Abstract
We showed previously that human malignant non-Hodgkin’s lymphomas (NHL) degrade extracellular matrix (ECM) components through the action of metalloproteinases and that elevated expression of matrix metalloproteinase-9 (MMP-9) and tissue inhibitor of metalloproteinase-1 (TIMP-1) correlated with a poor clinical outcome in patients with NHL. In the present study we sought to investigate whether there is any correlation between the expression of gelatinases (MMP-2 and MMP-9), TIMP-1, and the expression of cytokines and growth factors such as interleukin-1β (IL-1β), IL-6, IL-10, tumor necrosis factor (TNF-), transforming growth factor β (TGFβ), and basic fibroblast growth factor (bFGF) in human NHL. In lymphoma tissues obtained from 32 patients, elevated expression of IL-6 correlated significantly with elevated messenger RNA (mRNA) levels of MMP-9, MMP-2, and TIMP-1. Moreover, in human lymphoid cell lines of B- and T-cell origin (Raji, Jurkat, and NC-37), IL-6 stimulated production of MMP-9 and MMP-2 but not TIMP-1. In the Matrigel invasion assay IL-6 significantly upregulated transmigration of Raji and Jurkat cells, which in turn was inhibited by recombinant human TIMP-1 and anti-MMP-9 and MMP-2 antibodies. We postulate that IL-6 may play a role in the clinical aggressiveness of human NHL by stimulating MMP production.
HUMAN MALIGNANT non-Hodgkin’s lymphomas (NHL) represent a heterogeneous group of tumors, which vary in their biological aggressiveness and clinical course.1 We have shown previously that high-grade human NHL degrade extracellular matrix (ECM) components in vitro and that metalloproteinases play an important role in this phenomenon.2 We have also shown that matrix metalloproteinase-9 (MMP-9, Gelatinase B) and tissue inhibitor of metalloproteinase-1 (TIMP-1) are overexpressed in a subset of human high-grade NHL and this overexpression is associated with a poor clinical outcome for patients with these tumors.3-6
The MMPs are a family of zinc- and calcium-dependent proteolytic enzymes capable of degrading most ECM components.7 Their action in tissues is inhibited by specific tissue inhibitors (TIMPs).7 The majority of MMPs are secreted enzymes, but membrane-type matrix metalloproteinases (MT-MMPs) have also been described.8 Depending on their substrate specificity, MMPs are broadly divided into collagenases, stromelysins, and gelatinases.7 The latter group, consisting of Gelatinase A (MMP-2) and Gelatinase B (MMP-9), degrade denatured collagens (gelatin), native type IV and V collagens, and elastin.7MMPs have been implicated in tumor invasion and metastasis,9,10 and their role in the dissemination of hematologic malignancies, such as human NHL and acute myelogenous leukemia (AML), has recently been appreciated.2-4,11-14TIMP-1 has also been shown to be expressed by established neoplastic B-cell lines and high-grade NHL.3-6, 11, 15 In addition to and separately from its function as an MMP inhibitor, TIMP-1 has other functions, eg, it promotes the growth of a variety of cells, prevents apoptosis in B cells, and induces their differentiation.16-18 However, the mechanisms that control MMP and TIMP expression in human NHL are not known.
Under physiological and certain pathological conditions, regulation of MMP and TIMP expression involves several factors including steroid hormones, cellular oncogenes, growth factors, and cytokines.19 Different MMPs and TIMPs are stimulated by different factors and cell types respond differently to these various stimuli. For example, in immunological response, cell-type specific activation of various MMPs and TIMPs is mediated by many proinflammatory cytokines.20 In fibroblasts and macrophages, MMP-9 is upregulated by transforming growth factor β1 (TGFβ1), tumor necrosis factor α (TNF-α), basic fibroblast growth factor (bFGF), interleukin-1β (IL-1β), and lymphotoxin (LT).20-25 In T-lymphocytes, MMP-9 production is stimulated by IL-1, TNF-α, IL-2, and IL-4.26,27 It has also been reported that IL-2 and IL-4 stimulation of T-lymphocytes induces migration of these cells through Matrigel (Collaborative Biomedical Products, Bedford, MA), a model basement membrane.27 Interferon β-1b (IFNβ-1b) inhibits MMP-9 production and migration through Matrigel by T-lymphocytes.28 In a plasma cell line TNF-α and IL-1α (used in combination, but not alone) stimulated MMP-9 activity and had no effect on MMP-2.29 IL-6 had no effect on MMP-9 and MMP-2 production by myeloma cells, but IL-1β, TNF-α and Oncostatin M stimulated stromal cells in this tumoral environment to produce MMP-1.30 Expression of MMP-9 in Epstein-Barr virus (EBV)–transformed and tonsilar B lymphocytes is also induced by a combination of TNF-α and IL-1α.29 IL-10, IFNγ, and IL-4 downregulate MMP-9 production in monocytes and macrophages.31,32 In prostatic carcinoma cell lines, IL-10 and IL-4 (but not IL-6) downregulated MMP-2, but had no effect on MMP-9 production,33 whereas TGFβ1 stimulated production of MMP-9 and MMP-2 in cervical carcinoma cells.34 TIMP-1 production in fibroblasts, hepatocytes, and prostatic tumor cell lines was shown to be stimulated by IL-6, IL-1β, and IL-10.35-38 However, divergent regulation of MMP and TIMP expression is observed in different cell types.26,39,40 In T-lymphocytes, proinflammatory cytokines and chemokines differentially regulated proMMP-9 secretion but had no effect on TIMP-1 production.26 In mononuclear phagocytes, IL-10 inhibited secretion of MMP-9 and stimulated that of TIMP-1.40 In plasma cells, TIMP-1 production was induced by a combined treatment with TNF-α and IL-1β.29
Because human NHL are composed of a variety of malignant and non-neoplastic cells, all capable of MMP and TIMP production, the aim of this study was to examine which growth factors and cytokines are expressed in this complex environment and whether their overexpression and the expression of gelatinases (MMP-2 and MMP-9) and TIMP-1 are correlated. Finally, to establish a functional relationship, we evaluated whether cytokines stimulate lymphoid cell lines of T- and B-cell lineage to produce gelatinases and/or TIMP-1 and to degrade ECM in vitro.
MATERIALS AND METHODS
Specimen collection.
Twenty-one NHL tissue samples, as well as 1 hyperplastic tonsil, were received in the Department of Pathology at the Foothills Hospital, Calgary, Canada. Tissue in excess of that needed for diagnostic purposes was snap-frozen in liquid nitrogen and subsequently stored at −70°C. An assessment of the sample was done by histopathological examination of sections adjacent to the frozen tissue to determine whether the sample was representative of the rest of the specimen. Additionally, 1 NHL tissue was obtained from the Lethbridge Cancer Centre, Lethbridge, Canada and 10 NHL samples were received from The South-Western Oncology Group (SWOG) in Tucson, AZ. The samples were shipped on dry ice by courier to the Foothills Hospital, Pathology Department (Calgary, Alberta, Canada) and were received frozen. The lymphoma sample preparation in these other two centers was done similarly to that in the Foothills Hospital. The investigator performing reverse transcriptase-polymerase chain reaction (RT-PCR) was blinded with respect to histopathological classification until after the experiments were completed.
Diagnostic procedures and case description.
The cases were examined histopathologically and classified independently by two pathologists according to the Working Formulation (WF).1 Assessment was also done at a later date to classify the samples according to the REAL classification.41Assessment of lineage was done by flow cytometry and immunohistochemistry, as well as by Southern blot analysis for the presence of immunoglobulin heavy-chain and T-cell receptor beta gene rearrangements. All cases but one were of B-cell lineage. One large-cell lymphoma was of T-cell origin and was classified as a large-cell anaplastic NHL (case 10; Fig 1). Two cases (cases 1 and 2; Fig 1) were small lymphocytic NHL (WF and REAL), 3 (cases 3 to 5; Fig 1) follicular NHL (1 follicular, predominantly small cleaved cell in WF, follicular center, grade I in REAL; 1 follicular, mixed in WF, follicular center, grade II in REAL; and 1 follicular, predominantly large cell in WF, follicular center, grade III in REAL), 2 (cases 6 and 7; Fig 1) diffuse, small cleaved cell NHL in WF (marginal-zone lymphomas in REAL), 2 (cases 8 and 9 in Fig 1) diffuse-, mixed-, small- and large-cell NHL in WF (diffuse large B cell in REAL), 1 (case 11 in Fig 1) small, noncleaved NHL in WF (high-grade B-cell Burkitt’s-like in REAL), and 21 (cases 12 to 32 in Fig 1) large-cell immunoblastic in WF (diffuse, large B cell in REAL).
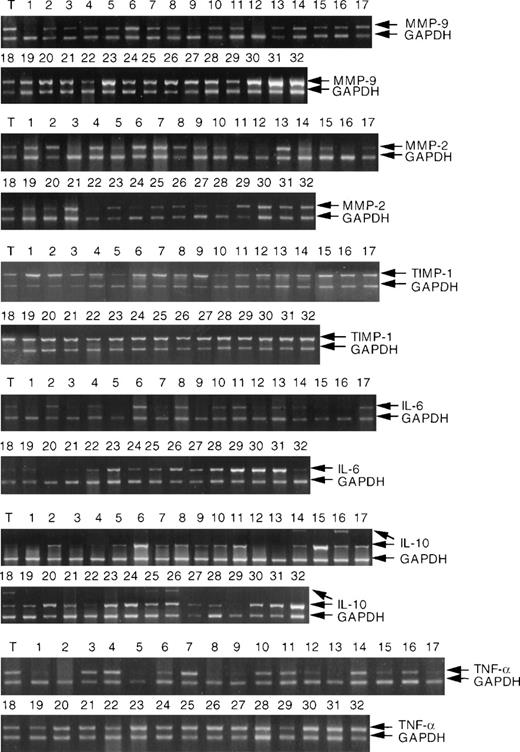
RT-PCR analysis of MMP-9, MMP-2, TIMP-1, IL-6, IL-10, and TNF- mRNA expression in NHL and tonsil (T). RNA extraction and RT-PCR analysis were performed as described in Materials and Methods. NHL are described (cases 1 to 32) in Materials and Methods. Each analysis was internally controlled by inclusion of GAPDH primers for the last 20 cycles of PCR.
RT-PCR analysis of MMP-9, MMP-2, TIMP-1, IL-6, IL-10, and TNF- mRNA expression in NHL and tonsil (T). RNA extraction and RT-PCR analysis were performed as described in Materials and Methods. NHL are described (cases 1 to 32) in Materials and Methods. Each analysis was internally controlled by inclusion of GAPDH primers for the last 20 cycles of PCR.
RNA extraction.
Total cellular RNA was extracted from tissues and cell lines by the acid guanidinium thiocyanate-phenol-chloroform extraction method, described by Chomczynski and Sacchi.42 RNA concentrations were determined using a Beckman DU65 spectrophotometer (Beckman Instruments, Fullerton, CA) (absorbance at 260 nm). The extent of protein contamination was checked by examining 260 nm/280 nm ratios.
RT reactions.
mRNA was converted to complementary DNA (cDNA) by RT reactions done in 0.5 mL tubes. Approximately 2 μg of RNA was added to each reaction tube. A master mixture was made with the following volume of reagents used per sample: 4 μL GIBCO-BRL (Rockville, MD) 5× First Strand Buffer [250 mmol/L Tris-HCl (pH 8.3 at room temperature), 375 mmol/L KCl, 15 mmol/L MgCl2], 2 μL N6 random oligonucleotides (100 pmol), 2 μL dNTP mixture (containing 10 mmol/L each of dATP, dGTP, dCTP, and dTTP at neutral pH), 2 μL GIBCO-BRL SuperScript RT RNAse H-RT, 0.2 μL of 1 mol/L dithiolthreitol (DTT) and 0.3 μL of ribonuclease inhibitor (RNAguard, Pharmacia). 10.5 μL of the master mixture was added to each tube and the final volume was made up to 20 μL with GIBCO water (ddH2O, RNAse-free). Subsequently, the samples were incubated at 42°C for 90 minutes using a Perkin Elmer-Cetus (Norwalk, CT) thermocycler. At the end of the incubation period, the samples were heated to 95°C for 5 minutes to inactivate the RT. Finally, the RT products were cooled to 4°C and stored at that temperature until use.
PCR.
Multiplex PCR were performed using a technique modeled after Wong et al.43 Fifty μL reaction volumes were used in PCR tubes with screw-cap lids containing volume-reducing inserts. Each reaction mixture contained 1 to 3 μL of RT product serving as template DNA. The volume of RT product put into the reaction was dictated by the volume of sample necessary to equalize the intensities of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) bands visualized during agarose gel electrophoresis. From 35.8 to 38.8 μL of GIBCO water was added to each tube depending on the volume of sample already present. Additional ingredients added to the master mixture included (per sample) 5 μL of 10× PCR reaction buffer (500 mmol/L KCl, 15 mmol/L MgCl2, and 100 mmol/L Tris-HCl), 1 μL dNTP mixture (10 mmol/L), and 1 μL each of the 5′ and 3′ starter primers (20 mmol/L). For all of the primer pairs except for IL-10 a “cold start” was performed. All of the reagents were kept on ice and Taq polymerase (0.2 μL per sample; Pharmacia, Piscataway, NJ) was added to the cold master mixture. 8.2 μL of the master mixture was then aliquoted into each reaction tube, and thermocycling was started using a Perkin Elmer-Cetus thermocycler. At 20 cycles, 1 μL of both 5′ and 3′ GAPDH primers were added. For all of the primers except IL-10, each PCR cycle consisted of a heat denaturation step at 94°C for 1 minute, a primer-annealing step at 55°C for 1 minute, and a strand elongation step at 72°C for 1 minute. The conditions for IL-10 PCR were based on those of Voorzanger et al.44 After a precycle at 95°C, a “hot start” was performed for primer specificity enhancement, meaning that Taq polymerase was not added to the master mixture, but instead was added during the first denaturation step (0.5 μL of Taq polymerase per tube). The denaturing step for the IL-10 reaction was conducted at 95°C for 30 seconds, with the annealing step at 60°C and an extension step at 72°C for 2 minutes. After the final cycle, the reaction was held at 72°C for 2 minutes. More than one PCR reaction had to be done for each set of primers so cDNA from one hyperplastic tonsil was run with each reaction as a control for differing reaction conditions. The conditions for the reactions were such that the plateau was not reached.
Gel electrophoresis and quantification.
Aliquots of PCR products (approximately 10 μL) were electrophoresed through 1.8% agarose gels containing 0.1 μg/μL ethidium bromide. Loading was equalized to the internal control mRNA (GAPDH) to give equivalent signals. Gels were illuminated with UV light and photographed using Polaroid film (Cambridge, MA). The intensities of the bands were quantified by computer densitometry. The photographs were scanned using a Hewlett-Packard (Palo Alto, CA) ScanJet Ink Scanner and DeskScan II software, then analyzed using the National Institutes of Health (NIH) (Bethesda, MD) Image program. Each band corresponded to a peak, the pixel density of which was proportional to the intensity of the ethidium bromide fluorescence signals. The final activity was defined as the ratio of specific primer/GAPDH intensity. For comparison of gels from different PCR reactions, but using the same primer, a control tonsil was run with each batch and gels were standardized to this sample. Because the mRNA levels in a sample would not change between PCR reactions but slightly different signals could result depending on the PCR conditions, different batches were adjusted to give equal target/GAPDH ratios in the control tonsil for all the batches. For instance, if PCR batch one had a target/GAPDH ratio of 1.8 for the control tonsil and PCR batch two had a ratio of 2.0 for the same sample, the ratios of all the samples in batch one would be multiplied by 1.11 to give equal tonsil activity in both batches.
Primers.
Sequences for human MMP-2, MMP-9, TIMP-1, IL-1β, IL-6, bFGF, TNF-α, and TGF-β were obtained from GenBank (National Center for Biotechnology Information, Bethesda, MD) and used to design primer pairs. The primers were separated by an intron and were chosen to have a 40% to 60% GC content. The sequence for the GAPDH primer pair was found in Wong et al43 and that for IL-10 was taken from Voorzanger et al.44 The optimum number of cycles for each primer was determined to keep signal amplification in the linear range. The primer sequences and the number of cycles used were as follows (name, sequence [5′→3′], cycle no.): MMP-2, GGCCCTGTCACTCCTGAGAT GGCATCCAGGTTATCGGGGA, 25; MMP-9, CAACATCACCTATTGGATCCCGGGTGTAGAGTCTCTCGCT, 25; TIMP-1, GCGGATCCAGCGCCCAGAGAGACACCTTAAGCTTCCACTCCGGGCAGGATT, 25; IL-1β TTCCCATTAGACAGCTGCACTGTTTGGGATCCACACTCTC, 32; IL-6, AAAATCTGCTCTGGTCTTCTGGGGTTTGCCGAGTAGACCTCA, 30; IL-10, AATGGCTCTAGAATGCACAGCTCAGCACTGAATGGCGAATTCTTTCTCAAGGGGCTGGGT, 30; bFGF, TCTTCCAATGTCTGCTAAGAGCTGTCCAGCAGTTTACACAGGACTGTT, 30; TNF-α, TCGAGTGACAAGCCCGTAGCAGAGCAATGACTCCAAAGTAGAC, 28; TGF-β, CCTGGACACCAACTATTGCTTCAGGACCTTGCTGTACTGCGTGTCCA 25; GAPDH, CGGAGTCAACGGATTTGGTCGTATAGCCTTCTCCATGGTGGTGAAGAC, 20.
Cell-conditioned media and cytokines.
The human cell lines Burkitt’s lymphoma (Raji), acute T-cell leukemia (Jurkat), and peripheral blood B lymphoblasts (NC-37) were obtained from the American Type Culture Collection (ATCC, Rockville, MD) and grown in 90% RPMI 1640 media supplemented with 10% fetal calf serum (GIBCO BRL Products, Burlington, Ontario, Canada). The cells were harvested at the exponential growth phase, washed 3× in serum-free Iscove’s modified Dulbecco’s medium (IMDM), aliquoted into sterile Eppendorf tubes at a concentration of 2 × 106cells/mL and incubated for 24 hours at 37°C and 5% CO2in the absence (control) or presence of cytokines (IL-6, IL-10, TNF-α). Human recombinant IL-6 (Genetics Institute, Cambridge, MA), IL-10, and TNF-α (R & D Systems Inc, Minneapolis, MN) were added at final concentrations of 100 ng/mL, 100 ng/mL, and 10 ng/mL, respectively. The cell-conditioned media (supernatants) were collected, concentrated 10-fold using the Centricon concentrator (Amicon, Beverly, MA) and analyzed by zymography and reverse zymography. Serum-free media conditioned by KG-1 and baby hamster kidney (BHK) cell lines, which secrete MMP-2 and MMP-9,2 12 were used as a positive control for zymographic analysis.
Zymographic analysis.
Gelatinolytic activities were assessed under nonreducing conditions using modified sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Fifteen μL of supernatants mixed with 5 μL of loading buffer (0.16 mol/L Tris-HC1, 50% glycerol, 8% SDS, and 0.08% bromophenol blue) were applied onto a 10% polyacrylamide gel copolymerized with 2 mg/mL gelatin (Sigma, Oakville, Ontario, Canada). Electrophoresis was performed using a mini-PROTEAN II electrophoresis system (Bio-Rad Laboratories, Mississauga, Ontario, Canada) under constant voltage (150 V) for 3 hours at 4°C. The gels were washed 3× for 20 minutes each with 2.5% Triton X-100 (Sigma) to remove the SDS and to allow the electrophoresed enzymes to renature, before being incubated in zymography buffer (5 mmol/L CaC12 and 50 mmol/L Tris-HC1, pH 7.5) for 18 hours at 37°C. The gels were then stained with 0.05% Coomassie brilliant blue G-250 (Sigma) in 2.5:1:7 ethanol:acetic acid:water and destained with 2:1:7 isopropanol:acetic acid:water. Prestained standard high-range (47 to 201 kD) protein markers (Bio-Rad) were used to determine the molecular weights of the gelatinases. Gels were laminated using BioDesign GelWrap (BioDesign Inc, Carmel, NY), photographed and scanned using a ScanJet 3c scanner and DeskScan II software (Hewlett Packard). The intensity of the bands in zymography was quantified using Scion Image for Windows software (Scion Corp, Frederick, MD).
Reverse zymography.
TIMP activities in cell-conditioned media were analyzed using reverse zymography as described by us previously.2 Briefly, the samples were electrophoresed in 0.1% SDS, 12% polyacrylamide gels containing 1 mg/mL gelatin. Conditioned medium from BHK cell line was added as a source of MMP-2. After electrophoresis the gels were washed overnight with 2.5% Triton X-100, incubated in 50 mmol/L Tris/HCl, pH 7.5, and 5 mmol/L CaCl2 for 16 to 18 hours at 37°C, then stained and destained. Dark blue bands against a pale blue background represent TIMP activity. Conditioned medium from BHK cells was used as a positive control for TIMP activities.
Matrigel invasion assay.
In vitro cell migration was determined in the Matrigel-based assay as described by us.45 Briefly, 13-mm polycarbonate filters of 8-μm pore size (Costar/Nucleopore, Toronto, Ontario, Canada) were coated with 25 μg of Matrigel. The lower compartments of the modified (blind well) Boyden chambers (Neuro Probe Inc, Gaithersburg, MD) were filled with IMDM conditioned by bone marrow fibroblasts and supplemented with 0.1% bovine serum albumin (BSA). Matrigel-coated filters were placed between the upper and lower compartments. Raji and Jurkat cells were suspended in IMDM/0.1% BSA at a concentration of 1.5 × 106 cells/mL then loaded onto the upper chambers. To assess whether cytokines modulate migration through Matrigel, cells were also preincubated at 37°C with 100 ng/mL IL-6 or IL-10 for 3 to 18 hours at 37°C in 5% CO2. Cells that had migrated through the Matrigel-coated filters were recovered from the lower compartments after 3 hours and counted using a Neubauer hemocytometer (VWR Scientific, Mississauga, Ontario, Canada). Percentage of cell invasion was calculated from the ratio of the number of cells recovered from the lower compartment to the total number of cells loaded in the upper compartment. Each experiment was performed using at least five chambers for each cell sample, and repeated at least 3×.
To further examine whether MMP-2 and/or MMP-9 are implicated in IL-6–stimulated invasion, the cells were incubated overnight with IL-6 and then treated for 1 hour with the following MMP inhibitors: 10 μg/mL anti-MMP-2 monoclonal antibody, 10 μg/mL anti–MMP-9 antibody (both from Oncogene Research Products, Cambridge, MA) and 10 μg/mL recombinant human TIMP-1 (gift from Dr A. Docherty, Celltech Pharmaceuticals, Slough, England) before loading onto Boyden chambers.
Statistical analysis.
Correlation between various measurements was established by Kendall’s rank correlation and Kruskal-Wallis rank sum tests. Significant differences between means of paired samples were determined using Student’s t-test (Microsoft Excel, Redmond, WA) and aP value < .05 was considered statistically significant.
RESULTS
Expression of cytokines, growth factors, MMPs, and TIMP-1 in NHL.
The results of RT-PCR analysis of the mRNA expressions of MMP-9, MMP-2, TIMP-1, IL-6, IL-10, and TNF-α in the group of 32 NHLs are shown in Fig 1. In some cases two distinct IL-10 bands were detected by RT-PCR analysis. We have established that both bands represent IL-10 transcripts (data not shown). The significance of this finding is uncertain and will be investigated further. Statistical analysis was based on the densitometric analysis of both bands. The data for each MMP, TIMP-1, cytokines, and growth factors are shown in Table1. High-grade NHL showed generally higher MMP-9, IL-6, and IL-10 levels than low-grade tumors. Considerable variation was, however, noted between the individual cases.
MMP, TIMP-1, Cytokine, and Growth Factor Expressions in NHL
| MMP-9 . | MMP-2 . | TIMP-1 . | IL-6 . | IL-10 . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIN | : | 0.189 | MIN | : | 0.129 | MIN | : | 0.495 | MIN | : | 0.000 | MIN | : | 0.032 |
| 1st Qu | : | 0.485 | 1st Qu | : | 0.337 | 1st Qu | : | 2.709 | 1st Qu | : | 0.457 | 1st Qu | : | 0.345 |
| MEDIAN | : | 1.094 | MEDIAN | : | 0.577 | MEDIAN | : | 3.796 | MEDIAN | : | 0.775 | MEDIAN | : | 0.573 |
| MEAN | : | 3.123 | MEAN | : | 0.708 | MEAN | : | 3.882 | MEAN | : | 1.112 | MEAN | : | 1.000 |
| 3rd Qu | : | 1.943 | 3rd Qu | : | 0.965 | 3rd Qu | : | 4.752 | 3rd Qu | : | 1.494 | 3rd Qu | : | 1.594 |
| MAX | : | 29.000 | MAX | : | 2.463 | MAX | : | 7.932 | MAX | : | 3.600 | MAX | : | 3.156 |
| MMP-9 . | MMP-2 . | TIMP-1 . | IL-6 . | IL-10 . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIN | : | 0.189 | MIN | : | 0.129 | MIN | : | 0.495 | MIN | : | 0.000 | MIN | : | 0.032 |
| 1st Qu | : | 0.485 | 1st Qu | : | 0.337 | 1st Qu | : | 2.709 | 1st Qu | : | 0.457 | 1st Qu | : | 0.345 |
| MEDIAN | : | 1.094 | MEDIAN | : | 0.577 | MEDIAN | : | 3.796 | MEDIAN | : | 0.775 | MEDIAN | : | 0.573 |
| MEAN | : | 3.123 | MEAN | : | 0.708 | MEAN | : | 3.882 | MEAN | : | 1.112 | MEAN | : | 1.000 |
| 3rd Qu | : | 1.943 | 3rd Qu | : | 0.965 | 3rd Qu | : | 4.752 | 3rd Qu | : | 1.494 | 3rd Qu | : | 1.594 |
| MAX | : | 29.000 | MAX | : | 2.463 | MAX | : | 7.932 | MAX | : | 3.600 | MAX | : | 3.156 |
| TNF-α . | IL-1β . | bFGF . | TGF-β . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MIN | : | 0.000 | MIN | : | 0.067 | MIN | : | 0.000 | MIN | : | 0.163 |
| 1st Qu | : | 0.521 | 1st Qu | : | 0.494 | 1st Qu | : | 0.654 | 1st Qu | : | 0.600 |
| MEDIAN | : | 0.847 | MEDIAN | : | 0.981 | MEDIAN | : | 1.602 | MEDIAN | : | 0.754 |
| MEAN | : | 1.068 | MEAN | : | 1.296 | MEAN | : | 1.750 | MEAN | : | 0.783 |
| 3rd Qu | : | 1.241 | 3rd Qu | : | 1.713 | 3rd Qu | : | 2.790 | 3rd Qu | : | 0.889 |
| MAX | : | 4.264 | MAX | : | 4.891 | MAX | : | 4.169 | MAX | : | 2.036 |
| TNF-α . | IL-1β . | bFGF . | TGF-β . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MIN | : | 0.000 | MIN | : | 0.067 | MIN | : | 0.000 | MIN | : | 0.163 |
| 1st Qu | : | 0.521 | 1st Qu | : | 0.494 | 1st Qu | : | 0.654 | 1st Qu | : | 0.600 |
| MEDIAN | : | 0.847 | MEDIAN | : | 0.981 | MEDIAN | : | 1.602 | MEDIAN | : | 0.754 |
| MEAN | : | 1.068 | MEAN | : | 1.296 | MEAN | : | 1.750 | MEAN | : | 0.783 |
| 3rd Qu | : | 1.241 | 3rd Qu | : | 1.713 | 3rd Qu | : | 2.790 | 3rd Qu | : | 0.889 |
| MAX | : | 4.264 | MAX | : | 4.891 | MAX | : | 4.169 | MAX | : | 2.036 |
MMP, TIMP-1, cytokine, and growth factor expressions were measured as described in the Materials and Methods. Minimum (min), maximum (max) values, median and mean are shown in the table.
Correlation between expression of cytokines, growth factors, and of MMPs and TIMP-1 in NHL.
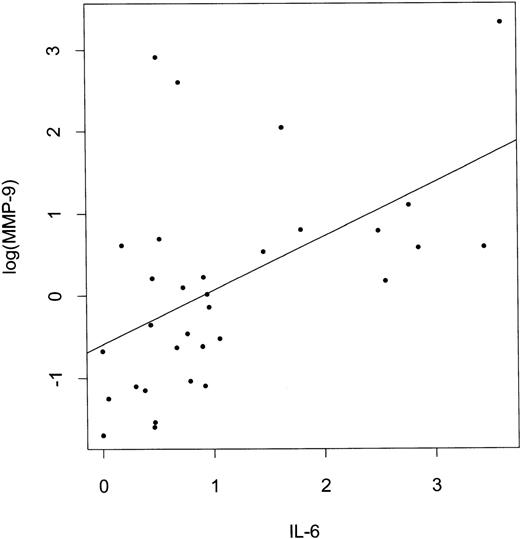
The Kendall’s rank correlation tau and P values are shown in Table 2. The strongest positive correlation was observed between RNA expression of MMP-9 and IL-6 (Fig2), followed by correlation between TIMP-1 and IL-6. MMP-9 expression also correlated with IL-10, and a weak correlation between MMP-9 and IL-1β was observed. MMP-2 RNA expression also correlated with IL-6 RNA levels, as well as with TGFβ and IL-1β. MMP-9, IL-6, and IL-10 expression correlated with NHL grade and was higher in the high-grade tumors. Expression of IL-6 correlated with IL-10 expression (correlation tau = .292, P = .019) and expression of both IL-6 and IL-10 correlated more significantly with MMP-9 expression than expression of each of these cytokines alone (tau=0.443, P = .0004).
Correlations Between Cytokine, Growth Factor, MMP, and TIMP-1 Expression in NHL
| . | IL-6 . | IL-10 . | TNF-α . | IL-1β . | bFGF . | TGFβ . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tau . | P . | Tau . | P . | Tau . | P . | Tau . | P . | Tau . | P . | Tau . | P . | |
| MMP-9 | 0.417 | .0008* | 0.335 | .0071* | 0.159 | .200 | 0.262 | .035* | 0.179 | .149 | 0.0685 | .581 |
| MMP-2 | 0.276 | .026* | −0.004 | .974 | 0.135 | .277 | 0.262 | .035* | 0.208 | .095 | 0.270 | .030* |
| TIMP-1 | 0.369 | .003* | 0.202 | .105 | 0.075 | .549 | 0.097 | .436 | 0.179 | .149 | 0.194 | .120 |
| . | IL-6 . | IL-10 . | TNF-α . | IL-1β . | bFGF . | TGFβ . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tau . | P . | Tau . | P . | Tau . | P . | Tau . | P . | Tau . | P . | Tau . | P . | |
| MMP-9 | 0.417 | .0008* | 0.335 | .0071* | 0.159 | .200 | 0.262 | .035* | 0.179 | .149 | 0.0685 | .581 |
| MMP-2 | 0.276 | .026* | −0.004 | .974 | 0.135 | .277 | 0.262 | .035* | 0.208 | .095 | 0.270 | .030* |
| TIMP-1 | 0.369 | .003* | 0.202 | .105 | 0.075 | .549 | 0.097 | .436 | 0.179 | .149 | 0.194 | .120 |
Abbreviation: tau, Kendall’s rank correlation tau.
Statistically significant P-value.
Correlation between log(MMP-9) and IL-6 expression in NHL. IL-6 and log(MMP-9) values represent densitometric measurements of the intensities of PCR bands shown in Fig 1 (methodology described in Materials and Methods).
Correlation between log(MMP-9) and IL-6 expression in NHL. IL-6 and log(MMP-9) values represent densitometric measurements of the intensities of PCR bands shown in Fig 1 (methodology described in Materials and Methods).
IL-6 stimulates MMP-2 and MMP-9 secretion in lymphoma cell lines.
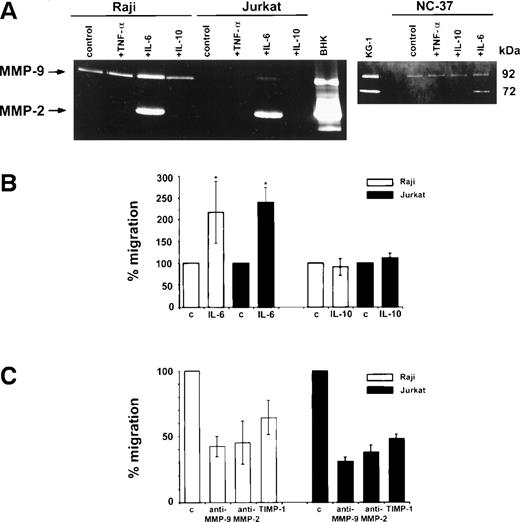
MMP-9 was found to be constitutively secreted into serum-free media by Raji and NC-37 cells but not by the Jurkat cell line and none of these cell lines secreted MMP-2 without a stimulus (Fig3A). However, after incubation with IL-6, both MMP-2 and MMP-9 activities were detected in supernatants of Raji, Jurkat, and NC-37 cells by gelatin zymography. Densitometric analysis showed that MMP-9 secretion by Raji cells was stimulated by IL-6 (1.75-fold). IL-10 and TNF-α did not stimulate MMP-9 or MMP-2 secretion by Raji, NC-37, or Jurkat cells.
Zymographic analysis of media conditioned by Raji, Jurkat, and NC-37 cell lines stimulated with cytokines and the in vitro invasion assay. (A) Zymograms of media conditioned by cells incubated for 16 hours at 37°C and 5% CO2 in serum-free IMDM in the absence (control) or presence of a cytokine. Zymography was performed using 10% polyacrylamide gels containing 2 mg/mL gelatin as described in Materials and Methods. The final concentrations of cytokines were 10 ng/mL TNF-, 100 ng/mL IL-6, and 100 ng/mL IL-10. Media conditioned by BHK and KG-1 cells were used as standards showing the positions of the 92 kD (MMP-9) and 72 kD (MMP-2) activities in the gels. (B) Matrigel invasion by Raji and Jurkat cells after stimulation with IL-6 and IL-10 or without (control) as described in Materials and Methods. The asterisks * indicate statistically significant differences in the percentage of migration (P = .046 for Raji and P= .0002 for Jurkat) in the presence of IL-6 versus control (without IL-6). (C) Effect of MMP inhibitors on the percentage of migration of IL-6–stimulated Raji and Jurkat cells. The final concentrations of anti–MMP-9 and anti–MMP-2 antibodies and recombinant TIMP-1, as well as the preincubation conditions are described in Materials and Methods. The basal migration of Raji and Jurkat cells was set at 100% (control) and the percentages of migration in the presence of the different inhibitors are represented as mean ± standard deviation relative to the control.
Zymographic analysis of media conditioned by Raji, Jurkat, and NC-37 cell lines stimulated with cytokines and the in vitro invasion assay. (A) Zymograms of media conditioned by cells incubated for 16 hours at 37°C and 5% CO2 in serum-free IMDM in the absence (control) or presence of a cytokine. Zymography was performed using 10% polyacrylamide gels containing 2 mg/mL gelatin as described in Materials and Methods. The final concentrations of cytokines were 10 ng/mL TNF-, 100 ng/mL IL-6, and 100 ng/mL IL-10. Media conditioned by BHK and KG-1 cells were used as standards showing the positions of the 92 kD (MMP-9) and 72 kD (MMP-2) activities in the gels. (B) Matrigel invasion by Raji and Jurkat cells after stimulation with IL-6 and IL-10 or without (control) as described in Materials and Methods. The asterisks * indicate statistically significant differences in the percentage of migration (P = .046 for Raji and P= .0002 for Jurkat) in the presence of IL-6 versus control (without IL-6). (C) Effect of MMP inhibitors on the percentage of migration of IL-6–stimulated Raji and Jurkat cells. The final concentrations of anti–MMP-9 and anti–MMP-2 antibodies and recombinant TIMP-1, as well as the preincubation conditions are described in Materials and Methods. The basal migration of Raji and Jurkat cells was set at 100% (control) and the percentages of migration in the presence of the different inhibitors are represented as mean ± standard deviation relative to the control.
IL-6–stimulated Matrigel invasion by lymphoma cell lines is inhibited by recombinant TIMP-1 and anti-MMP-9 and MMP-2 antibodies.
After a 16-hour incubation with IL-6, both Raji and Jurkat cells showed increased in vitro invasion of Matrigel in comparison to the control (Fig 3B). The percentage of Raji and Jurkat cells migrating through the Matrigel increased significantly (P = .046 and P = .0002, respectively) after incubation with IL-6, but not after incubation with IL-10.
Moreover, IL-6–stimulated Matrigel invasion by Raji cells was significantly reduced by anti–MMP-9 and anti–MMP-2 antibodies and by recombinant human TIMP-1 to 42%, 45%, and 64%, respectively, relative to the control (Fig 3C). The migration of Jurkat cells was also inhibited by these inhibitors to 31%, 38%, and 48%, respectively, relative to the control (Fig 3C).
IL-6 induces MMP-9 and MMP-2 transcripts in lymphoid cell lines and has no effect on TIMP-1 production by these cells.
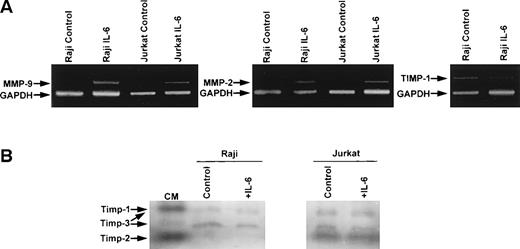
RT-PCR analysis of RNA extracted from Raji and Jurkat cell lines showed induction of MMP-9 and MMP-2 mRNA in cells stimulated with IL-6 (Fig4A). Neither MMP-9 nor MMP-2 transcripts were detected in unstimulated Raji or Jurkat cell lines, whereas both transcripts were observed after 6 hours of IL-6 stimulation. TIMP-1 transcripts were detected in Raji and Jurkat cells, but no induction of TIMP-1 mRNA was observed with IL-6 (Fig 4A). Reverse zymography showed TIMP-1 activity in both Raji and Jurkat cells (Fig 4B). In addition to TIMP-1 protein, TIMP-2 was also detected in Jurkat cells and TIMP-3 was present in both cell lines. No induction of TIMP activity was observed after treatment with IL-6 (Fig 4B).
RT-PCR and reverse zymographic analyses of RNA extracted from Raji and Jurkat cells and media conditioned by Raji and Jurkat cell lines stimulated with IL-6. (A) RNA extracted from Raji and Jurkat cell lines was analyzed for the expression of MMP-9, MMP-2, and TIMP-1 by RT-PCR as described in the Materials and Methods. Cells were cultured in serum-free media with and without IL-6 (100 ng/mL). Each analysis was internally controlled by inclusion of GAPDH primers. (B) Reverse zymogram of conditioned media from Raji and Jurkat cell lines stimulated 100 ng/mL IL-6. Conditioned medium (CM) from BHK cells was used as a positive control for TIMP activities.
RT-PCR and reverse zymographic analyses of RNA extracted from Raji and Jurkat cells and media conditioned by Raji and Jurkat cell lines stimulated with IL-6. (A) RNA extracted from Raji and Jurkat cell lines was analyzed for the expression of MMP-9, MMP-2, and TIMP-1 by RT-PCR as described in the Materials and Methods. Cells were cultured in serum-free media with and without IL-6 (100 ng/mL). Each analysis was internally controlled by inclusion of GAPDH primers. (B) Reverse zymogram of conditioned media from Raji and Jurkat cell lines stimulated 100 ng/mL IL-6. Conditioned medium (CM) from BHK cells was used as a positive control for TIMP activities.
DISCUSSION
There is growing evidence of the complexity of the mechanisms regulating MMP and TIMP expression and their activity. This is due not only to the fact that different cell types respond differently to stimulation by cytokines and growth factors, but also due to formation of TIMP/MMP complexes, which once formed, may assume new biological activity.46 Because MMP-9 and TIMP-1 had previously been shown by us to be overexpressed in a subset of human high-grade NHL, which is associated with a poor clinical outcome,3-6 in this study we decided to evaluate NHL for the coexpression of cytokines and growth factors. Our primary goal was to look for possible associations between overexpression of MMP-9 and TIMP-1 in relation to the expression of cytokines and growth factors in lymphoma tissue obtained from NHL patients. Assessment of the expression of IL-1β, IL-6, IL-10, TNF-α, TGFβ, and bFGF mRNA in the lymphoma tissue showed the strongest positive correlation between IL-6 and MMP-9, followed by correlations between the expression of TIMP-1 and IL-6, MMP-9 and IL-10, and MMP-2 and IL-6. This is an important finding in the setting of human NHL because the overexpression of both IL-6 and IL-10 in these hematological malignancies has been postulated to adversely affect prognosis.47-49 In this study the levels of IL-6 transcripts also correlated with the levels of IL-10 transcripts, supporting a hypothesis that in NHL these two cytokines may act as cooperative factors.44 IL-6 has been implicated in the pathogenesis of many human diseases50-52; however, its role in MMP and TIMP regulation in human malignant lymphoid cells has not been described. Recently, it has been shown that IL-6 has no effect on MMP-2 and MMP-9 production by myeloma cells.30IL-10 is known to stimulate TIMP-1 and inhibit MMP production by human mononuclear phagocytes,31, 40 but in our study no correlation between IL-10 and TIMP-1 production in NHLs was found.
We next investigated the functional role of IL-6 and IL-10 in the induction of gelatinases (MMP-2 and MMP-9) in cultures of human B- and T-cell lines (Raji, NC-37, and Jurkat) and evaluated whether stimulation with these cytokines would lead to increased ECM degradation. We chose lymphoma cell lines that in our previous studies showed low or undetectable secretion of MMP-2 and MMP-9 and low or no ability to destroy ECM in vitro.2 These cells produce only very small amounts of IL-10 and IL-6 constitutively, and Raji cells have been shown to have a small number of IL-6 receptors.53-56 In the present study, Raji, Jurkat, and NC-37 cell lines were stimulated with IL-6, IL-10, and TNF-α, because we have recently reported that this last cytokine greatly increases both MMP-9 and MMP-2 secretion by bone marrow CD34+cells.57 Because plasma levels of these cytokines vary considerably in different non-Hodgkin’s lymphomas and other hematological states47, 48, 58 the concentrations of these cytokines for our in vitro experiments were chosen based on this fact and according to previous reports.26 57-59 We found that IL-6 stimulated production of both MMP-2 and MMP-9 by all three lymphoid cell lines, indicating that this cytokine may play a direct role in the stimulation of MMP production by malignant lymphomas. Moreover, IL-6, but not IL-10, significantly increased Matrigel invasion by these lymphoid cell lines and this Matrigel invasion was significantly reduced by recombinant human TIMP-1 and anti–MMP-9 and MMP-2 antibodies, suggesting that induction of gelatinases leads to ECM degradation by these cells. Inhibition of Matrigel invasion with both anti–MMP-9 and MMP-2 antibodies indicates that both gelatinases may mediate lymphoid cell transmigration through basement membranes.
The role of IL-6 in the pathophysiology of NHL has always been thought to be related to its growth-factor activity. This is the first report suggesting that in NHL IL-6 also induces MMP. In large-cell NHL, IL-6 has been shown to be produced by non-neoplastic cells, whereas the malignant lymphocytes expressed the IL-6 receptor.60Expression of IL-6 mRNA in the present study was measured in lymphoma tissues containing malignant lymphocytes as well as non-neoplastic cells, the latter including reactive lymphocytes, macrophages, endothelial cells, and fibroblasts. It is therefore likely that non-neoplastic cells were responsible for IL-6 production whereas the malignant lymphocytes responded to the stimulus. This is in agreement with our previous findings that MMP-9 is produced by large lymphoma cells2 3 and with the current observation that after stimulation with IL-6, gelatinase production increases in cultures of lymphoma cell lines. Our present study suggests that in NHL, IL-6 may have a dual function. In addition to its growth-promoting function, it may act through the stimulation of MMP production, increasing the ability of lymphoma cells to destroy ECM and spread throughout the body. In consequence, human NHL that overexpress both IL-6 and MMP may have more aggressive biological behavior.
TIMP-1 expression in NHL tissue correlated with IL-6 expression, but IL-6 did not increase TIMP-1 production in lymphoid cell lines. It is possible that in the NHL microenvironment IL-6 induces TIMP-1 production by cells other than lymphoid, eg, macrophages or fibroblasts.25,35 This concept is consistent with our previous study showing that, in lymphoma tissue, TIMP-1 is produced by stromal cells.5 Production of TIMP-1 by these cells may in turn protect B-lymphoid cells from apoptosis and induce their differentiation.17,18 These effects may explain the association between high TIMP-1 levels and the adverse prognosis of NHL.6 We propose that in patients with NHL, IL-6 may induce production of both gelatinases and TIMP-1 and both may adversely affect the clinical outcome, although they act through different mechanisms.
Taken together, these data indicate a causal relationship between increased IL-6 production in lymphomas and expression of MMP-2, MMP-9, and TIMP-1. The relationship between IL-10 and MMP-9 expression observed in human NHL was not apparent in the lymphoma cell lines studied. This likely reflects a requirement for IL-10 to act in combination with other growth factors and cytokines in the tumor microenvironment to induce MMP-9 transcription. In this study we evaluated MMP and TIMP-1 in lymphoma tissues and lymphoid cell lines. By comparison, normal human lymphoid cells such as T lymphocytes constitutively express MMP-9, and both MMP-2 and MMP-9 are induced by T-cell activation.61 Moreover, mixed T-lymphocytes (isolated from human blood) were shown to transmigrate across Matrigel, a process that is MMP-dependent and stimulated by IL-2.61Inducible MMP production by T-lymphocytes and macrophages has been implicated in various immunological processes; however, different mechanisms may operate in normal lymphocytes and neoplastic lymphoid cells.20 For example, MMP-9 and TIMP-1 production in T-cell lymphoma was shown to be induced during T-lymphoma/endothelial cell contact through the intercellular adhesion molecule-1/LFA-1 interaction.62 The mechanisms of MMP and TIMP regulation in normal and neoplastic cells of B origin are unknown. Based on studies of nonlymphoid cells it has been proposed that cell-specific basal and inducible expression of MMP and TIMP is regulated on the molecular level by several control elements. The family of Activator Protein-1 (AP-1) transcription factors has long been thought to play a major role in the transcriptional activation of MMP and TIMP promoters.63,64 Transactivation by cytokines and growth factors requires specific interactions of AP-1 with othercis-acting elements (eg, Ets/PEA3 binding sites) and these complex interactions control the transcription of MMP and TIMP in response to particular inducers and repressors.63 64 The molecular mechanisms of MMP-2 and MMP-9 induction by IL-6 are unknown, and further studies are needed to elucidate the transcriptional regulatory pathways in NHL.
It is of interest that in some lymphoma cell lines, MMP-9 is induced by EBV latent membrane protein 1, and IL-6 receptor in Burkitt’s lymphoma is upregulated by EBV infection in vitro.65, 66 Some of the Burkitt’s lymphoma cell lines were also described previously to overexpress MMP-9.11 A common pathogenetic link may exist between EBV infection, overexpression of IL-6 and its receptor, induction of MMP-9, and development of lymphoid malignancies. This would be of special importance in the lymphoproliferative disorders occurring in an immunocompromised host (eg, post-transplantation, human immunovirus [HIV] infection).
In conclusion, our findings imply the existence of a previously undescribed mechanism operating in the pathogenesis of human NHL. This may lead to new therapeutic approaches in the treatment of these disorders.
ACKNOWLEDGMENT
We acknowledge Maria Cobuhat and Adrian Dobrowsky (University of Alberta) and Ms Anita Martin (University of Calgary) for technical help; Lawrence S. Urbanski and Andrea L. Stabbler for assistance in computer imaging, as well as Susan Watson for her secretarial assistance.
Supported by a grant from the Medical Research Council of Canada to A.E.K. (MT-12706) and a grant from Canadian Blood Services R & D to A.J-W. D.R.E. is supported by the Norfolk and Norwich Big C Appeal.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal