cell signaling. To determine whether TNFα-induced ROS in U937, we monitored ROS formation by oxidation of DCF, a standard indicator of intracellular oxidation.20 TNFα stimulation induced a highly reproducible and significant change in DCF fluorescence, first detectable between 5 and 8 minutes (Fig 4). The plateau in ROS formation was transient, peaking at 2-fold increase in mean fluorescence intensity at 15 minutes, and declined thereafter, even in the continuous presence of hormone. Various concentrations of antioxidants were used in preliminary studies to identify the smallest concentrations that could suppress TNFα-induced DCF fluorescence (not shown). We found that 2% (vol/vol) DMSO, 4 mmol/L vitamin C, and 7.5 mmol/L NAC were sufficient to significantly block the inducible ROS formation in U937 cells (Fig 4).
TNF induces intracellular oxidation in U937. DCF-DA–loaded cell cultures were left untreated or stimulated with 20 ng/mL TNF in the absense or presence of pretreatment with antioxidant (2% [vol/vol] DMSO, 7.5 mmol/L NAC, or 4 mmol/L vitamin C; determined to be effective concentrations in preliminary experiments). Mean fluorescence intensity, each point representing 104 cells, is plotted as a function of time. The error bars represent SD from 3 independent experiments. Statistical analysis of the TNF-only stimulated cells (at 12, 15, and 18 minutes) versus unstimulated or versus TNF plus antioxidant, yields P < .0001.
TNF induces intracellular oxidation in U937. DCF-DA–loaded cell cultures were left untreated or stimulated with 20 ng/mL TNF in the absense or presence of pretreatment with antioxidant (2% [vol/vol] DMSO, 7.5 mmol/L NAC, or 4 mmol/L vitamin C; determined to be effective concentrations in preliminary experiments). Mean fluorescence intensity, each point representing 104 cells, is plotted as a function of time. The error bars represent SD from 3 independent experiments. Statistical analysis of the TNF-only stimulated cells (at 12, 15, and 18 minutes) versus unstimulated or versus TNF plus antioxidant, yields P < .0001.
Antioxidants block IL-8 gene expression.
Identification of the minimum effective dose of antioxidants enabled us to test whether TNFα stimulation of IL-8 gene expression is mediated by ROS. To determine whether antioxidants interfere with TNFα-induced IL-8 expression, IL-8 protein secretion from DMSO-pretreated cells was measured by ELISA. The presence of 2% DMSO significantly interfered with over 90% of inducible IL-8 secretion without affecting cell viability or cell number (Fig5A).
TNF stimulation of IL-8 is dependent on intracellular oxidation. (A) Inducible IL-8 protein secretion is sensitive to DMSO. Triplicate cultures of U937 cultures were untreated (control) or stimulated with 20 ng/mL TNF in the absence or presence of 2% (vol/vol) DMSO. At indicated times, cell culture supernatants were harvested and immunoreactive IL-8 determined by ELISA. Shown is the mean ± SD of n = 3 independent experiments (for both time points shown, P < .0001 for nonpretreated vpretreated with 2% (vol/vol) DMSO). (B) DMSO causes a dose-dependent inhibition of TNF-inducible IL-8 mRNA. Cells were unstimulated/stimulated with 20 ng/mL TNF (1 hour) in the absense/presence of DMSO of indicated concentrations (vol/vol). Cells were lysed and the total RNA subject to Northern analysis by hybridization with a hIL-8 cDNA probe (top) and an 18S cDNA probe (bottom) as an internal control. (C) DMSO causes a dose-dependent inhibition of TNF-inducible IL-8 promoter activity. Triplicate cell cultures were transfected with the –162 IL-8/LUC reporter plasmid and an SV40/alkaline phosphatase plasmid as an internal control. Cells were unstimulated/stimulated with 20 ng/mL TNF in the absence or presence of indicated concentrations of DMSO (in % [vol/vol]). Fold-induction of the Luciferase activity of stimulated (calculated from unstimulated) cells is shown (P < .0001 for stimulated v pretreated with 2% (vol/vol) DMSO before stimulation.
TNF stimulation of IL-8 is dependent on intracellular oxidation. (A) Inducible IL-8 protein secretion is sensitive to DMSO. Triplicate cultures of U937 cultures were untreated (control) or stimulated with 20 ng/mL TNF in the absence or presence of 2% (vol/vol) DMSO. At indicated times, cell culture supernatants were harvested and immunoreactive IL-8 determined by ELISA. Shown is the mean ± SD of n = 3 independent experiments (for both time points shown, P < .0001 for nonpretreated vpretreated with 2% (vol/vol) DMSO). (B) DMSO causes a dose-dependent inhibition of TNF-inducible IL-8 mRNA. Cells were unstimulated/stimulated with 20 ng/mL TNF (1 hour) in the absense/presence of DMSO of indicated concentrations (vol/vol). Cells were lysed and the total RNA subject to Northern analysis by hybridization with a hIL-8 cDNA probe (top) and an 18S cDNA probe (bottom) as an internal control. (C) DMSO causes a dose-dependent inhibition of TNF-inducible IL-8 promoter activity. Triplicate cell cultures were transfected with the –162 IL-8/LUC reporter plasmid and an SV40/alkaline phosphatase plasmid as an internal control. Cells were unstimulated/stimulated with 20 ng/mL TNF in the absence or presence of indicated concentrations of DMSO (in % [vol/vol]). Fold-induction of the Luciferase activity of stimulated (calculated from unstimulated) cells is shown (P < .0001 for stimulated v pretreated with 2% (vol/vol) DMSO before stimulation.
Northern blot analysis to assay changes in endogenous IL-8 expression was next performed in U937 cells stimulated in the presence of increasing concentrations of DMSO. Compared with TNFα alone, treatment with 0.4% DMSO inhibited IL-8 mRNA induction by 35% and treatment with 2% DMSO inhibited IL-8 induction by 85% (Fig 5B). This apparently was not a nonspecific effect because steady state levels of 18S RNA and total cell number were unchanged (not shown).
To determine whether the antioxidant effect influenced IL-8 gene expression at the transcriptional level, the same experiment was conducted in U937 cells transiently transfected with the –162 hIL-8/LUC reporter gene. A similar inhibition was seen; treatment with 0.4% DMSO inhibited TNFα-inducible IL-8 induction by 40%, and treatment with 2% DMSO inhibited IL-8 induction by 90% (Fig 5C). These data indicate the antioxidant effect occurs by interference of TNFα-inducible transcription.
Identification of antioxidant sensitive site on the IL-8 promoter.
The effect of DMSO on transcription could be through interference of the AP-1 or NF-κB activities. Transient transfections of hIL-8/LUC 5′ deletions were next tested to localize the DMSO effect. All of the 5′ deletions that were TNFα inducible, including the –99 hIL-8/LUC (that contains only the NF-κB site), were potently inhibited by DMSO (over 90%) and therefore antioxidant sensitive (Fig 6A). Site mutations of NF-κB, AP-1, and NF-IL6 were similarly tested (Fig 6B). All mutations containing the NF-κB site were inhibited more than 80%, while mutation at the NF-κB site abolished both inducibility by TNFα and sensitivity to DMSO. Finally, multimers of each element were tested (Fig 6C). No significant inhibition of reporter gene activity driven by NF-IL6 or AP-1 was seen; only the NF-κB site was inhibited by treatment with 2% DMSO (90% inhibition). Taken together, these data indicate antioxidant effect is predominantly mediated by interference with NF-κB transcriptional activity.
Antioxidant effect maps to the NF-κB element. (A) Serial 5′ deletions of the IL-8 promoter analyzed for their DMSO-sensitivity. Triplicate cell cultures were transfected with the indicated IL-8/LUC reporter plasmids and an SV40/alkaline phosphatase plasmid as an internal control. Cells were unstimulated/stimulated with 20 ng/mL TNF in the absence or presence of 2% (vol/vol) DMSO. Fold-induction of the Luciferase activity of stimulated cells is shown (P < .0001 for –99, -162, -1498: nonpretreated vpretreated with 2% (vol/vol) DMSO). (B) DMSO-sensitivity of IL-8 promoter point mutations. Triplicate cell cultures were transfected with the indicated IL-8/LUC reporter plasmids and an SV40/alkaline phosphatase plasmid as an internal control. Cells were unstimulated/stimulated with 20 ng/mL TNF in the absence/presence of 2% (vol/vol) DMSO. Fold-induction of the Luciferase activity of stimulated cells is shown (P < .0001 for nonpretreatedv DMSO-pretreated, except ▵NF-κB). (C) DMSO effects on IL-8 multimers. Multimers of NF-κB, NF-IL6, and AP-1 sites ligated upstream of an inert hIL-8 TATA box were analyzed for their DMSO sensitivity. Triplicate cell cultures were transfected with the indicated IL-8/LUC reporter plasmids and a SV40/alkaline phosphatase plasmid as an internal control. Cells were untreated or stimulated with 20 ng/mL TNF in the absence or presence of 2% (vol/vol) DMSO. Fold-induction of stimulated Luciferase activity is shown (for NF-κB multimer nonpretreated v DMSO-pretreated, P < .0001).
Antioxidant effect maps to the NF-κB element. (A) Serial 5′ deletions of the IL-8 promoter analyzed for their DMSO-sensitivity. Triplicate cell cultures were transfected with the indicated IL-8/LUC reporter plasmids and an SV40/alkaline phosphatase plasmid as an internal control. Cells were unstimulated/stimulated with 20 ng/mL TNF in the absence or presence of 2% (vol/vol) DMSO. Fold-induction of the Luciferase activity of stimulated cells is shown (P < .0001 for –99, -162, -1498: nonpretreated vpretreated with 2% (vol/vol) DMSO). (B) DMSO-sensitivity of IL-8 promoter point mutations. Triplicate cell cultures were transfected with the indicated IL-8/LUC reporter plasmids and an SV40/alkaline phosphatase plasmid as an internal control. Cells were unstimulated/stimulated with 20 ng/mL TNF in the absence/presence of 2% (vol/vol) DMSO. Fold-induction of the Luciferase activity of stimulated cells is shown (P < .0001 for nonpretreatedv DMSO-pretreated, except ▵NF-κB). (C) DMSO effects on IL-8 multimers. Multimers of NF-κB, NF-IL6, and AP-1 sites ligated upstream of an inert hIL-8 TATA box were analyzed for their DMSO sensitivity. Triplicate cell cultures were transfected with the indicated IL-8/LUC reporter plasmids and a SV40/alkaline phosphatase plasmid as an internal control. Cells were untreated or stimulated with 20 ng/mL TNF in the absence or presence of 2% (vol/vol) DMSO. Fold-induction of stimulated Luciferase activity is shown (for NF-κB multimer nonpretreated v DMSO-pretreated, P < .0001).
We tested whether TNFα-inducible proteolysis of IκBα is affected by antioxidant concentrations effective in our experiments. Western immunoblots were performed to measure changes in IκBα abundance in DMSO-pretreated cells at various doses of TNFα (Fig 7A). Surprisingly, IκBα was rapidly proteolyzed equivalently in the DMSO-pretreated cells. This indicated that DMSO effect was apparently not mediated by influencing NF-κB translocation. To further show this, sucrose cushion-purified nuclear extracts were assayed for steady state changes in Rel A by Western immunoblot assay (Fig 7B). Untreated nuclei contain very low levels of Rel A, whereas nuclear Rel A abundance is strongly induced after TNFα-treatment; these data indicate pretreatment with 2% DMSO does not change nuclear Rel A abundance.
DMSO effect is independent of NF-κB binding and translocation. (A) Two percent (vol/vol) DMSO pretreatment has no effect on IκB proteolysis. Cells were stimulated with the indicated concentrations of TNF, in the absence or presence of 1 hour pretreatment with 2% (vol/vol) DMSO. Top panel, Western immunoblot with anti-IκB antibody; bottom panel, Western immunoblot with an anti-β–actin antibody (internal control). (B) Two percent (vol/vol) DMSO pretreatment has no effect on Rel A nuclear translocation. Cells were stimulated with 20 ng/mL TNF, in the absence or presence of 1 hour pretreatment with 2% (vol/vol) DMSO. Nuclei were purified over sucrose cushion and tested by Western immunoblot with anti-Rel A antibody. Sixty-five kD Rel A is strongly induced by TNF in the absence or presence of DMSO. A nonspecific band serves here as an internal control for protein loading (control). (C) Effect of TNF on NF-κB binding in the presence of DMSO. Cells were stimulated with increasing concentrations of TNF (indicated at top), in the absence or presence of 1 hour pretreatment with 2% (vol/vol) DMSO. Shown is EMSA analysis of nuclear extracts for binding to radiolabeled NF-κB. C1/C2 binding increases proportionally with TNF dose. 0.032 ng/mL TNF yields a 7-fold weaker signal than 20 ng/mL (360,351 v 2,645,248 arbitrary units [a.u.]). In contrast, 2% (vol/vol) DMSO pretreatment does not reduce NF-κB binding (2,691,962 a.u.). For nuclear extracts, stimulated with 20 ng/mL TNF, various concentrations of protein were used to determine assay linearity with protein input (compare lanes 7, 6, and 4). (D) DMSO inhibits IL-8 promoter induction independently of TNF-induced changes in NF-κB Rel A DNA binding. Luciferase induction of the IL-8 promoter by the indicated amounts of TNF. Triplicate cell cultures were transfected with the –162 IL-8/LUC reporter plasmid and an SV40/alkaline phosphatase plasmid as an internal control. Cells were unstimulated/stimulated with the indicated doses (in ng/mL) TNF. The fold-induction of Luciferase activity of stimulated cells is shown (P < .005 for all pairwise comparisons). 0.032 ng/mL TNF activates the NF-κB–dependent promoter 7-fold weaker than 20 ng/mL. Two percent (vol/vol) DMSO pretreatment reduces TNF-inducible promoter activity by 8-fold. (E) Abundance of TNF-inducible NF-κB Rel A binding is not changed by pretreatment with promoter-inhibitory doses of DMSO. Cells unstimulated/stimulated with 20 ng/mL TNF for 15 minutes in the absence or presence of 2% (vol/vol) DMSO. After treatment, nuclear extracts were analyzed for NF-κB binding by microaffinity isolation. Shown is the Western immunoblot with rabbit antihuman Rel A polyclonal antibody (NS, nonspecific).
DMSO effect is independent of NF-κB binding and translocation. (A) Two percent (vol/vol) DMSO pretreatment has no effect on IκB proteolysis. Cells were stimulated with the indicated concentrations of TNF, in the absence or presence of 1 hour pretreatment with 2% (vol/vol) DMSO. Top panel, Western immunoblot with anti-IκB antibody; bottom panel, Western immunoblot with an anti-β–actin antibody (internal control). (B) Two percent (vol/vol) DMSO pretreatment has no effect on Rel A nuclear translocation. Cells were stimulated with 20 ng/mL TNF, in the absence or presence of 1 hour pretreatment with 2% (vol/vol) DMSO. Nuclei were purified over sucrose cushion and tested by Western immunoblot with anti-Rel A antibody. Sixty-five kD Rel A is strongly induced by TNF in the absence or presence of DMSO. A nonspecific band serves here as an internal control for protein loading (control). (C) Effect of TNF on NF-κB binding in the presence of DMSO. Cells were stimulated with increasing concentrations of TNF (indicated at top), in the absence or presence of 1 hour pretreatment with 2% (vol/vol) DMSO. Shown is EMSA analysis of nuclear extracts for binding to radiolabeled NF-κB. C1/C2 binding increases proportionally with TNF dose. 0.032 ng/mL TNF yields a 7-fold weaker signal than 20 ng/mL (360,351 v 2,645,248 arbitrary units [a.u.]). In contrast, 2% (vol/vol) DMSO pretreatment does not reduce NF-κB binding (2,691,962 a.u.). For nuclear extracts, stimulated with 20 ng/mL TNF, various concentrations of protein were used to determine assay linearity with protein input (compare lanes 7, 6, and 4). (D) DMSO inhibits IL-8 promoter induction independently of TNF-induced changes in NF-κB Rel A DNA binding. Luciferase induction of the IL-8 promoter by the indicated amounts of TNF. Triplicate cell cultures were transfected with the –162 IL-8/LUC reporter plasmid and an SV40/alkaline phosphatase plasmid as an internal control. Cells were unstimulated/stimulated with the indicated doses (in ng/mL) TNF. The fold-induction of Luciferase activity of stimulated cells is shown (P < .005 for all pairwise comparisons). 0.032 ng/mL TNF activates the NF-κB–dependent promoter 7-fold weaker than 20 ng/mL. Two percent (vol/vol) DMSO pretreatment reduces TNF-inducible promoter activity by 8-fold. (E) Abundance of TNF-inducible NF-κB Rel A binding is not changed by pretreatment with promoter-inhibitory doses of DMSO. Cells unstimulated/stimulated with 20 ng/mL TNF for 15 minutes in the absence or presence of 2% (vol/vol) DMSO. After treatment, nuclear extracts were analyzed for NF-κB binding by microaffinity isolation. Shown is the Western immunoblot with rabbit antihuman Rel A polyclonal antibody (NS, nonspecific).
Activation of NF-κB DNA binding activity has been reported to be antioxidant-sensitive in some cell systems.17,25,27 28 To determine whether DMSO treatment interfered with Rel A binding, sucrose cushion purified nuclear extracts were analyzed by EMSA (Fig 7C). Under EMSA conditions shown, a linear relationship was observed between TNFα dose (up to 20 ng/mL) and C1/C2 binding (cf, lanes 1 to 4). Also, a linear relationship of C1/C2 binding was observed as a function of input nuclear proteins (cf, lanes 7, 6, and 4). However, pretreatment with DMSO had no detectable influence on the magnitude of inducible NF-κB DNA binding.
These data indicate that DMSO influenced NF-κB–dependent transcription without influencing its DNA binding. To verify this surprising result, transfection studies were conducted to determine the transcriptional dose-response relationship (as for the DNA binding experiment in Fig 7C). As shown in Fig 7D, 2% DMSO inhibited the transcriptional induction of IL-8 by 90%, to a level produced by 0.032 ng/mL TNFα. However, the amount of NF-κB binding activity in the presence of 2% DMSO is not reduced accordingly (cf, Fig 7C), indicating the transcriptional inhibition is mechanistically separate from inhibition of DNA binding. To further exclude the potential possibility that DMSO interfered with selective recruitment of Rel A transactivator on the NF-κB site that might not be detected in EMSA, we performed a 2-step microaffinity isolation/Western immunoblot. In this assay, biotinylated NF-κB binding site is used to pull down NF-κB proteins that are subsequently detected by Western. We have previously shown that this assay detects NF-κB members in a sequence-specific fashion.7 As shown in Fig 7E, TNFα strongly induces 65-kD Rel A binding; the abundance of Rel A is not influenced by pretreatment with 2% DMSO. These data indicate the antioxidant DMSO selectively blocks TNFα-inducible NF-κB transcription without affecting IκBα proteolysis, Rel A translocation, or NF-κB binding activity.
Antioxidant inhibition of IL-8 expression occurs after IκBα proteolysis.
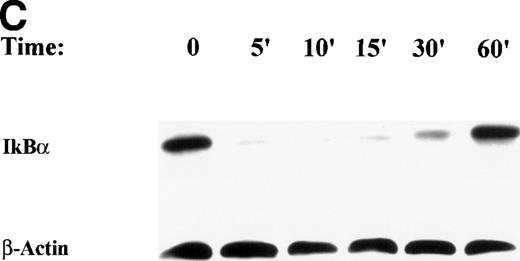
After TNFα treatment, IκBα proteolysis was complete within 5 minutes (Fig 3C), whereas ROS production was delayed 8 to 15 minutes (Fig 4). This suggests that temporally, the requirement for ROS production in NF-κB–activated IL-8 transcription may be after NF-κB translocates. If so, posttreatment with DMSO (relative to TNF stimulation) would still interfere with IL-8 gene expression. U937 cells were pre or posttreated with 2% DMSO relative to a 1-hour stimulation with TNFα. IL-8 gene expression was quantitated by Northern blot (Fig 8A). We found that delaying DMSO treatment up to 15 minutes after TNFα stimulation produced a similar, significant inhibition of IL-8 mRNA induction. Again, the DMSO effect was independent of IκBα proteolysis (Fig 8B) or changes in NF-κB DNA binding (Fig 8C). These data strongly argue that the antioxidant sensitive pathway is separate and distinct from NF-κB translocation.
Antioxidant effect at times subsequent to IκB proteolysis. (A) Effect of posttreatment on TNF-inducible IL-8 mRNA accumulation. Cells were unstimulated or stimulated with 20 ng/mL TNF in the absence or presence of 2% (vol/vol) DMSO. DMSO was administered at the indicated times (before [-] or after [+] TNF administration). (B) Posttreatment with 2% (vol/vol) DMSO has no effect on IκB proteolysis. Cells were unstimulated or stimulated with 20 ng/mL TNF in the absence or presence of 2% (vol/vol) DMSO. DMSO was administered at the indicated times (preceding [-] or succeeding [+] TNF administration). Top panel, Western immunoblot with anti-IκB antibody; bottom panel, Western immunoblot with an anti-β–actin antibody (internal control). (C) Pre or posttreatment with 2% (vol/vol) DMSO has no effect on Rel A DNA binding in EMSA. Cells were unstimulated/stimulated with 20 ng/mL TNF in the absence or presence of 2% (vol/vol) DMSO. DMSO was administered at the indicated times (before [-] or after [+] TNF administration). Nuclei were isolated and subjected to EMSA analysis with a radiolabeled NF-κB site. Only bound complexes are shown.
Antioxidant effect at times subsequent to IκB proteolysis. (A) Effect of posttreatment on TNF-inducible IL-8 mRNA accumulation. Cells were unstimulated or stimulated with 20 ng/mL TNF in the absence or presence of 2% (vol/vol) DMSO. DMSO was administered at the indicated times (before [-] or after [+] TNF administration). (B) Posttreatment with 2% (vol/vol) DMSO has no effect on IκB proteolysis. Cells were unstimulated or stimulated with 20 ng/mL TNF in the absence or presence of 2% (vol/vol) DMSO. DMSO was administered at the indicated times (preceding [-] or succeeding [+] TNF administration). Top panel, Western immunoblot with anti-IκB antibody; bottom panel, Western immunoblot with an anti-β–actin antibody (internal control). (C) Pre or posttreatment with 2% (vol/vol) DMSO has no effect on Rel A DNA binding in EMSA. Cells were unstimulated/stimulated with 20 ng/mL TNF in the absence or presence of 2% (vol/vol) DMSO. DMSO was administered at the indicated times (before [-] or after [+] TNF administration). Nuclei were isolated and subjected to EMSA analysis with a radiolabeled NF-κB site. Only bound complexes are shown.
Antioxidants NAC and vitamin C also selectively affect NF-κB transcription.
To exclude nonspecific effects of DMSO, other chemically unrelated antioxidants NAC and vitamin C were tested for ability to interfere with TNFα-inducible NF-κB transcriptional activity. Transient transfection assays using the NF-κB multimer /LUC reporter indicated that doses of antioxidants at concentrations that block ROS formation (7.5 mmol/L NAC, 4 mmol/L vitamin C, see Fig 4) similarly significantly block NF-κB–dependent transcription (60% by NAC, 85% by vitamin C). These antioxidant effects are independent from changes in NF-κB binding as measured in EMSA (not shown). Taken together, these data indicate the requirement of an ROS-dependent activation pathway of NF-κB that is distinct from the nuclear translocation pathway responsible for inducible DNA-binding.
DISCUSSION
In a variety of pathophysiological states initiated by infectious or inflammatory agents, TNFα secretion is responsible for activating the cytokine cascade required for appropriate cellular responses in the target tissue. An important cellular target of TNFα is the circulating monocyte, wherein TNFα induces a program of phenotypic changes required for phagocytosis and by inducing the secretion of other inflammatory mediators, allowing for the recruitment of neutrophils into the target tissue. Specifically, production of IL-8 allows for the activation, demargination, and chemotaxis of neutrophils into the inflamed tissue. This study provides additional mechanistic information to the observations of others where TNFα has been shown to induce IL-8 secretion in whole blood, the majority of which is derived from monocytes.2
We observe here that TNFα activates IL-8 expression through the participation of 2 regulatory factors. Although both AP-1 and NF-κB are required for maximal IL-8 gene expression, only NF-κB is truly TNFα inducible. In unstimulated cells, NF-κB is maintained in an inactive state in the cytoplasm through association with the IκB inhibitors. Dissociation of Rel A from IκB is a prerequisite for Rel A cytoplasmic-to-nuclear translocation.29,30 This is accomplished through a 2-step mechanism, where first the IκBα inhibitor is phosphorylated at serine residues 32 and 36 by the ubiquitous IκB kinase, IKK,31 and second the phospho-IκBα is polyubiquitinated and proteolyzed through the 26S proteasome.32 Our data shows that IκBα proteolysis is a consequence of TNFα stimulation in U937 cells and occurs coincidentally with Rel A translocation. The Rel A NF-κB subunit is the major TNFα-inducible transactivator of the IL-8 promoter in a variety of cell types, including epithelial,7fibrosarcoma,10 and histiocytic cells (this study). Consistent with these findings, in U937 cells, our antibody interference assays indicate that Rel A is largely responsible for the strongly inducible C1 and C2 complexes (Fig 3A), and microaffinity isolation/Western immunoblot assays indicate Rel A binding is strongly induced after TNFα treatment (Fig 7E).
Although Rel A translocation has been thought to be necessary and sufficient for transcriptional activation of IL-8, our observations show that pretreatment with antioxidants dissociates the 2 processes (of translocation and transcriptional activation). Our conclusions are based on the lack of antioxidant effect on inducible changes in steady state Rel A abundance in the nuclear compartment (by Western blot) and the measurement of Rel A binding in microaffinity capture and EMSA assays (Fig 7). It is important to highlight that changes in Rel A binding detected by EMSA correlate linearly with NF-κB transcription over the TNFα dose-response curve (Fig 7). In this experiment, 0.032 ng/mL TNFα activates the NF-κB–dependent promoter 7-fold weaker than 20 ng/mL and yields a corresponding 7-fold weaker signal in EMSA analysis. In sharp contrast, 2% (vol/vol)-DMSO pretreatment reduces TNFα-inducible promoter activity by 8-fold; this would be expected to reduce NF-κB binding by a similar extent (7-fold to 8-fold). However, because no such reduction in NF-κB is observed, we interpret the antioxidant effect is clearly independent of changes in Rel A DNA binding activity.
Our study indicates that ROS production is a necessary prerequisite for IL-8 production by TNFα through a requirement for the transcriptional function of NF-κB. TNFα induces ROS production in numerous independent assays, including depletion of antioxidant pools,33 elicitation of Electron Paramagnetic Resonance-detectable 2,2,6,6,-tetramethyl-1-piperidine-n-oxyl decay,34 by 5’,5’-dimethylpyrroline-N-oxide spin trapping,35 thiobarbituric acid-detectable lipid peroxidation,36 and DCF oxidation (this study). Importantly, our data indicates that TNFα induces IκBα proteolysis and ROS production in U937 cells with discrete kinetics. In these cells, IκBα proteolysis, Rel A translocation, and NF-κB binding occur unmeasurably rapidly (within 5 minutes); however, the kinetics of ROS production are delayed, being first detectable at 8 minutes, with a peak 15 minutes after stimulation. Although this apparent delay may be the consequence of the kinetics of DCF oxidation, this explanation is unlikely for 2 reasons. First, DCF oxidation indicates within seconds an immediate and steep increase in oxidant levels in response to H202-treatment (data not shown and Bass et al20), which shows that the oxidation lag in response to TNFα is not a detection artifact. Second, addition of antioxidant (up to) 15 minutes after TNFα administration still blocks inducible IL-8 transcription (Fig 8). These observations strongly argue that the ROS transcriptional activation pathway is distinct (temporally and mechanistically) from that involved in NF-κB translocation.
Although it is widely appreciated that the transcription factor, NF-κB, is activated by pharmacologic doses of oxidants,37the administration of extracellular oxidants may not faithfully reproduce the kinetics, magnitude, or subcellular compartmentalization of ROS produced as a consequence of hormone receptor activation. For example, addition of extracellular H202 to Jurkat T cells, HeLa cervical carcinoma, L6 skeletal muscle, and other cells is sufficient to induce NF-κB translocation.17,37However, the degree of intracellular oxidation required for NF-κB translocation by extracellular H202 is at least an order of magnitude more than that produced by TNFα (data not shown; Schmid et al38); these results, therefore, are of uncertain relevance to hormone-induced signaling.
Conversely, other studies have shown that antioxidants inhibit inducible NF-κB binding. In murine macrophage cells, 1% DMSO inhibits lipopolysaccharide (LPS)-induced NF-κB translocation.39 In another study, almost complete inhibition of TNFα-induced Rel A translocation was produced with 10 mmol/L NAC in synovial fibroblasts.40 Previously reported studies of antioxidant inhibition of stimulus-dependent NF-κB translocation have been conducted using 20 mmol/L NAC (or higher) and 100 μmol/L pyrrolidine dithiocarbamate.17,25 27 In these studies, antioxidant pretreatment dramatically decreased PMA-, TNFα-or H2O2-induced NF-κB binding in several cell types, including Jurkat T cells, Ltk-mouse fibroblasts, 70Z/3 mouse pre B cells. The antioxidant effect on NF-κB translocation, therefore, appears to be cell-type–dependent and stimulus-dependent.
As a specific example, NAC concentrations that inhibited TNFα-induced NF-κB binding in Jurkat T cells fail to inhibit it in endothelial cells.36 Also, although IL-1β caused ROS formation and antioxidant-sensitive NF-κB translocation in 70Z/3 lymphoid cells, in ovarian carcinoma (OVCAR-3) epithelial cells, IL-1β failed to cause detectable ROS formation and activates NF-κB translocation in antioxidant-insensitive manner.41These studies indicate the existence of ROS-dependent pathway(s) for NF-κB translocation are found in a cell-type restricted manner. Our observations indicate the presence of an antioxidant-sensitive signalling pathway in U937 cells; this pathway is sensitive to low doses of antioxidants that inhibit inducible, but not constitutive, ROS production and functions at a level independently of NF-κB translocation.
Along with this study, several lines of evidence are consistent with the existence of an independent NF-κB activating pathway. For example, translocation of NF-κB has been shown insufficient for IL-1β or TNFα-induced NF-κB–dependent transcription in airway epithelial cells.42 In that study, NF-κB–dependent transcription, but not translocation, was blocked by pretreatment with protein kinase inhibitors of the p38 and MAP kinases. However, the relationship of the MAP kinase cascade to ROS production was unexplored and will require further investigation. In another study, DMSO interfered with LPS-induced liver cytokine expression through a mechanism that could not be readily explained by the slight attenuation DMSO caused on NF-κB translocation.43 We speculate that the ROS signalling pathway could affect posttranslational modification of Rel A, the recruitment of coactivators to the IL-8 promoter, or the assembly of an NF-κB driven preinitiation complex.7 One inducible event that is clearly distinct from the nuclear translocation and DNA binding of Rel A is the phosphorylation of Rel A in response to TNFα,44 LPS,45 and PMA.46Intriguingly, LPS-induced phosphorylation of Rel A was completely blocked by an antioxidant.45 Further definition of this pathway will be required experimentally.
In U937, ROS may be important general second messenger signals for cytokine production. Treatment with the potent NF-κB activating agent, LPS, also increases intracellular ROS production.47At low concentrations, the antioxidants pyrrolidine dithiocarbonate (PDTC) and NAC completely blocked LPS-inducible ROS formation, without significant inhibition of NF-κB binding (less than 20%).47 The oxidant pathway may be mediating transcriptional activation in several receptor-mediated pathways that activate NF-κB. It will be interesting to compare the requirement of other NF-κB activating cytokines on ROS signalling.
Antioxidants have been shown effective in reducing IL-8 secretion and the severity of septic shock48 and airway inflammation49 in humans. Identification of the component of signal transduction that is sensitive to antioxidants will open the door to more selective treatment of inflammatory disorders without occurrence of side effects that would arise from the complete deactivation of the TNFα signaling cascade.
ACKNOWLEDGMENT
The authors thank D. Wang for the gift of 18S plasmid and the current and previous members of the Brasier Lab for valuable suggestions and ideas.
Supported in part by Grant No. 1 R01 55630 from the National Heart, Lung and Blood Institute (to A.R.B.), Grant No. 1 R01 AI40218 from the National Institute of Allergy and Infectious Diseases (to A.R.B.), and Grant No. ES06676 from the National Institute of Environmental Health Sciences (to R.S. Lloyd). A.R.B. is an Established Investigator of the American Heart Association.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Allan R. Brasier, MD, M.R.B. 8.138, University of Texas Medical Branch, 301 University Blvd, Galveston, TX 77555-1060; e-mail: arbrasie@utmb.edu.

![Fig. 4. TNF induces intracellular oxidation in U937. DCF-DA–loaded cell cultures were left untreated or stimulated with 20 ng/mL TNF in the absense or presence of pretreatment with antioxidant (2% [vol/vol] DMSO, 7.5 mmol/L NAC, or 4 mmol/L vitamin C; determined to be effective concentrations in preliminary experiments). Mean fluorescence intensity, each point representing 104 cells, is plotted as a function of time. The error bars represent SD from 3 independent experiments. Statistical analysis of the TNF-only stimulated cells (at 12, 15, and 18 minutes) versus unstimulated or versus TNF plus antioxidant, yields P < .0001.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/94/6/10.1182_blood.v94.6.1878/5/m_blod41803004x.jpeg?Expires=1769099902&Signature=CiI0CWBuuv5vz0YUDoCGfylLWjxAUuk976hFrf46CUmvp~jTlOBZlkcw-QCrmdhrvx3vLAJlSaaucDB~hUEoiSR-e1~kg92hVKHVuXFJGlLDyCMDrppMRcopfW7NISfyhgyJMa-IL9epTRvmeL~OJVaEX2g~kYaOIEkR44J4nbJpWW3n8608xCN0kro0mjPDd4M4q7nH7SOmoiX~C0TNWBo1pHwOg5JYK0698SUxTUDLX68wxqOQ~F7mUucSPldvCQVmJU6vNoe1EedIVZ2oQMUqzVtc8fE21cS1DBWg8BB74oobSrCxY9BlEHwYBxE-MRveP16bXJuD5pp6ARIQig__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 5. TNF stimulation of IL-8 is dependent on intracellular oxidation. (A) Inducible IL-8 protein secretion is sensitive to DMSO. Triplicate cultures of U937 cultures were untreated (control) or stimulated with 20 ng/mL TNF in the absence or presence of 2% (vol/vol) DMSO. At indicated times, cell culture supernatants were harvested and immunoreactive IL-8 determined by ELISA. Shown is the mean ± SD of n = 3 independent experiments (for both time points shown, P < .0001 for nonpretreated vpretreated with 2% (vol/vol) DMSO). (B) DMSO causes a dose-dependent inhibition of TNF-inducible IL-8 mRNA. Cells were unstimulated/stimulated with 20 ng/mL TNF (1 hour) in the absense/presence of DMSO of indicated concentrations (vol/vol). Cells were lysed and the total RNA subject to Northern analysis by hybridization with a hIL-8 cDNA probe (top) and an 18S cDNA probe (bottom) as an internal control. (C) DMSO causes a dose-dependent inhibition of TNF-inducible IL-8 promoter activity. Triplicate cell cultures were transfected with the –162 IL-8/LUC reporter plasmid and an SV40/alkaline phosphatase plasmid as an internal control. Cells were unstimulated/stimulated with 20 ng/mL TNF in the absence or presence of indicated concentrations of DMSO (in % [vol/vol]). Fold-induction of the Luciferase activity of stimulated (calculated from unstimulated) cells is shown (P < .0001 for stimulated v pretreated with 2% (vol/vol) DMSO before stimulation.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/94/6/10.1182_blood.v94.6.1878/5/m_blod41803005ax.jpeg?Expires=1769099902&Signature=Y8dBPz3MzJxIJvOAMUyA0pgD4paV-2qDevjkBJoUdqz6dB8bkH2nb9UaEs8EBsuqiZlTibsL0w-wfhKNw8Tyy0tZVz2gk~vpiLuih-Vdl4JykS6nxrBiPUPrHknpqAJ9-P4k4ae1jEhXJiTwwKnGZWoU80LjKLDekpWLxYxnlP16JdKG45f7Bp78QyPSwrZPRtiHF12CSVTHoDzI7t3RVNrEOpv8aIEiBtpoiFMXuB-iuzLFCw42BbGQEvh~5PBeE61bsF3Utegv07zFlstU4XzGIWTYG3ojcQkSa1Om3cRB-pEH~usIHgPUIX~1g35~ncYVBi2C-se6189cPHa3AQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 5. TNF stimulation of IL-8 is dependent on intracellular oxidation. (A) Inducible IL-8 protein secretion is sensitive to DMSO. Triplicate cultures of U937 cultures were untreated (control) or stimulated with 20 ng/mL TNF in the absence or presence of 2% (vol/vol) DMSO. At indicated times, cell culture supernatants were harvested and immunoreactive IL-8 determined by ELISA. Shown is the mean ± SD of n = 3 independent experiments (for both time points shown, P < .0001 for nonpretreated vpretreated with 2% (vol/vol) DMSO). (B) DMSO causes a dose-dependent inhibition of TNF-inducible IL-8 mRNA. Cells were unstimulated/stimulated with 20 ng/mL TNF (1 hour) in the absense/presence of DMSO of indicated concentrations (vol/vol). Cells were lysed and the total RNA subject to Northern analysis by hybridization with a hIL-8 cDNA probe (top) and an 18S cDNA probe (bottom) as an internal control. (C) DMSO causes a dose-dependent inhibition of TNF-inducible IL-8 promoter activity. Triplicate cell cultures were transfected with the –162 IL-8/LUC reporter plasmid and an SV40/alkaline phosphatase plasmid as an internal control. Cells were unstimulated/stimulated with 20 ng/mL TNF in the absence or presence of indicated concentrations of DMSO (in % [vol/vol]). Fold-induction of the Luciferase activity of stimulated (calculated from unstimulated) cells is shown (P < .0001 for stimulated v pretreated with 2% (vol/vol) DMSO before stimulation.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/94/6/10.1182_blood.v94.6.1878/5/m_blod41803005bw.jpeg?Expires=1769099902&Signature=TKkrG6q3w2KGBkDZaslmTIghUSTR~8-AmPM-JcJnYgzUkoO~T4Gme~bIgtaQIg881AOE3VJr9nfy4weX1elbTDWMVu7FD4ODl42VGgZYQek1dUaKz3d5gWMfexDBvHlM8sav~21pQqQjCaac81qeGXjqJgZDu5Cn2K2myPzl1tT2jR69yQTlFv3wxUrlmr3RjarvY7qerHa6~2zd2VdZZaikAQMSZjbJ5TQUoqcATsPQ3QfPAUbC~sCAuCamrbaolNgNdXut1MIGpujwIL4InFPR2ovNpFYbNbJmwyWlrxYTxDhw9dLc1ixvii~cI26r4GozVUPwZep698ezo1NGUg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 5. TNF stimulation of IL-8 is dependent on intracellular oxidation. (A) Inducible IL-8 protein secretion is sensitive to DMSO. Triplicate cultures of U937 cultures were untreated (control) or stimulated with 20 ng/mL TNF in the absence or presence of 2% (vol/vol) DMSO. At indicated times, cell culture supernatants were harvested and immunoreactive IL-8 determined by ELISA. Shown is the mean ± SD of n = 3 independent experiments (for both time points shown, P < .0001 for nonpretreated vpretreated with 2% (vol/vol) DMSO). (B) DMSO causes a dose-dependent inhibition of TNF-inducible IL-8 mRNA. Cells were unstimulated/stimulated with 20 ng/mL TNF (1 hour) in the absense/presence of DMSO of indicated concentrations (vol/vol). Cells were lysed and the total RNA subject to Northern analysis by hybridization with a hIL-8 cDNA probe (top) and an 18S cDNA probe (bottom) as an internal control. (C) DMSO causes a dose-dependent inhibition of TNF-inducible IL-8 promoter activity. Triplicate cell cultures were transfected with the –162 IL-8/LUC reporter plasmid and an SV40/alkaline phosphatase plasmid as an internal control. Cells were unstimulated/stimulated with 20 ng/mL TNF in the absence or presence of indicated concentrations of DMSO (in % [vol/vol]). Fold-induction of the Luciferase activity of stimulated (calculated from unstimulated) cells is shown (P < .0001 for stimulated v pretreated with 2% (vol/vol) DMSO before stimulation.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/94/6/10.1182_blood.v94.6.1878/5/m_blod41803005cx.jpeg?Expires=1769099902&Signature=2p-GN-MJczGvARzlKOFOJjwnOcqBFp5-iyU1bvYI4c6Q3aezhPnEUnbmihOsoxRXA5dCjoUGc3VUFAw8O67q-KAtiYUGhVOorlxQstqAK3pOUZZ-k7QrDccxsqt8y1VHJ-bXm730vtbCjUlcWf6nDQThvSrGDHgzNSp~6JBqP36JBzWaJfr0Wmc2gm3K7jtkxafLvM1OiYxlxhepcSX3GmML~TFCduPwNxXhZX0FnEr50MbCvjijwXrn3mg8hS3Vsk4sZ3JFjUx2KCCEWjHIvDHicFB7KVbPUXRAk6qps4XYc7lcGP1lMmm8bA-wjMobo3H6AlAlXOAZUK33YUz0Pg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 7. DMSO effect is independent of NF-κB binding and translocation. (A) Two percent (vol/vol) DMSO pretreatment has no effect on IκB proteolysis. Cells were stimulated with the indicated concentrations of TNF, in the absence or presence of 1 hour pretreatment with 2% (vol/vol) DMSO. Top panel, Western immunoblot with anti-IκB antibody; bottom panel, Western immunoblot with an anti-β–actin antibody (internal control). (B) Two percent (vol/vol) DMSO pretreatment has no effect on Rel A nuclear translocation. Cells were stimulated with 20 ng/mL TNF, in the absence or presence of 1 hour pretreatment with 2% (vol/vol) DMSO. Nuclei were purified over sucrose cushion and tested by Western immunoblot with anti-Rel A antibody. Sixty-five kD Rel A is strongly induced by TNF in the absence or presence of DMSO. A nonspecific band serves here as an internal control for protein loading (control). (C) Effect of TNF on NF-κB binding in the presence of DMSO. Cells were stimulated with increasing concentrations of TNF (indicated at top), in the absence or presence of 1 hour pretreatment with 2% (vol/vol) DMSO. Shown is EMSA analysis of nuclear extracts for binding to radiolabeled NF-κB. C1/C2 binding increases proportionally with TNF dose. 0.032 ng/mL TNF yields a 7-fold weaker signal than 20 ng/mL (360,351 v 2,645,248 arbitrary units [a.u.]). In contrast, 2% (vol/vol) DMSO pretreatment does not reduce NF-κB binding (2,691,962 a.u.). For nuclear extracts, stimulated with 20 ng/mL TNF, various concentrations of protein were used to determine assay linearity with protein input (compare lanes 7, 6, and 4). (D) DMSO inhibits IL-8 promoter induction independently of TNF-induced changes in NF-κB Rel A DNA binding. Luciferase induction of the IL-8 promoter by the indicated amounts of TNF. Triplicate cell cultures were transfected with the –162 IL-8/LUC reporter plasmid and an SV40/alkaline phosphatase plasmid as an internal control. Cells were unstimulated/stimulated with the indicated doses (in ng/mL) TNF. The fold-induction of Luciferase activity of stimulated cells is shown (P < .005 for all pairwise comparisons). 0.032 ng/mL TNF activates the NF-κB–dependent promoter 7-fold weaker than 20 ng/mL. Two percent (vol/vol) DMSO pretreatment reduces TNF-inducible promoter activity by 8-fold. (E) Abundance of TNF-inducible NF-κB Rel A binding is not changed by pretreatment with promoter-inhibitory doses of DMSO. Cells unstimulated/stimulated with 20 ng/mL TNF for 15 minutes in the absence or presence of 2% (vol/vol) DMSO. After treatment, nuclear extracts were analyzed for NF-κB binding by microaffinity isolation. Shown is the Western immunoblot with rabbit antihuman Rel A polyclonal antibody (NS, nonspecific).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/94/6/10.1182_blood.v94.6.1878/5/m_blod41803007aw.jpeg?Expires=1769099902&Signature=BxeH~lV3eRXhKPEjpPNu35HkkcMz2hMecgUL-JkuSMWbNz8r4URSDHBYW94iMNvIy23OTvT6ORspvW8cg7HxtRs7E2864k852FiRIoNYByQFbD690JxsDs273~kYtzsFAOyMAITs3oUKhqiZ1j~o6a4CALtq8r0AojrHb2B6kDRscVl7DT05v3qLwV5nxvoqgNaW-AY1jrq63AIGHoNoohQUclvSwYB8pjGhlRXXedqq0IzljgZ9dtMSDMtiRT0YiadMZDZIBjRcf5Ct3qqpeS-yHjFFZ90edOF46Ud-zEg7VUcEeAP4sh-uv6nnxJMt4mq9lAHkjByJzDpBv5EGWg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 7. DMSO effect is independent of NF-κB binding and translocation. (A) Two percent (vol/vol) DMSO pretreatment has no effect on IκB proteolysis. Cells were stimulated with the indicated concentrations of TNF, in the absence or presence of 1 hour pretreatment with 2% (vol/vol) DMSO. Top panel, Western immunoblot with anti-IκB antibody; bottom panel, Western immunoblot with an anti-β–actin antibody (internal control). (B) Two percent (vol/vol) DMSO pretreatment has no effect on Rel A nuclear translocation. Cells were stimulated with 20 ng/mL TNF, in the absence or presence of 1 hour pretreatment with 2% (vol/vol) DMSO. Nuclei were purified over sucrose cushion and tested by Western immunoblot with anti-Rel A antibody. Sixty-five kD Rel A is strongly induced by TNF in the absence or presence of DMSO. A nonspecific band serves here as an internal control for protein loading (control). (C) Effect of TNF on NF-κB binding in the presence of DMSO. Cells were stimulated with increasing concentrations of TNF (indicated at top), in the absence or presence of 1 hour pretreatment with 2% (vol/vol) DMSO. Shown is EMSA analysis of nuclear extracts for binding to radiolabeled NF-κB. C1/C2 binding increases proportionally with TNF dose. 0.032 ng/mL TNF yields a 7-fold weaker signal than 20 ng/mL (360,351 v 2,645,248 arbitrary units [a.u.]). In contrast, 2% (vol/vol) DMSO pretreatment does not reduce NF-κB binding (2,691,962 a.u.). For nuclear extracts, stimulated with 20 ng/mL TNF, various concentrations of protein were used to determine assay linearity with protein input (compare lanes 7, 6, and 4). (D) DMSO inhibits IL-8 promoter induction independently of TNF-induced changes in NF-κB Rel A DNA binding. Luciferase induction of the IL-8 promoter by the indicated amounts of TNF. Triplicate cell cultures were transfected with the –162 IL-8/LUC reporter plasmid and an SV40/alkaline phosphatase plasmid as an internal control. Cells were unstimulated/stimulated with the indicated doses (in ng/mL) TNF. The fold-induction of Luciferase activity of stimulated cells is shown (P < .005 for all pairwise comparisons). 0.032 ng/mL TNF activates the NF-κB–dependent promoter 7-fold weaker than 20 ng/mL. Two percent (vol/vol) DMSO pretreatment reduces TNF-inducible promoter activity by 8-fold. (E) Abundance of TNF-inducible NF-κB Rel A binding is not changed by pretreatment with promoter-inhibitory doses of DMSO. Cells unstimulated/stimulated with 20 ng/mL TNF for 15 minutes in the absence or presence of 2% (vol/vol) DMSO. After treatment, nuclear extracts were analyzed for NF-κB binding by microaffinity isolation. Shown is the Western immunoblot with rabbit antihuman Rel A polyclonal antibody (NS, nonspecific).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/94/6/10.1182_blood.v94.6.1878/5/m_blod41803007bw.jpeg?Expires=1769099902&Signature=QaMxg3sGkejR4Uje5JD9cNLYE1TWFaMg3GMRn2snyNU1078~BtmbTFLOrbEaQQiJgBD8YGPtFubQD9sxCTU4tMlHIPzuErtNwhn-jK~1OK5o0dsgVAgg73QeRsth1Gn1k-mewfx3Hyh5EsbG82qh-QFNTckIpUf~ta8skFme7jQCkKbqq323CsL~BrcAefUa-BZU0M078nZfFigWe5LTr43CHnrze8Lzv~a59U9kcTB1oTSR5JzZ1lAz-hF1Sw794iABOHlh50XcCRDVtG~EEryF3RH7385RlcfSnq6H0SjjlSxoeITfN~paEwLO3ZTLNpgjWFan2MAEq-NeV4vEdg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 7. DMSO effect is independent of NF-κB binding and translocation. (A) Two percent (vol/vol) DMSO pretreatment has no effect on IκB proteolysis. Cells were stimulated with the indicated concentrations of TNF, in the absence or presence of 1 hour pretreatment with 2% (vol/vol) DMSO. Top panel, Western immunoblot with anti-IκB antibody; bottom panel, Western immunoblot with an anti-β–actin antibody (internal control). (B) Two percent (vol/vol) DMSO pretreatment has no effect on Rel A nuclear translocation. Cells were stimulated with 20 ng/mL TNF, in the absence or presence of 1 hour pretreatment with 2% (vol/vol) DMSO. Nuclei were purified over sucrose cushion and tested by Western immunoblot with anti-Rel A antibody. Sixty-five kD Rel A is strongly induced by TNF in the absence or presence of DMSO. A nonspecific band serves here as an internal control for protein loading (control). (C) Effect of TNF on NF-κB binding in the presence of DMSO. Cells were stimulated with increasing concentrations of TNF (indicated at top), in the absence or presence of 1 hour pretreatment with 2% (vol/vol) DMSO. Shown is EMSA analysis of nuclear extracts for binding to radiolabeled NF-κB. C1/C2 binding increases proportionally with TNF dose. 0.032 ng/mL TNF yields a 7-fold weaker signal than 20 ng/mL (360,351 v 2,645,248 arbitrary units [a.u.]). In contrast, 2% (vol/vol) DMSO pretreatment does not reduce NF-κB binding (2,691,962 a.u.). For nuclear extracts, stimulated with 20 ng/mL TNF, various concentrations of protein were used to determine assay linearity with protein input (compare lanes 7, 6, and 4). (D) DMSO inhibits IL-8 promoter induction independently of TNF-induced changes in NF-κB Rel A DNA binding. Luciferase induction of the IL-8 promoter by the indicated amounts of TNF. Triplicate cell cultures were transfected with the –162 IL-8/LUC reporter plasmid and an SV40/alkaline phosphatase plasmid as an internal control. Cells were unstimulated/stimulated with the indicated doses (in ng/mL) TNF. The fold-induction of Luciferase activity of stimulated cells is shown (P < .005 for all pairwise comparisons). 0.032 ng/mL TNF activates the NF-κB–dependent promoter 7-fold weaker than 20 ng/mL. Two percent (vol/vol) DMSO pretreatment reduces TNF-inducible promoter activity by 8-fold. (E) Abundance of TNF-inducible NF-κB Rel A binding is not changed by pretreatment with promoter-inhibitory doses of DMSO. Cells unstimulated/stimulated with 20 ng/mL TNF for 15 minutes in the absence or presence of 2% (vol/vol) DMSO. After treatment, nuclear extracts were analyzed for NF-κB binding by microaffinity isolation. Shown is the Western immunoblot with rabbit antihuman Rel A polyclonal antibody (NS, nonspecific).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/94/6/10.1182_blood.v94.6.1878/5/m_blod41803007cw.jpeg?Expires=1769099902&Signature=RO5VGOtQLIqdUCqJBP5b7rQZZrmVE3M8zmLDKXVi9~o-j863EUHXUpopoOQthIp8IVN36mceCTwWu2dc5p3M9uu0iaLTWK33GXQnrnf4IxkJf7Fqg4McA7Ih8bynZAC4nSyARtSyOu5QCzFkstlw1uIZUaL6eilwOv8uxMVxOZ8D3HDixGa2KmlUDIPCJu0JdyI3w85V89BIPN5Eoxf0ix~iAWkVq3cWiABl2zOjqpqp4Yba2Ruf5sfj8EMG91W77~IkIH26VWzqpKq19bfybl-UESyMVsMGmQgJhq9Uz-vP2jalu7Nw-jLqeuTbYWpmh5DMV9UlYjKjjK7zYJ0tag__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 7. DMSO effect is independent of NF-κB binding and translocation. (A) Two percent (vol/vol) DMSO pretreatment has no effect on IκB proteolysis. Cells were stimulated with the indicated concentrations of TNF, in the absence or presence of 1 hour pretreatment with 2% (vol/vol) DMSO. Top panel, Western immunoblot with anti-IκB antibody; bottom panel, Western immunoblot with an anti-β–actin antibody (internal control). (B) Two percent (vol/vol) DMSO pretreatment has no effect on Rel A nuclear translocation. Cells were stimulated with 20 ng/mL TNF, in the absence or presence of 1 hour pretreatment with 2% (vol/vol) DMSO. Nuclei were purified over sucrose cushion and tested by Western immunoblot with anti-Rel A antibody. Sixty-five kD Rel A is strongly induced by TNF in the absence or presence of DMSO. A nonspecific band serves here as an internal control for protein loading (control). (C) Effect of TNF on NF-κB binding in the presence of DMSO. Cells were stimulated with increasing concentrations of TNF (indicated at top), in the absence or presence of 1 hour pretreatment with 2% (vol/vol) DMSO. Shown is EMSA analysis of nuclear extracts for binding to radiolabeled NF-κB. C1/C2 binding increases proportionally with TNF dose. 0.032 ng/mL TNF yields a 7-fold weaker signal than 20 ng/mL (360,351 v 2,645,248 arbitrary units [a.u.]). In contrast, 2% (vol/vol) DMSO pretreatment does not reduce NF-κB binding (2,691,962 a.u.). For nuclear extracts, stimulated with 20 ng/mL TNF, various concentrations of protein were used to determine assay linearity with protein input (compare lanes 7, 6, and 4). (D) DMSO inhibits IL-8 promoter induction independently of TNF-induced changes in NF-κB Rel A DNA binding. Luciferase induction of the IL-8 promoter by the indicated amounts of TNF. Triplicate cell cultures were transfected with the –162 IL-8/LUC reporter plasmid and an SV40/alkaline phosphatase plasmid as an internal control. Cells were unstimulated/stimulated with the indicated doses (in ng/mL) TNF. The fold-induction of Luciferase activity of stimulated cells is shown (P < .005 for all pairwise comparisons). 0.032 ng/mL TNF activates the NF-κB–dependent promoter 7-fold weaker than 20 ng/mL. Two percent (vol/vol) DMSO pretreatment reduces TNF-inducible promoter activity by 8-fold. (E) Abundance of TNF-inducible NF-κB Rel A binding is not changed by pretreatment with promoter-inhibitory doses of DMSO. Cells unstimulated/stimulated with 20 ng/mL TNF for 15 minutes in the absence or presence of 2% (vol/vol) DMSO. After treatment, nuclear extracts were analyzed for NF-κB binding by microaffinity isolation. Shown is the Western immunoblot with rabbit antihuman Rel A polyclonal antibody (NS, nonspecific).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/94/6/10.1182_blood.v94.6.1878/5/m_blod41803007dx.jpeg?Expires=1769099902&Signature=EOMYp507VIelDnyY9xBUJ7TQCUI1t66RaqCbaWKgBssXG2XCNFkJQqHN-cCvPqmnJliJSN19wQK5j1AspgXhsxJ7PJ~92t0D0abSiB3XSnnIP-x6fEFXvshGeVEKpe~i-rEmcc~fiVWGYUYiMOL2posfWcmavyyLMXGIfDTrS0SOpYsJR-Kmml~qc~cYRMK5EeSF6w-I~6qdnbfTkXmi6c-RiPLQ8ZQT86rg~sAf6jVEIagALmdHQ9ytFIcFgQKVo4eSP7ziGUhG6xf1Af9LvDlu86N8WupUHCAL28W4LYBISaMbXNtlAY8eT5T-pcBWXCWeCHwva9kM~7BTXi-6Hw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 7. DMSO effect is independent of NF-κB binding and translocation. (A) Two percent (vol/vol) DMSO pretreatment has no effect on IκB proteolysis. Cells were stimulated with the indicated concentrations of TNF, in the absence or presence of 1 hour pretreatment with 2% (vol/vol) DMSO. Top panel, Western immunoblot with anti-IκB antibody; bottom panel, Western immunoblot with an anti-β–actin antibody (internal control). (B) Two percent (vol/vol) DMSO pretreatment has no effect on Rel A nuclear translocation. Cells were stimulated with 20 ng/mL TNF, in the absence or presence of 1 hour pretreatment with 2% (vol/vol) DMSO. Nuclei were purified over sucrose cushion and tested by Western immunoblot with anti-Rel A antibody. Sixty-five kD Rel A is strongly induced by TNF in the absence or presence of DMSO. A nonspecific band serves here as an internal control for protein loading (control). (C) Effect of TNF on NF-κB binding in the presence of DMSO. Cells were stimulated with increasing concentrations of TNF (indicated at top), in the absence or presence of 1 hour pretreatment with 2% (vol/vol) DMSO. Shown is EMSA analysis of nuclear extracts for binding to radiolabeled NF-κB. C1/C2 binding increases proportionally with TNF dose. 0.032 ng/mL TNF yields a 7-fold weaker signal than 20 ng/mL (360,351 v 2,645,248 arbitrary units [a.u.]). In contrast, 2% (vol/vol) DMSO pretreatment does not reduce NF-κB binding (2,691,962 a.u.). For nuclear extracts, stimulated with 20 ng/mL TNF, various concentrations of protein were used to determine assay linearity with protein input (compare lanes 7, 6, and 4). (D) DMSO inhibits IL-8 promoter induction independently of TNF-induced changes in NF-κB Rel A DNA binding. Luciferase induction of the IL-8 promoter by the indicated amounts of TNF. Triplicate cell cultures were transfected with the –162 IL-8/LUC reporter plasmid and an SV40/alkaline phosphatase plasmid as an internal control. Cells were unstimulated/stimulated with the indicated doses (in ng/mL) TNF. The fold-induction of Luciferase activity of stimulated cells is shown (P < .005 for all pairwise comparisons). 0.032 ng/mL TNF activates the NF-κB–dependent promoter 7-fold weaker than 20 ng/mL. Two percent (vol/vol) DMSO pretreatment reduces TNF-inducible promoter activity by 8-fold. (E) Abundance of TNF-inducible NF-κB Rel A binding is not changed by pretreatment with promoter-inhibitory doses of DMSO. Cells unstimulated/stimulated with 20 ng/mL TNF for 15 minutes in the absence or presence of 2% (vol/vol) DMSO. After treatment, nuclear extracts were analyzed for NF-κB binding by microaffinity isolation. Shown is the Western immunoblot with rabbit antihuman Rel A polyclonal antibody (NS, nonspecific).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/94/6/10.1182_blood.v94.6.1878/5/m_blod41803007ew.jpeg?Expires=1769099902&Signature=YWzwf1X9uBWoHQ9N3FXlD3POnVvGu-svJDlcFkCmzhNZW9DjWrmwd2wm8KqVM51rIKQaoZyfiYzDOZ7LbFY3C~TF4oH8CR21ZtYn6RhAHC11SXgGQ7E8h1sBi1hE-ohPpnj8syt7SgQyEmr6EdOunyiKugjJa-~OSqDr1zZJwDVC1AmcodNmwRZJKIJTS9ifFJSyCvqdiL5HnndKajIVboQl22SIYYYDcavroZGUtawiYOiVpt9N4kSaHCmptRKLdamfDv6x8R1v~te4hERxoAmZnon5y6S8LHZFcqb~yJmAxuP7YbsR20sVP-4k88z4azxWklWmtcXNf6ve6xDP4A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 8. Antioxidant effect at times subsequent to IκB proteolysis. (A) Effect of posttreatment on TNF-inducible IL-8 mRNA accumulation. Cells were unstimulated or stimulated with 20 ng/mL TNF in the absence or presence of 2% (vol/vol) DMSO. DMSO was administered at the indicated times (before [-] or after [+] TNF administration). (B) Posttreatment with 2% (vol/vol) DMSO has no effect on IκB proteolysis. Cells were unstimulated or stimulated with 20 ng/mL TNF in the absence or presence of 2% (vol/vol) DMSO. DMSO was administered at the indicated times (preceding [-] or succeeding [+] TNF administration). Top panel, Western immunoblot with anti-IκB antibody; bottom panel, Western immunoblot with an anti-β–actin antibody (internal control). (C) Pre or posttreatment with 2% (vol/vol) DMSO has no effect on Rel A DNA binding in EMSA. Cells were unstimulated/stimulated with 20 ng/mL TNF in the absence or presence of 2% (vol/vol) DMSO. DMSO was administered at the indicated times (before [-] or after [+] TNF administration). Nuclei were isolated and subjected to EMSA analysis with a radiolabeled NF-κB site. Only bound complexes are shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/94/6/10.1182_blood.v94.6.1878/5/m_blod41803008aw.jpeg?Expires=1769099902&Signature=YxEHab6Sum3Z-Rtu03qoZ3lEfxFVBUmQBoLZYrAvOcDjwZNL2q7XO5EO-Ue~LkP7e9xX18-zkRf~e1Lo5tYVm5~p6Eb6tsgFIrRr2vyKIv5ioINzy7g~NfNdcO4OwCccqegMgLVNSee5wKI9HdiejsPn4tFK-q5A15l2rsRZ-XwMvrgs0xtIkM1gY5N1S-ucPi~xGB9PntCtEzAi44Jm1NzRo0NAdmULY3woM5Ip5Gb4S68uGXg7d4OT98onAxt6mwy79Dra6FHREs9ezu~EZXKTHazJJ1081-h5NV0vSWnqCWWNuwiKwMjdzAARrjp1B9P3vCXi2dXasLEIA-P9Ug__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 8. Antioxidant effect at times subsequent to IκB proteolysis. (A) Effect of posttreatment on TNF-inducible IL-8 mRNA accumulation. Cells were unstimulated or stimulated with 20 ng/mL TNF in the absence or presence of 2% (vol/vol) DMSO. DMSO was administered at the indicated times (before [-] or after [+] TNF administration). (B) Posttreatment with 2% (vol/vol) DMSO has no effect on IκB proteolysis. Cells were unstimulated or stimulated with 20 ng/mL TNF in the absence or presence of 2% (vol/vol) DMSO. DMSO was administered at the indicated times (preceding [-] or succeeding [+] TNF administration). Top panel, Western immunoblot with anti-IκB antibody; bottom panel, Western immunoblot with an anti-β–actin antibody (internal control). (C) Pre or posttreatment with 2% (vol/vol) DMSO has no effect on Rel A DNA binding in EMSA. Cells were unstimulated/stimulated with 20 ng/mL TNF in the absence or presence of 2% (vol/vol) DMSO. DMSO was administered at the indicated times (before [-] or after [+] TNF administration). Nuclei were isolated and subjected to EMSA analysis with a radiolabeled NF-κB site. Only bound complexes are shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/94/6/10.1182_blood.v94.6.1878/5/m_blod41803008bw.jpeg?Expires=1769099902&Signature=SYj7DpwR1Y~v~bL5diVkMEuHDtxCz0epRQINCOYkuNzdOgchK6ELee~JaRsZVUPtuD48IPdxK-pnZQcVfBd~lmURTjzQHYMkXJ8Fvv7nkmSciBgoM~PM8jvE6yJvKClOqRpG3RZ1rLTdSnzQHbQd4DQDMQ3KmOhfEuG17fqBrq6hBEPC7~yMKvuTnat~aQHKeI1-WaEIT8rmo2gpCCoWGG7m4OOZUw2kz1L3imtIUeegI8jtpkmsiZ1S81PDFnCQ~yOACOmtRM-IQXzOqsOo6owzrJeudA~j3AcX3eoTgO1wOKplGoNWbxaE~tRkeRthae501fVcQlVhrZeY2xMoJw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 8. Antioxidant effect at times subsequent to IκB proteolysis. (A) Effect of posttreatment on TNF-inducible IL-8 mRNA accumulation. Cells were unstimulated or stimulated with 20 ng/mL TNF in the absence or presence of 2% (vol/vol) DMSO. DMSO was administered at the indicated times (before [-] or after [+] TNF administration). (B) Posttreatment with 2% (vol/vol) DMSO has no effect on IκB proteolysis. Cells were unstimulated or stimulated with 20 ng/mL TNF in the absence or presence of 2% (vol/vol) DMSO. DMSO was administered at the indicated times (preceding [-] or succeeding [+] TNF administration). Top panel, Western immunoblot with anti-IκB antibody; bottom panel, Western immunoblot with an anti-β–actin antibody (internal control). (C) Pre or posttreatment with 2% (vol/vol) DMSO has no effect on Rel A DNA binding in EMSA. Cells were unstimulated/stimulated with 20 ng/mL TNF in the absence or presence of 2% (vol/vol) DMSO. DMSO was administered at the indicated times (before [-] or after [+] TNF administration). Nuclei were isolated and subjected to EMSA analysis with a radiolabeled NF-κB site. Only bound complexes are shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/94/6/10.1182_blood.v94.6.1878/5/m_blod41803008cw.jpeg?Expires=1769099902&Signature=lrPpZFKEQ8NAinqCHDe9kqflEze8XV4DKsWYwK8cjl4X7ROqJTYrHMmSODmAgiVm9Jv~cK-sVoT8hcFTr~woG8p-8zfMHJXFxIBAoMZVY3SqhK19It8JaDFLbRViOV469opn9gSAf~~HjeXZVWkRKK-1TPXEngk~AjQtwwLC1XYSI6XQ9eFNxN7WN8dXDBTtjyQhGXXs2gSDh0cgyfFYz7kDoB-r3TgGPopjoJmq~DfVbC-udZ5-KMpSoi77pkq5EOOJhXftYwXhvzkNxf8BtRw71K524srgYNXDzNlRM-m3lw~pncEkvnI~DrVmgx7C5nPM5pcCeqxfDQh~EekDMA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal