Optimization of mobilization, harvest, and transduction of hematopoietic stem cells is critical to successful stem cell gene therapy. We evaluated the utility of a novel protocol involving Flt3-ligand (Flt3-L) and granulocyte colony-stimulating factor (G-CSF) mobilization of peripheral blood stem cells and retrovirus transduction using hematopoietic growth factors to introduce a reporter gene, murine CD24 (mCD24), into hematopoietic stem cells in nonhuman primates. Rhesus macaques were treated with Flt3-L (200 μg/kg) and G-CSF (20 μg/kg) for 7 days and autologous CD34+ peripheral blood stem cells harvested by leukapheresis. CD34+ cells were transduced with an MFGS-based retrovirus vector encoding mCD24 using 4 daily transductions with centrifugations in the presence of Flt3-L (100 ng/mL), human stem cell factor (50 ng/mL), and PIXY321 (50 ng/mL) in serum-free medium. An important and novel feature of this study is that enhanced in vivo engraftment of transduced stem cells was achieved by conditioning the animals with a low-morbidity regimen of sublethal irradiation (320 to 400 cGy) on the day of transplantation. Engraftment was monitored sequentially in the bone marrow and blood using both multiparameter flow cytometry and semi-quantitative DNA polymerase chain reaction (PCR). Our data show successful and persistent engraftment of transduced primitive progenitors capable of giving rise to marked cells of multiple hematopoietic lineages, including granulocytes, monocytes, and B and T lymphocytes. At 4 to 6 weeks posttransplantation, 47% ± 32% (n = 4) of granulocytes expressed mCD24 antigen at the cell surface. Peak in vivo levels of genetically modified peripheral blood lymphocytes approached 35% ± 22% (n = 4) as assessed both by flow cytometry and PCR 6 to 10 weeks posttransplantation. In addition, naı̈ve (CD45RA+and CD62L+) CD4+ and CD8+cells were the predominant phenotype of the marked CD3+ T cells detected at early time points. A high level of marking persisted at between 10% and 15% of peripheral blood leukocytes for 4 months and at lower levels past 6 months in some animals. A cytotoxic T-lymphocyte response against mCD24 was detected in only 1 animal. This degree of persistent long-lived, high-level gene marking of multiple hematopoietic lineages, including naı̈ve T cells, using a nonablative marrow conditioning regimen represents an important step toward the ultimate goal of high-level permanent transduced gene expression in stem cells.
THE DEVELOPMENT OF effective stem cell gene therapy protocols is central to the eventual success of this therapeutic modality. Until very recently, the use of retrovirus vectors to transduce primate hematopoietic progenitor cells has resulted in disappointingly low levels of in vivo engraftment.1-3 Efforts to improve this strategy have included development of novel transduction protocols using long-term culture of unfractionated marrow4,5 and delineation of growth factors required during ex vivo culture to enhance transduction while preserving long-term engraftment potential.6-9
Prolonged ex vivo expansion of genetically modified cells using interleukin-3 (IL-3), IL-6, and stem cell factor (SCF) allows enhanced proliferation of cells with enhancement of transduction of cycling cells, but has a negative effect on engraftment.6Transduction of long-term cultured unfractionated marrow has resulted in poor levels of engraftment in human trials.10 Another issue is that many hematopoietic stem cell gene transfer studies have relied on vectors encoding the neomycin resistance gene, where molecular techniques or antibiotic selection may be used to determine both in vivo and in vitro transduction efficiency.11,12There has been concern that neomycin might be toxic or that it may induce an immune response. Alternative gene-marking strategies have included the introduction of DNA that encodes for surface antigens that can be detected by flow cytometry.7 13-15 This has facilitated the use of flow cytometry to determine transduction efficiency and level of gene expression in individual cells. In addition, such transduced surface markers allow use of flow cytometry for enrichment of transduced populations.
In an attempt to optimize stem cell gene therapy strategies, we chose to use retrovirus vectors encoding for surface antigens as a rapid means to identify and track genetically labeled cells. Further optimization of this preclinical protocol includes the use of the combination of Flt3-ligand (Flt3-L) and granulocyte colony-stimulating factor (G-CSF) to mobilize CD34+ peripheral blood stem cells (PBSCs). Transduction of selected CD34+ cells was evaluated using 2 MFGS-based retrovirus vectors encoding human truncated nerve growth factor receptor (htNGFR)14 or the mCD24 surface antigen,13 in the presence of growth factors and facilitated by centrifugation. As a further modification, animals were irradiated with a low-morbidity sublethal regimen (320 to 400 cGy) for pretransplant marrow conditioning to enhance engraftment. This approach was based on observations in the murine model that this assists in the engraftment of congeneic hematopoietic progenitors without the significant increase in the morbidity and mortality associated with lethal irradiation.16 In those murine studies, the engraftment facilitating effect of low-dose irradiation was potentiated by administration of G-CSF to the animals for a few days before the irradiation,16 an effect that may be reproduced by the growth-factor mobilization regimens required for harvest of PBSC. Subsequent to autologous PBSC transplants, flow cytometry and polymerase chain reaction (PCR) were used to monitor the frequency of labeled cells in various subsets of peripheral blood. Our data show that using this protocol, including sublethal irradiation, we were able to achieve successful and persistent engraftment of transduced stem cells, resulting in gene expression in multiple hematopoietic lineages. Peak in vivo levels of granulocyte marking exceeded 60% in some animals at 3 to 6 weeks posttransplantation, and labeled cells (between 5% and 10%) were detectable for at least 6 months. This clearly shows that levels of gene marking sufficient for clinical applications of stem cell gene therapy can be attained using an optimized mobilization, transduction, and nonablative conditioning regimen. Subsequent to the conduct of our studies, 2 reports have appeared recently demonstrating signficant levels of long-term engraftment of transduced stem cells in rhesus macaques6and in baboons.7 Our studies complement those published studies by demonstrating, in addition, that a nonablative sublethal marrow irradiation conditioning is sufficient to ensure engraftment of transduced long-term repopulating stem cells.
MATERIALS AND METHODS
Animals.
Male rhesus monkeys, Macaca mulatta, mean weight 6.2 ± 0.2 kg, were housed in individual stainless steel cages in conventional holding rooms at the Veterinary Resources Department at University of Maryland Greenebaum Cancer Center animal facilities accredited by the American Association for Accreditation of Laboratory Animal Care. Research was conducted according to the principles enunciated in the Guide for the Care and Use of Laboratory Animals, prepared by the Institute of Laboratory Animal Resources, National Research Council.17
Rhesus leukapheresis procedure.
The leukapheresis protocol was a modification of that developed through the efforts of H. Cullis and R. Donahue18 performed under general anesthesia. Briefly, collection was accomplished using a small S25A separation chamber and a shunt chamber in the place of a collection chamber. A standard apheresis kit was installed in the CS3000+ blood separator (Baxter Healthcare Corp, Deerfield, IL). After autoprime, the roller clamps to the acid citrate dextrose (ACD), saline, and vent prime lines were closed to prevent hemodiluting the donor when using halt/irrigate. The return line was modified by tightly rolling and taping a 150-mL transfer pack and sterile-docking a male luer to the shortened outlet line. A blood component recipient set with a 170-μm filter and drip chamber was spiked into the modified 150-mL transfer pack and connected to the packed red blood cell line using a needle lock device. The blood component recipient set was connected to an 18-gauge Angiocath (Becton Dickinson, Franklin Lakes, NJ) placed in the saphenous vein of the donor. Hemostats were placed both on the standard, unused, return line and the inlet for the ACD line present on the draw line. The apheresis kit was primed with autologous blood that had been collected in citrate phosphate dextrose 3 weeks before the leukapheresis procedure. The donor received a dose of 100 U/kg heparin immediately before the procedure. The inlet line was connected to an indwelling 6.5-French catheter placed in the femoral artery. Blood was processed at the rate of 12 mL/min in automatic mode for a total of 3.0 times the animal’s calculated blood volume.
At the completion of the procedure, the product was collected and 4 mL of citrate phosphate dextrose (CPD) was added. Peripheral blood mononuclear cells (PBMNC) were collected by leukapheresis on day 7 of growth-factor administration. The collection day was selected based on the kinetic profile noted herein for CD34+ cells and colony-forming cells (CFCs) mobilized by the combined Flt3-L/G-CSF administration. Cells counts were performed on a Sysmex K-4500 (Long Grove, IL). The number of PBMNCs, CD34+ cells, and CFCs processed for each animal was evaluated, and the mean and standard error of the mean (SEM) were then determined. The total number of PBMNCs, CD34+ cells, or CFCs collected in the apheresis product was based on the complete blood cell count of the leukapheresis product multiplied by the total percentage of lymphocytes and monocytes, CD34+ cells, or CFCs within the product and the volume of the leukapheresis product.
Cytokine-induced mobilization, irradiation, and transplantation.
After baseline evaluations, the animals (n = 5) were administered Flt3-L (200 μg/kg/d) (Mobist; a gift from Immunex Corp, Seattle, WA) and G-CSF (20 μg/kg/d) (Neupogen; Amgen, Thousand Oaks, CA) in subcutaneous, 1-mL bolus injections once a day for 7 consecutive days up to the day of apheresis. After the apheresis collection of CD34+ cells, 2 animals continued to receive Flt3-Ligand/G-CSF for 3 additional days (10 days total) to follow the level of mobilization with continued administration of these agents. One animal was removed from the study after the leukapheresis procedure due to pulmonary complications associated with the induction of anesthesia required for that procedure. Those respiratory problems were detected following a difficult time inducing anesthesia before initiating the apheresis procedure. Anesthetic and respiratory problems continued during and after the apheresis. The attending veterinarian recommended removal from the study.
Monkeys, after a prehabituation period, were unilaterally irradiated in Lucite restraining chairs with 250 kVp X-radiation at 13 cGy/min in the posterior-anterior position, rotated 180° at the mid-dose (160 or 200 cGy) to the anterior-posterior position for completion of the total (320 or 400 cGy) midline tissue exposure. Dosimetry was performed using paired 0.5-mL ionization chambers, with calibration factors traceable to the National Institute of Standards and Technology. Animals were transfused with the 4-day cultured and transduced autologous CD34+ cells within 2 hours to 20 hours after the sublethal x-irradiation.19 This dose of radiation was well tolerated in that the animals continued to feed normally and none of the animals developed obvious infection. Additional details regarding the response to this dose of irradiation are provided in the Results section.
Hematologic recovery after irradiation and transplantation.
Peripheral blood was obtained from the saphenous vein of anesthetized primates (Ketaset, 10 mg/kg intramuscular; Fort Dodge Laboratories, Fort Dodge, IA) for complete blood counts and clonogenic assays. Assessment of hematologic evaluations has been previously described.19 Animals subjected to irradiation conditioning received clinical support, which consisted of antibiotics and/or fresh irradiated whole blood and fluids as needed. An antibiotic regimen was initiated prophylactically when the white blood cell count (WBC) was <1,000/μL and continued daily until the WBC was >1,000/μL for 3 consecutive days and an absolute neutrophil count (ANC) >500/μL had been attained. Gentamicin (Lyphomed, Deerfield, IL) (10 mg once daily) and Baytril (Bayer Corp, Shawnee Mission, KS) (10 mg once daily) were administered intramuscularly. Standard care for animals subjected to marrow-suppressive therapies was that fresh, irradiated (1,500 cGy60Co) whole blood (approximately 30 mL/transfusion) from a random donor pool (monkeys weighing >10 kg) would be administered when the platelet count was <20,000/μL and the hematocrit level was <18%. However, as will be noted in the Results, none of the animals required transfusions for irradiation-related marrow suppression.
Retrovirus vectors, immunoselection, and transduction of CD34+ cells.
For these studies, mCD24 antigen cDNA13 or htNGFR cDNA14 were each inserted into the MFGS retrovirus vector plasmid without any internal promotors or selective markers, and amphotropic producer clones selected by transfecting the ψ-CRIP packaging line and screening as previously described.20,21The titer of supernatant from the MFGS-mCD24 producer line was over 106 infective particles/mL, while the titer of the MFGS-htNGFR producer was more than half a log lower. Supernatant from both lines was negative for replication competent retrovirus. As previously described, the virus supernatant used for transduction was harvested over an 8-hour period from confluent producer cell cultures in the same serum-free medium (described below) used to culture the CD34+ cells.21
The apheresis product was subjected to ammonium chloride lysis of erythrocytes and the CD34+ cells were recovered by positive immunoselection using the Ceprate LC-234-Biotin Kit (CellPro, Inc, Bothell, WA) according to the manufacturer’s instructions. CD34+ cells were enumerated using flow cytometry employing the same biotin-labeled antibody as used in the Ceprate kit for primary labeling followed by secondary labeling with fluorescein isothiocyanate (FITC)- or phycoerythrin (PE)-conjugated streptavidin (Becton Dickinson, San Jose, CA). Culture of CD34+ cells was initiated by seeding 0.8 × 106 cells per well into 6-well culture plates containing 5 mL of CD34 cell growth media (X-VIVO 10 media; BioWhittaker, Walkersville, MD) supplemented with 1% human serum albumin (HSA; Sigma, St Louis, MO) and 3 recombinant human growth factors (50 ng/mL PIXY321 [IL-3/granulocyte-macrophage (GM)-CSF fusion protein], 100 ng/mL Flt3-L [both PIXY321 and Flt3-L are gifts from Immunex Corp], and 50 ng/mL SCF [R&D Systems, Minneapolis, MN]).
The first transduction occurred at 16 hours (the morning after initiation of the culture). The CD34+ cells from all the wells were pooled, pelleted by centrifugation, and resuspended in an equal volume of a 1:1 mixture of neat, serum-free viral supernatant and fresh serum-free culture medium, where the final mixture contained 6 μg/mL protamine (Sigma, St Louis, MO), 1% HSA, and the 3 growth factors at the concentrations noted above. The cells were redistributed at 5 mL per well to the same 6-well plates.
The covered and tape-sealed plates were then centrifuged in flat-bed carriers for 20 minutes at 1,200g at 32°C (spinocculation). About 4 mL of the supernatant was removed from each well and the cells immediately resuspended in situ in a new aliquot of diluted viral supernatant and centrifuged as before. This was repeated a third time and after the third centrifugation, cells were not diluted or disturbed while plates were incubated for another 3 to 4 hours at 37°C and 7% CO2. Transduction media was then exchanged for fresh PBSC growth media and the CD34+ cells were then incubated overnight for 18 hours and the transduction procedure repeated on a second, third, and fourth day. After the fourth day of transduction, the autologous transduced CD34+ cells were washed free of medium, suspended in saline with 5% autologous serum, and injected intravenously into the irradiated animals. Small aliquots of transduced or control nontransduced (naı̈ve) CD34+ cells were maintained in liquid or semisolid agarose culture for further in vitro analysis.20 21
Colony assays.
Both transduced and naı̈ve CD34+ cells were plated into a solution of Iscove’s modified Dulbecco’s medium (IMDM) with 20% fetal calf serum (FCS), 2% low-melt agarose (Sea Kem; FMC, Rockland, ME). Specifically, cells were added to 3.5 mL of IMDM (GIBCO-BRL, Gaithersburg, MD) containing 20 ng/mL GM-CSF (R&D Systems), 50 ng/mL SCF (R&D Systems), 40 ng/mL G-CSF (Amgen), 2 U/mL Epogen (Amgen), and 20 ng/mL IL-3 (R&D Systems). Then, 0.4 mL of 2% agarose at approximately 40°C was added to the cell suspension and mixed well. Working rapidly before the agarose gelled, 1 mL/well was plated out into a 12-well plate. Cultures were incubated at 37°C for 10 to 14 days, counted, and stained. Agarose cultures were fixed using Citrate-Acetone-Methanol for 3 minutes at room temperature, and then washed 3 times with 1X TRIS-buffered saline (TBS). Agarose discs were then air-dried under airflow (overnight) and then washed again until colorless. For antibody staining, discs were first blocked with 100 μg/mL rat IgG (Pierce, Rockford, IL) in TBS/1% bovine serum albumin (BSA) for 30 minutes at room temperature. The block solution was aspirated, and 0.5 mL of rat anti-mouse mCD24 (10 μg/mL) (Pharmingen, San Diego, CA) in TBS/1% BSA was added and then incubated for 60 minutes at room temperature. Discs were then washed 4 times with TBS/1% BSA. Then 0.5 mL/well of Streptavidin-conjugated alkaline phosphatase (10 μg/mL) (Pierce) in TBS/1% BSA was added and incubated for 30 minutes at room temperature, followed by 5 washes. To develop the color reaction, 1 mL of Fast Red Substrate (Sigma) was added to each disc, and incubated for 45 minutes at room temperature, followed by 3 washes with distilled water. Agarose discs were then transferred to microscope slides and allowed to air dry. Hematoxylin was used to counterstain. Positive colonies developed a pink hue in addition to the blue color of the hematoxylin stain, while negative colonies were colored only by the hematoxylin stain.
Limiting dilution long-term culture-initiating cells (LTC-IC).
Bone marrow stroma was prepared by trypsinizing (0.05% Trypsin-0.53 mmol/L EDTA [GIBCO-BRL]) adherent stromal cells from 10- to 14-day-old primary rhesus bone marrow cultures in LTBMC media (Stem Cell Technologies, Vancouver, Canada) with the addition of 10−6 mol/L hydrocortisone (Sigma). Cells were gamma irradiated (15 Gy) and placed in flat-bottom 96-well plates at a concentration of 1 × 105 cells/mL. Irradiated stroma was maintained for up to 10 days at 37°C in LTBMC media before use. CD34+ cells were added to the wells at concentrations of 1, 3, 10, 30, 60, 120, and 240 cells per well, with 24 replicate wells for each condition. Plates were incubated at 37°C with 5% CO2 for an additional 5 weeks. Media exchanges were performed weekly. After 5 weeks, nonadherent cells were removed and 100 μL of 0.05% trypsin was added to each well. After 10 minutes at 37°C, adherent cells were collected via vigorous pipeting of each well and subsequent washing with an additional 200 μL of media. Adherent cells were pooled with the nonadherent cells and washed 3 times for each well. Supernatant was discarded, and the cells were resuspended in 200 μL of complete methylcellulose media (Stem Cell Technologies) with the addition of IL-3 (20 ng/mL), SCF (50 ng/mL), erythropoietin (3 U/mL), and GM-CSF (30 ng/mL), and replated in 96-well plates. Plates were returned to 37°C and scored at 14 and 21 days. Wells were scored as positive on the basis of an identifiable colony of more than 20 cells. The fraction of nonresponding wells was the number of wells in which no colony(s) grew after 3 weeks in methylcellulose and growth factors.22 LTC-IC frequency was estimated using the maximum likelihood analysis.23
PCR.
PCR analysis of the ψ packaging sequence was performed on colonies derived from LTC-IC cultures. Colonies were harvested into 0.2-mL PCR tubes containing 50 μL of phosphate-buffered saline. Colonies were spun down and the supernatant carefully removed using a separate stuffed sterile tip for each colony. Twenty-five microliters of lysis buffer (50 mmol/L KCL, 10 mmol/L Tris, 2.5 mmol/L MgCl, .5% Tween 20, 0.5% NP40, Proteinase K 20 μg/mL) was added to each colony-containing tube and samples were incubated for 1 hour at 55°C, followed by a 15-minute incubation at 95°C. One tenth or 2.5 μL of lysate was used in each PCR reaction. In general, 10 colonies or more were analyzed for each condition.
The PCR reaction was conducted in a 25-μL vol, in the M.J. Research PT-200 DNA Engine (M.J. Research, Watertown, MA). The cycling parameters were an initial denaturation of 92°C for 30 seconds, followed by 40 cycles of 92°C for 15 seconds, 64°C for 15 seconds, 72°C for 15 seconds, and a final extension of 72°C for 5 minutes. The primers (5′ CGC AAC CCT GGG AGA CGT CC) (3′ CGT CTC CTA CCA GAA CCA CAT ATC C) (GIBCO-BRL) produce a 134-bp product. Products were run on a 2.5% agarose gel containing ethidium bromide.
Semi-quantitative PCR analysis of mCD24 in the transduced progenitor cell product and on neutrophils purified from peripheral blood was performed on genomic DNA isolated using QIAmp Tissue kit (Quiagen, Stanford, CA). Detection was performed in 2 steps. In the first step, a pair of oligonucleotide primers were used to amplify vector sequence from genomic DNA. A labeled DNA probe produced by PCR from vector plasmid using oligonucleotide primers internal to the first primer set was then used to detect and quantify vector specific PCR product derived from the genomic DNA. The reaction to produce the mCD24 PCR-labeled probe from vector plasmid was performed in a 50-μL vol, with the following cycling parameters: Initial 3-minute denaturation at 96°C, followed by 40 cycles of 95°C for 30 seconds, 60°C for 30 seconds, 72°C for 30 seconds, and a final extension of 72°C for 5 minutes. Primers used for probe generation (mCD24-5′ TTA CTG CAA CAA AAC ATC TG) (mCD24-3′ AGA GAG AGC CAG GAC ACC AG) were used in a reaction containing fluoresceinated d-UTP, to generate a fluorescein-labeled probe that was purified on a PCR Select II column (5-Prime-3 Prime, Boulder, CO). The gene encoding mCD24 appears to be a single copy and intronless. Therefore, serial dilutions (2-fold) of wild-type mouse DNA into a background of rhesus DNA were generated to determine the mCD24 copy number as a control for experimental samples. The PCR to detect genomic mCD24 (external primers) used the following primers (5′ AGC GGC CAT GGG CAG AGC GAT GG) (3′ AGC ATC CCT AAC AGT AGA GAT ATA G) and the parameters were an initial 3-minute denaturation at 96°C, followed by 26 cycles of 95°C for 30 seconds, 60°C for 30 seconds, 72°C for 30 seconds, and a final extension of 72°C for 5 minutes. Twenty to 40 μL of the PCR reaction products were applied directly to Nytran Plus (Schleicher & Schuell, Keene, NH) via Manifold II slot apparatus (Schleicher & Schuell), followed by detection using the fluoresceinated probe with an ECL probe-amp protocol (ECL, Buckinghamshire, UK), including prehybridization, hybridization, peroxidase-labeled antifluorescein antibody incubation, and signal detection. It is important to note that in our unpublished studies (N.W.-T. and H.L.M., October 1997) the CD24 antigen coding sequence appears to be contained within 1 exon in both mouse and rhesus. We also found that rhesus CD24 coding sequence is identical to mouse in the regions defined by the outer primer set, but differed sufficiently in internal sequence that the probe created by the inner primers only detected the mouse sequence. These factors allowed use of known ratios of rhesus to mouse cells to prepare control DNA for the PCR reaction to establish standard curves used to evaluate copy number of transduced mCD24 sequence in rhesus blood cells.
To confirm the results of the blotting analysis described above, some samples of frozen pellets of unfractionated peripheral blood leukocytes subsequently were analyzed using a highly quantitative real-time PCR detection of the ψ packaging sequence using TaqMan reagents for fluorogenic 5′ nuclease assay analyzed on an ABI PRISM 7700 Sequence Detection System (PE Applied Biosystems, Foster City, CA). The PCR primers for the real-time amplification analysis overlapped with, but were distinct from, the ψ packaging sequence primers noted just above: 5′ GG AGA CGT CCC AGG GAC TTC and 3′ CCA GAA CCA CAT ATC CCT CCT CTA. The probe labeled with fluorescent reporter and quencher was: Fam-CCG TTT TTG TGG CCC GAC CTG A-Tamra. Amplification was performed using manufacturer-provided reagents following the standard recommended amplification conditions recommended by the manufacturer. K562 cells transduced to contain known copy numbers of MFGS vector as determined by flow cytometry were used as standards.
Flow cytometric analysis and cell sorting.
Antibodies used for immunophenotyping of rhesus cells included anti-CD3 (6G12) (kindly provided by J. Wong, Massachusetts General Hospital, Boston, MA),24 anti-CD4 (OKT4) (Ortho Diagnostics, Raritan, NJ), anti-CD8 (Leu-2a) (Becton Dickinson), anti-CD45RA (Becton Dickinson), anti-CD62L (Becton Dickinson), anti-CD34 (QBend-10) (Immunotech, Westbrook, ME), anti-CD56 (Becton Dickinson), anti-CD20 (Becton Dickinson), and anti-CD38 (OKT-10) (Ortho Diagnostics). Cells were stained according to the manufacturer’s instructions for staining and whole-blood lysis (Becton Dickinson). Four-color flow cytometric analysis of the cells was performed using a FACS Vantage (Becton Dickinson). For assays evaluating transduced expression of surface mCD24 or htNGFR, negative fluorescence gates were defined using fluorescent isotype-matched mouse nonspecific Ig. Furthermore, each assay used blood cells from control animals that were not transplanted to establish background signal by labeling with fluorescent antibody specific for mCD24 or htNGFR. Any positive background fluorescence detected in these control blood cells was subtracted from the signal detected in the blood cells from experimental animals to obtain the values reported in the Results section, with any resultant negative values reported as 0% positive. The background signal for mCD24 in control animal blood cells was consistently less than 1% or 2% of cells in the positive region, while that for htNGFR was higher, approaching 5% in some analyses. This hNGFR signal in control cells could represent a low level of expression of endogenous primate NGFR in peripheral blood cells.
Cytotoxic T-lymphocyte (CTL) assays.
PBMNC isolated from CPT vacutainer (Falcon/Becton Dickinson) tubes shipped overnight were resuspended in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 10 mm HEPES, 2 mmol/L L-glutamine, 50 IU/mL penicillin, and 50 μg/mL streptomycin in R10, then cultured with stimulator cells expressing mCD24 or htNGFR (gamma irradiated, 10,000 rads) at a responder to stimulator ratio of 10:1 in R10 medium. Stimulator cells consisted of transduced autologous, herpes papio-transformed B-lymphoblastoid cell lines (hp B-LCL) that had been developed from each animal in the study. Each lymphoblastoid cell line was transduced with the MFGS-htNGFR or MFGS-mCD24 retrovirus vector and sorted using a FACSVantage (Becton Dickinson) to obtain high-expressing cell lines. Recombinant IL-2 (donated by M. Gately, Hoffman LaRoche, Nutley, NJ) was added to a final concentration of 20 U/mL on day 4 of culture. CTL assays were performed 10 to 14 days after stimulation.
Target cells consisted of autologous or allogeneic hp B-LCL stably expressing mCD24 or htNGFR. Nontransduced hp B-LCL were used as negative controls. On the day of assay these were labeled with51Chromium (DuPont-NEN, Wilmington, DE), 100 μCi per 106 cells. Target cells (104 cells/well) were dispensed in duplicate for each effector:target (E:T) ratio into 96-well U-bottom plates (CoStar, Cambridge, MA). Cold-target inhibition was used to decrease background lysis. Cold targets consisted of unlabeled nontransduced autologous hp B-LCL and were used at a cold:hot target ratio of 15:1. Chromium release was assayed after a 5-hour incubation at 37°C in a 5% CO2 incubator. Plates were spun at 1,000 rpm for 7 minutes at 4°C, after which 30 μL of supernatant was harvested from each well into wells of a Luma Plate-96 (Packard, Meridan, CT) and allowed to dry overnight. Emitted radioactivity was measured in a 1450 MicroBetaPlus Liquid Scintillation Counter (Wallac, Turku, Finland). Spontaneous release was measured from wells containing only target cells and medium. Maximum release was measured from wells containing target cells and 0.1% Triton X-100 (Sigma). The percentage specific cytotoxicity was calculated as follows: [(Test Release − Spontaneous Release)/(Maximum Release − Spontaneous Release)] × 100. Spontaneous release of target cells was less than 25% in all assays. Effector to target ratios for which background lysis of control targets exceeded 20% were excluded from analysis. A specific lysis of greater than 10% seen at more than one E:T ratio as compared with a control nontransducedhp B-LCL was interpreted as significant.
RESULTS
Combination Flt3-L/G-CSF for mobilization of PBSCs.
PBSCs were mobilized via treatment of normal rhesus macaques with combined administration of Flt3-L and G-CSF for 7 consecutive days. The Flt3-L/G-CSF combination induced a significant increase in WBC and MNCs from respective baseline values of 6,600/μL and 4,600/μL to respective peak values of 53,000/μL and 13,100/μL at day 7 of administration, the day of apheresis (Fig 1). The CFC/mL (mean ± SEM) increased from a baseline value of 140/mL (±60 SEM) to 12,754/mL (±4,935 SEM) at day 7 of Flt3-L/G-CSF administration. After apheresis, the number of CFC/mL decreased to 5,690/mL. The 2 animals that continued to receive Flt3-L/G-CSF continued to mobilize CFCs. CFC/mL increased over the next 2 days to a peak of 14,042/mL (±7,599 SEM) on day 10, the final day of Flt3-L/G-CSF administration before irradiation of the animals in preparation for transfusion of the retrovirus-transduced cells. Peripheral blood CD34+ cell analysis was performed on 3 of the 5 animals administered Flt3-L/G-CSF at baseline and on day 7, the day of apheresis. Circulating CD34+ cells increased from a baseline value of 3/μL to 69/μL (±12.9 SEM) the day of apheresis.
Peripheral WBC and analysis of CFC for the 5 normal primates (mean ± SEM) administered Flt3-L and G-CSF. These growth factors were administered for 7 days (days 0-6) before apheresis on day 6. One animal was lost to the study after the apheresis and 2 animals continued to receive these growth factors for 3 additional days (days 7-9). Because the kinetics of changes in WBC and CFC did not differ in the animals receiving the additional days of growth factors, the data shown at each day are pooled from all the animals available for analysis on that day.
Peripheral WBC and analysis of CFC for the 5 normal primates (mean ± SEM) administered Flt3-L and G-CSF. These growth factors were administered for 7 days (days 0-6) before apheresis on day 6. One animal was lost to the study after the apheresis and 2 animals continued to receive these growth factors for 3 additional days (days 7-9). Because the kinetics of changes in WBC and CFC did not differ in the animals receiving the additional days of growth factors, the data shown at each day are pooled from all the animals available for analysis on that day.
The leukapheresis cellular product, CFC, and CD34+ cells.
A single, large-volume (3X blood volume) apheresis was performed on each animal on the seventh day of the Flt3-L/G-CSF administration (study day 6). A WBC count of 389.7 × 103/μL (±73.2 SEM), CFC/mL of 102,192 (±37,592 SEM), and a CD34+ cell count of 446.8/μL (±21.7 SEM) characterized the apheresis product. The PBMNC collection efficiency for the procedure was 0.35. The Flt3-L/G-CSF-induced mobilization and apheresis procedure provided sufficient CD34+ cells (range, 14.5 to 16.8 × 106) to initiate immunoselection and subsequent in vitro culture for gene transduction. The in vitro transduction protocol resulted in a 3.5-fold expansion in cell number by the day of transplantation. As noted in Table 1, 73 ± 51.4 × 106 cells, of which greater than 70% were CD34+, were reinfused into the animals after in vitro transduction, representing a mean of 12 × 106CD34+ cells/kg. These data show that Flt3-L and G-CSF treatment was effective at mobilizing stem cells to the peripheral compartment for subsequent harvesting via leukapheresis and immune selection.
Apheresis Product Cell Count
| Animal No. . | PBMNC in Apheresis Product . | CD34+Cells Immune Selected . | Yield of CD34+ Cells as of % PBMNC . | Transduced CD34+ Cells Infused . |
|---|---|---|---|---|
| 4657 | 1.45 × 1010 | 22.6 × 106 | 0.155 | 98 × 106 |
| 4630 | 2.6 × 1010 | 12.56 × 106 | 0.048 | (died) |
| 4694 | 1.7 × 1010 | 48.25 × 106 | 0.28 | 121 × 106 |
| 4649 | 1.2 × 1010 | 11.705 × 106 | 0.097 | 70 × 106 |
| J546 | 0.84 × 1010 | 10.9 × 106 | 0.13 | 2.5 × 106 |
| Mean ± SD | 1.56 ± 0.66 × 1010 | 21.2 ± 15.8 × 106 | 0.142 ± 0.08 | 73 ± 51.4 × 106 |
| Animal No. . | PBMNC in Apheresis Product . | CD34+Cells Immune Selected . | Yield of CD34+ Cells as of % PBMNC . | Transduced CD34+ Cells Infused . |
|---|---|---|---|---|
| 4657 | 1.45 × 1010 | 22.6 × 106 | 0.155 | 98 × 106 |
| 4630 | 2.6 × 1010 | 12.56 × 106 | 0.048 | (died) |
| 4694 | 1.7 × 1010 | 48.25 × 106 | 0.28 | 121 × 106 |
| 4649 | 1.2 × 1010 | 11.705 × 106 | 0.097 | 70 × 106 |
| J546 | 0.84 × 1010 | 10.9 × 106 | 0.13 | 2.5 × 106 |
| Mean ± SD | 1.56 ± 0.66 × 1010 | 21.2 ± 15.8 × 106 | 0.142 ± 0.08 | 73 ± 51.4 × 106 |
CD34+ cells placed in culture after immune selection from the apheresis product, and the number of cells transfused after 4 days of ex vivo culture and transduction.
Retrovirus transduction.
With animals 4630, 4657, and 4690, one half of the CD34-selected PBSC were transduced with MFGS-mCD24 and the other half transduced with MFGS-htNGFR. All of the CD34-selected PBSC from animals no. 4649 and J546 were transduced with only MFGS-mCD24. Flow cytometric analysis was used to assess the efficiency of transduction of CD34+cells. As shown in Table 2, the MFGS-mCD24 vector resulted in 43% ± 13% (n = 5) transduction of CD34+cells and the MFGS-htNGFR vectors resulted in 21% ± 2% (n = 2) transduction of CD34+ cells after 4 days of ex vivo transduction using spinocculation (infection performed using 3 sequential centrifugations of cells with vector each day for 4 consecutive days) and exogenous growth factors to optimize gene delivery, as assessed by flow cytometry. It is important to note that transduction efficiency was determined on cells that were CD34+ after transduction and not the bulk cell population.
Flow Cytometric Evaluation of Transduction Efficiency in the Ex Vivo–Cultured CD34+ Cells After Four Days of Spinocculation-Enhanced Transduction
| Animal No. . | CD34+ Cell Transduction Efficiency (%) . | |
|---|---|---|
| htNGFR+ Cells . | mCD24+ Cells . | |
| 4657 | 21 | 43 |
| 4630* | 23 | 61 |
| 4694 | 19 | 41 |
| 4649 | ND | 47 |
| J546 | ND | 25 |
| Mean ± SD | 21 ± 2 | 43 ± 13 |
| Animal No. . | CD34+ Cell Transduction Efficiency (%) . | |
|---|---|---|
| htNGFR+ Cells . | mCD24+ Cells . | |
| 4657 | 21 | 43 |
| 4630* | 23 | 61 |
| 4694 | 19 | 41 |
| 4649 | ND | 47 |
| J546 | ND | 25 |
| Mean ± SD | 21 ± 2 | 43 ± 13 |
Abbreviation: ND, not determined.
Animal died before reinfusion.
Before autologous transplantation of these cells, an aliquot of cells was also used to establish both colony-forming unit (CFU) assays and limiting dilution LTC-ICs to evaluate potential toxicity of the transduction protocol as well as the efficiency of gene transduction in primitive cells. As indicated in Materials and Methods, colonies expressing the transduced surface antigen in the CFU assays were assessed using an in situ labeling technique using enzyme-conjugated antibody to either mCD24 or htNGFR. An example of visual detection of mCD24+ colonies in a CFU assay is shown in Fig 2 where positive colonies have a pink hue. The frequency of transduced CFU was markedly different when the 2 vectors were compared, with overall higher transduction efficiency observed with the MFGS-mCD24 vector (Table 3). Although variations were observed in the transduction efficiency of myeloid and erythroid CFU, the overall observation was that the MFGS-mCD24 achieved higher levels of transduction efficiency in both myeloid and erythroid progeny than the MFGS-htNGFR vector (Table 3). Interestingly, when LTC-IC were analyzed, 2 observations were striking. Firstly, the transduction conditions resulted in an increase in the yield of LTC-IC compared with CD34+ cells analyzed before transduction as obtained in a limiting dilution assay, suggesting that the transduction protocol which included a 4-day culture period resulted in an in vitro expansion of LTC-IC. However, the frequency of LTC-IC in PBSC populations, even without the transduction protocol, was generally lower than what we have observed in bone marrow CD34+ cells (an approximate 8-fold fewer LTC-IC in PBSCs compared with bone marrow). Furthermore, a concerning aspect was the fact that no htNGFR+ LTC-IC could be detected via PCR in either of the 2 animals treated with this protocol (Table 4). Based on these data, a high level of background staining for htNGFR in flow cytometry analysis, and poor levels of in vivo engraftment of htNGFR-transduced cells, subsequent experiments were performed using only MFGS-mCD24 transduction of CD34+ progenitor cells.
Photomicrograph showing mCD24 expression in cells in colonies derived from transduced primate CFC using in situ labeling in agarose with anti-mCD24 followed by Streptavidin-conjugated alkaline phosphatase and development of the positive pink color reaction using Fast Red Substrate. In this photograph, colonies are counterstained blue-black with hematoxylin. Pink colonies are those that contain cells expressing the mCD24 transduced gene. In this low-magnification photomicrograph, 3 mCD24-positive pink colonies (P) and 2 of the several negative blue colonies (N) are indicated. No pink colonies were seen in any colony assays performed using progenitors that were not transduced (not shown). For actual visual scoring it is easier to count the total number of pink colonies in an agarose disk before counterstaining, where negative colonies show no color, and then score total colonies in the same disk after counterstaining.
Photomicrograph showing mCD24 expression in cells in colonies derived from transduced primate CFC using in situ labeling in agarose with anti-mCD24 followed by Streptavidin-conjugated alkaline phosphatase and development of the positive pink color reaction using Fast Red Substrate. In this photograph, colonies are counterstained blue-black with hematoxylin. Pink colonies are those that contain cells expressing the mCD24 transduced gene. In this low-magnification photomicrograph, 3 mCD24-positive pink colonies (P) and 2 of the several negative blue colonies (N) are indicated. No pink colonies were seen in any colony assays performed using progenitors that were not transduced (not shown). For actual visual scoring it is easier to count the total number of pink colonies in an agarose disk before counterstaining, where negative colonies show no color, and then score total colonies in the same disk after counterstaining.
Transduction Efficiency of CFU as Determined by Specific Antibody Staining of Colonies Derived From the Ex Vivo–Transduced CD34+ Cells
| Animal No. . | Transduced Marker . | Colony Type . | Marker Positive CFU/Total CFU . | % of CFU Transduced . |
|---|---|---|---|---|
| 4657 | mCD24 | CFU-GM | 89/324 | 27.5 |
| GEMM | 155/190 | 81.6 | ||
| BFU-e | 2/4 | 50 | ||
| CFU-e | 69/104 | 66.3 | ||
| htNGFR | CFU-GM | 12/196 | 6.1 | |
| GEMM | 22/119 | 18.5 | ||
| BFU-e | 3/22 | 13.6 | ||
| CFU-e | 38/101 | 37.6 | ||
| 4649 | mCD24 | CFU-GM | 66/170 | 39 |
| 4694 | mCD24 | CFU-GM | 40/173 | 23 |
| J546 | mCD24 | CFU-GM | 11/89 | 12.5 |
| 4630 | mCD24 | CFU-GM | 23/121 | 19 |
| Animal No. . | Transduced Marker . | Colony Type . | Marker Positive CFU/Total CFU . | % of CFU Transduced . |
|---|---|---|---|---|
| 4657 | mCD24 | CFU-GM | 89/324 | 27.5 |
| GEMM | 155/190 | 81.6 | ||
| BFU-e | 2/4 | 50 | ||
| CFU-e | 69/104 | 66.3 | ||
| htNGFR | CFU-GM | 12/196 | 6.1 | |
| GEMM | 22/119 | 18.5 | ||
| BFU-e | 3/22 | 13.6 | ||
| CFU-e | 38/101 | 37.6 | ||
| 4649 | mCD24 | CFU-GM | 66/170 | 39 |
| 4694 | mCD24 | CFU-GM | 40/173 | 23 |
| J546 | mCD24 | CFU-GM | 11/89 | 12.5 |
| 4630 | mCD24 | CFU-GM | 23/121 | 19 |
Abbreviations: CFU-GM, colony-forming unit-granulocyte, macrophage; GEMM, granulocyte, erythroid, macrophage, megakaryocyte; BFU-e, burst-forming unit-erythroid; CFU-e, colony-forming unit-erythroid.
Limit Dilution LTC-IC Evaluation of Transduction Efficacy and Toxicity of MFGS-mCD24 and MFGS-htNGFR Vectors
| LTC-IC Frequency and MFGS-mCD24 Transduction of LTC-IC . | |||
|---|---|---|---|
| Animal No. . | LTC-IC Frequency: Nontransduced PBSC . | LTC-IC Frequency: Transduced PBSC . | % of mCD24+ LTC-IC Among Transduced PBSC . |
| 4630 | 1 in 2,380 | 1 in 5,479 | 60 |
| 4694 | 1 in 558 | 1 in 421 | 40 |
| LTC-IC Frequency and MFGS-htNGFR Transduction of LTC-IC | |||
| Animal No. | LTC-IC Frequency: Nontransduced PBSC | LTC-IC Frequency: Transduced PBSC | % of htNGFR+ LTC-IC Among Transduced PBSC |
| 4630 | 1 in 2,380 | 1 in 20,000 | 0 |
| 4694 | 1 in 558 | 1 in 729 | 0 |
| LTC-IC Frequency and MFGS-mCD24 Transduction of LTC-IC . | |||
|---|---|---|---|
| Animal No. . | LTC-IC Frequency: Nontransduced PBSC . | LTC-IC Frequency: Transduced PBSC . | % of mCD24+ LTC-IC Among Transduced PBSC . |
| 4630 | 1 in 2,380 | 1 in 5,479 | 60 |
| 4694 | 1 in 558 | 1 in 421 | 40 |
| LTC-IC Frequency and MFGS-htNGFR Transduction of LTC-IC | |||
| Animal No. | LTC-IC Frequency: Nontransduced PBSC | LTC-IC Frequency: Transduced PBSC | % of htNGFR+ LTC-IC Among Transduced PBSC |
| 4630 | 1 in 2,380 | 1 in 20,000 | 0 |
| 4694 | 1 in 558 | 1 in 729 | 0 |
Hematologic response to irradiation and to the transplantation of cultured/transduced cells.
Animals exposed to 320 to 400 cGy total body irradiation (TBI) and subsequently transplanted with ex vivo cultured and transduced CD34+ progenitor cells experienced a modest degree of myelosuppression. The animals experienced afebrile neutropenia (ANC >500/μL) for a period of 10 days with a neutrophil nadir of approximately 100/μL. On average, the animals required 20.8 days to reach a neutrophil count of ≥500/μL, and were administered antibiotics for an average of 15.5 days. The antibiotics were administered as part of the prophylactic regimen for neutropenia and not because infection was evident in any of the animals. Only 1 of 4 animals experienced thrombocytopenia of <28,000/μL (minimum in that animal of 10,000/μL). The period of thrombocytopenia for the group averaged 2.5 days with a mean platelet nadir of 43,000/μL. None of the animals required transfusions. Also, during the 2 weeks after sublethal irradiation conditioning the animals continued to feed and maintain their weight. There were no immediate or delayed reactions to the intravenous administration of the autologous transduced CD34+ progenitor cells.
In vivo engraftment of labeled granulocytes, monocytes, and lymphocytes as assessed by flow cytometry.
Sequential samples of buffy coats were examined by flow cytometry for the surface expression of htNGFR or mCD24 to evaluate transduction efficiency. This procedure allowed good separation between mCD24+ and mCD24− cells (Fig 3). However, the data generated in 2 animals with the htNGFR vector suggested minimal expression of that marker that did not persist for more than a few weeks (data not shown). These data, in addition to the failure to detect htNGFR+LTC-IC, suggested that this marker was not efficiently transduced into the most primitive cells using the MFGS-htNGFR vector. This vector was not used to transduce PBSC from the last 2 animals studied and only the evaluation of expression of mCD24 is reviewed below.
Flow cytometry detection of mCD24+ cells in the white blood cell buffy coat fraction of peripheral blood at about 6 weeks posttransplant with CD34+ PBSC transduced with MFGS-mCD24 (shaded histograms). The erythrocytes in a sample of whole blood were lysed and the leukocytes labeled with anti-mCD24 as outlined in Materials and Methods. The level of background labeling is delineated by similar analysis of whole blood leukocytes from a nontransplanted control animal (open histograms).
Flow cytometry detection of mCD24+ cells in the white blood cell buffy coat fraction of peripheral blood at about 6 weeks posttransplant with CD34+ PBSC transduced with MFGS-mCD24 (shaded histograms). The erythrocytes in a sample of whole blood were lysed and the leukocytes labeled with anti-mCD24 as outlined in Materials and Methods. The level of background labeling is delineated by similar analysis of whole blood leukocytes from a nontransplanted control animal (open histograms).
The mCD24 expression by the whole peripheral blood leukocyte fraction and in specific leukocyte subsets is shown in Fig 4 (see page 2277) over 24 weeks after transplantation of transduced PBSC in 4 animals. To detect transduction of the multipotent progenitor cells that would be expected to give rise to multiple lineages of leukocytes, we specifically evaluated expression of mCD24 in granulocytes, monocytes, and lymphocyte subsets. Because of poor cross-reactivity of anti-human granulocyte antigens to rhesus granulocytes, we used the FSC/SSC profile (forward vside scatter) to delineate granulocyte subsets. Anti-CD14 was used to evaluate monocytes, while anti-CD2 and anti-CD20 were used to evaluate the T- and B-lymphocyte subsets, respectively. Because of differences in autofluorescence background, the negative quadrants were established separately for granulocytes, monocytes, and lymphocytes in stained samples from a nontransplanted control animal (data not shown). In addition, as outlined in Materials and Methods, background fluorescence seen with blood from control animals using fluorescent anti-mCD24 specific antibody was subtracted from the signal detected with blood cells from experimental animals in reporting the data.
Flow cytometry detection of mCD24 expression in different peripheral blood leukocyte subsets at selected times after transplantation with autologous CD34+ PBSC transduced with MFGS-mCD24. Shown are the whole leukocyte fraction (TOTAL; magenta squares), granulocytes (GRANS; light blue diamonds), monocytes (CD14; dark blue squares), T lymphocytes (CD2; green circles), and B lymphocytes (CD20; red triangles).
Flow cytometry detection of mCD24 expression in different peripheral blood leukocyte subsets at selected times after transplantation with autologous CD34+ PBSC transduced with MFGS-mCD24. Shown are the whole leukocyte fraction (TOTAL; magenta squares), granulocytes (GRANS; light blue diamonds), monocytes (CD14; dark blue squares), T lymphocytes (CD2; green circles), and B lymphocytes (CD20; red triangles).
In general, peak levels of mCD24 detected in the whole-leukocyte fraction peaked between 6 and 8 weeks, ranging from 6% to over 60% of leukocytes in the 4 animals. The highest percentages of marker positive cells were seen by flow cytometry at times when the animals were still mildly to moderately cytopenic, making it difficult to obtain large numbers of cells at those times for quantitative vector DNA analysis. The presence of mCD24 was first evident in circulating granulocytes at about 2 to 3 weeks (not shown), which is predicted based on the kinetics of myeloid progenitors in vivo. Interestingly, the mCD24 expressing granulocytes persisted at a level above background for 24 weeks, which would suggest that we had potentially labeled a more primitive, longer-lived progenitor subset. Peak granulocyte marking was 47% ± 32% (n = 4) and in 2 of the animals granulocyte marking was greater than 60% at 5 weeks, though these very high levels of marking did not persist. Monocytes examined for mCD24+ staining at sequential time points also indicated a persistent level of gene marking over the 24-week period, peaking at over 20% in 2 of the animals and at over 5% in the other 2 animals. Similar to the observations seen with granulocytes, monocytes expressing mCD24 were evident at the earliest time points examined and persisted throughout the observation period. In 3 of the 4 animals, greater than 5% of monocytes expressed mCD24 surface antigen at 24 weeks posttransplant. As expected, the marking of lymphocyte subsets generally was low at 3 to 5 weeks posttransplant, with the level of marking increasing at subsequent time points. Peak levels of lymphocyte marking approached 35% ± 22% (n = 4). As with granulocytes and monocytes, there was persistence of marking of lymphocytes through the 24 weeks shown in Fig4. More details about the marking of lymphocyte subsets will be noted below.
In vivo engraftment as assessed by semi-quantitative PCR.
Semi-quantitative PCR detection of mCD24 sequence was used to further evaluate the presence of genetically labeled cells in vivo. The frequency of labeled cells was determined in leukocyte pellets, and quantitated using mouse DNA to generate a standard PCR curve for mCD24 DNA sequence. A representative slot blot for animal no. 4657 is shown in Fig 5. Data from sequential time points from the four transplanted animals is summarized in Table 5. The frequency of labeled cells detected using semi-quantitative PCR was in a similar range to that detected using flow cytometry. The detection of more than 1 copy per cell may indicate the presence of more than 1 integrated retrovirus sequence in some cells. As shown in Table 5, high levels of gene marking were detected in animals 4694, 4649, and 4657, with kinetics that showed peak engraftment between 4 and 6 weeks posttransplantation. Durable engraftment at levels greater than 4% of leukocytes was evident. Animal no. J546 showed a low level of gene marking compared with the other three animals, and no signal was detected by PCR after week 12 posttransplantation. The gene marking detectable using either PCR or flow cytometry for the most part showed a reasonable correlation between the two methods. This observation would suggest that gene expression was persistent during the study period. Subsequent to these analyses, some stored samples of unfractionated blood leukocyte DNA were analyzed also using real time PCR detection of the ψ packaging sequence using TaqMan reagents for fluorogenic 5′ nuclease assay analyzed on an ABI PRISM 7700 Sequence Detection System as indicated in Materials and Methods. This analysis was consistent with the analysis obtained by the PCR slot-blot detection of mCD24 sequence, except that the absolute value for copy number was about half to one third of the values obtained from the slot blots (data not shown). It is not clear whether the lower values obtained using the real-time PCR system more accurately represents the correct copy number of vector sequence or whether this was a result of previous handling and storage of DNA samples that had been analyzed by the first method.
Semiquantitative PCR detection of mCD24 sequence in peripheral blood leukocyte genomic DNA from animal no. 4657 at sequential time points after autologous transplantation of CD34+ cells transduced with a retrovirus vector encoding mCD24. As outlined in the Materials and Methods and Results text, a semi-quantitative PCR standard curve was generated, using a method that allowed accurate quantitation of copy number over a range of 2 copies per cell down to about 0.005 copies per cells (about 0.5% of cells). The standard curve was generated using 2-fold serial dilutions of wild-type mouse DNA into a background of naı̈ve control rhesus DNA. Signal as shown in this auto-exposure was obtained by probing a slot blot of the PCR product with a labeled probe specific to mCD24 with detection of the signal by enhanced chemiluminescence detection on x-ray film.
Semiquantitative PCR detection of mCD24 sequence in peripheral blood leukocyte genomic DNA from animal no. 4657 at sequential time points after autologous transplantation of CD34+ cells transduced with a retrovirus vector encoding mCD24. As outlined in the Materials and Methods and Results text, a semi-quantitative PCR standard curve was generated, using a method that allowed accurate quantitation of copy number over a range of 2 copies per cell down to about 0.005 copies per cells (about 0.5% of cells). The standard curve was generated using 2-fold serial dilutions of wild-type mouse DNA into a background of naı̈ve control rhesus DNA. Signal as shown in this auto-exposure was obtained by probing a slot blot of the PCR product with a labeled probe specific to mCD24 with detection of the signal by enhanced chemiluminescence detection on x-ray film.
Semiquantitative PCR Analysis of mCD24 Sequence Copy Number in Genomic DNA of Peripheral Blood Leukocytes From Animals Transplanted With Autologous CD34+ PBSC Transduced Ex Vivo With MFGS Retrovirus Vector Encoding mCD24
| 4694 . | 4649 . | 4657 . | J546 . | ||||
|---|---|---|---|---|---|---|---|
| Week . | PCR . | Week . | PCR . | Week . | PCR . | Week . | PCR . |
| 4 | 0.468 | 4 | 0.348 | 4 | 0.858 | 4 | 0.109 |
| 6 | 0.117 | 5 | 0.191 | 6 | 1.434 | 5 | 0.304 |
| 9 | 0.034 | 6.5 | 0.065 | 8 | 1.455 | 8 | 0.159 |
| 14 | 0.045 | 7.5 | 0.025 | 13 | 0.982 | 12 | 0.084 |
| 8.5 | 0.023 | 17 | 0.054 | 16 | 0 | ||
| 9.5 | 0.047 | 22 | 0.920 | 20 | 0 | ||
| 10.5 | 0.018 | ||||||
| 11.5 | 0.053 | ||||||
| 16 | 0.010 | ||||||
| 19 | 0.008 | ||||||
| 21 | 0.010 | ||||||
| 24 | 0.005 | ||||||
| 28 | 0.004 | ||||||
| 4694 . | 4649 . | 4657 . | J546 . | ||||
|---|---|---|---|---|---|---|---|
| Week . | PCR . | Week . | PCR . | Week . | PCR . | Week . | PCR . |
| 4 | 0.468 | 4 | 0.348 | 4 | 0.858 | 4 | 0.109 |
| 6 | 0.117 | 5 | 0.191 | 6 | 1.434 | 5 | 0.304 |
| 9 | 0.034 | 6.5 | 0.065 | 8 | 1.455 | 8 | 0.159 |
| 14 | 0.045 | 7.5 | 0.025 | 13 | 0.982 | 12 | 0.084 |
| 8.5 | 0.023 | 17 | 0.054 | 16 | 0 | ||
| 9.5 | 0.047 | 22 | 0.920 | 20 | 0 | ||
| 10.5 | 0.018 | ||||||
| 11.5 | 0.053 | ||||||
| 16 | 0.010 | ||||||
| 19 | 0.008 | ||||||
| 21 | 0.010 | ||||||
| 24 | 0.005 | ||||||
| 28 | 0.004 | ||||||
The copy number indicated is relative to the control standards as indicated in Fig 5 and in Materials and Methods. The level of accurate quantitative detection of signal as the assay is set up is a copy number of about 0.005 (marking of about 0.5% of cells).
Gene marking of T-cell subsets.
The introduction of a marker gene by transduction into a true T-cell progenitor would be expected to result in both CD4+ and CD8+ progeny (and natural killer [NK] cells) expressing the gene. We examined the kinetics and frequency of the mCD24 antigen in CD4+ and CD8+ subsets and found that the peak expression of mCD24 in CD4+ and CD8+subsets was not necessarily coincident, although both subsets were labeled with a similar degree of efficiency (Fig 6).
Flow cytometry detection of mCD24 expression by peripheral blood CD3+CD4+ (▪) and CD3+CD8+ (◍) lymphocytes at selected times after transplantation with autologous CD34+ PBSC transduced with MFGS-mCD24.
Flow cytometry detection of mCD24 expression by peripheral blood CD3+CD4+ (▪) and CD3+CD8+ (◍) lymphocytes at selected times after transplantation with autologous CD34+ PBSC transduced with MFGS-mCD24.
As an additional parameter of the ability to transduce genes into primitive progenitors capable of giving rise to T lymphocytes in vivo, we examined expression of mCD24+ in peripheral blood CD3+ T lymphocytes having the CD45RA+CD62L+ phenotype of naı̈ve (nonmemory) lymphocytes (R1 gate, Fig 7, top panel, left dot plot). Remarkably, at the earliest time points that T-cell recovery began (about 3 weeks), we observed that the majority of such naı̈ve (CD3+ CD45RA+ CD62L+) cells were mCD24+ (Fig 7, top panel, right dot plot). This finding is consistent with the maturation of a transduced T-lymphocyte progenitor into a naı̈ve T lymphocyte, and suggests that the gene transduction protocol used in this study facilitated the introduction of the mCD24 marker gene into T-cell progenitors. The converse analysis was performed by gating on CD3+ cells that were either mCD24+ or mCD24− (R4 or R3 gate, respectively, in Fig 7, bottom panel, left dot plot). It is evident that the predominant phenotype of mCD24+ cells (gate R4) is that of naı̈ve T cells (CD3+ CD45RA+CD62L+), whereas the predominant phenotype of mCD24− cells (gate R3) is that of subsets of memory T cells (Fig 7, bottom panel, right upper and right lower dot plots, respectively). In addition, the presence of mCD24+naı̈ve T cells would imply that expression of this transduced gene used is not toxic or inhibitory to the T-cell differentiation processes.
Flow cytometry detection of mCD24 expression by peripheral blood CD3+ naı̈ve and memory T lymphocytes at about 3 to 4 weeks after transplantation with autologous CD34+ PBSC transduced with MFGS-mCD24. For all of the dot-plot analyses shown in this figure, an initial collection gate was established to analyze only CD3+ T lymphocytes. For subsequent analyses, naı̈ve T lymphocytes are defined as those simultaneously expressing CD62L (L-selectin Leu8) and CD45RA surface antigens. In all of the dot plots divided into 4 quadrants by lines defining positive from negative labeling by the indicated antibody, the numbers in those quadrants refer to the percent of the cells analyzed in that plot that fall within the indicated quadrant. In the top boxed panel a naı̈ve T-lymphocyte subset is defined in the lefthand dot plot by the R1 gate. When naı̈ve T cells in this R1 gate are further analyzed for expression of the transduced mCD24 gene product (MUCD24 on the vertical axis) in the righthand dot plot of the top boxed panel, almost all such cells are shown to be mCD24+. The FSC on the horizontal axis of this dot plot refers to forward scatter, a rough measure of cell size, and this parameter is used here merely to allow the presentation of the data in a dot-plot format. The converse analysis is shown in 3 dot plots contained within the bottom boxed panel of this figure. The lefthand dot plot in the bottom panel defines analysis gates containing mCD24+ T lymphocytes (R4) or mCD24− T lymphocytes (R3). As indicated by the arrows pointing from R4 or R3, cells contained within either of these gates were further analyzed to delineate naı̈ve T cells from memory T-cell subsets.
Flow cytometry detection of mCD24 expression by peripheral blood CD3+ naı̈ve and memory T lymphocytes at about 3 to 4 weeks after transplantation with autologous CD34+ PBSC transduced with MFGS-mCD24. For all of the dot-plot analyses shown in this figure, an initial collection gate was established to analyze only CD3+ T lymphocytes. For subsequent analyses, naı̈ve T lymphocytes are defined as those simultaneously expressing CD62L (L-selectin Leu8) and CD45RA surface antigens. In all of the dot plots divided into 4 quadrants by lines defining positive from negative labeling by the indicated antibody, the numbers in those quadrants refer to the percent of the cells analyzed in that plot that fall within the indicated quadrant. In the top boxed panel a naı̈ve T-lymphocyte subset is defined in the lefthand dot plot by the R1 gate. When naı̈ve T cells in this R1 gate are further analyzed for expression of the transduced mCD24 gene product (MUCD24 on the vertical axis) in the righthand dot plot of the top boxed panel, almost all such cells are shown to be mCD24+. The FSC on the horizontal axis of this dot plot refers to forward scatter, a rough measure of cell size, and this parameter is used here merely to allow the presentation of the data in a dot-plot format. The converse analysis is shown in 3 dot plots contained within the bottom boxed panel of this figure. The lefthand dot plot in the bottom panel defines analysis gates containing mCD24+ T lymphocytes (R4) or mCD24− T lymphocytes (R3). As indicated by the arrows pointing from R4 or R3, cells contained within either of these gates were further analyzed to delineate naı̈ve T cells from memory T-cell subsets.
Using a combination of anti-mCD24 and anti-CD20, we were able to detect gene marking in peripheral blood B lymphocytes. The kinetics of labeling of B cells varied between the 4 animals, and in animals no. 4649 and 4694 (Fig 4) we observed the highest level of B-cell labeling (about 40% of CD20+ cells) at 8 to 10 weeks posttransplantation.
In addition, we examined if NK progenitors had been labeled. In macaques, there is a distinct population of CD3−CD8+ NK cells. Flow cytometric analysis of this subset of cells showed high, early labeling of peripheral NK cells with the mCD24 antigen. The kinetics of labeled cells followed a similar trend as the granulocytes, which may reflect the inherent differences with respect to turnover rates for NK cells in CD3+ T lymphocytes. These data show clearly that CD4+ and CD8+ T lymphocytes, CD20+B lymphocytes, and NK cell progenitors can all be efficiently labeled, resulting in moderate levels of gene marking in circulating lymphocyte populations (data not shown for NK cells).
Gene marking of hematopoietic progenitors.
While in all 4 of the animals there was a high level of marking of some or all leukocyte subsets for at least 4 months, by 6 months there was persistence of marking but a significant decrease in the level of marking of some or all leukocyte subsets by both PCR and flow cytometry. Examination of bone marrow aspirates in 3 of the 4 animals at these late time points showed a low level of gene marking (<5%) (n = 3) in CD34+ cells, consistent with the declining levels of marking seen in the peripheral blood analysis. Interestingly, a minor fraction (<1%) of CD34+CD38−cells remained mCD24+ as determined by flow cytometry (data not shown). This suggests that a small number of phenotypically very primitive hematopoietic progenitors capable of maintaining hematopoiesis for more than 6 months were transduced ex vivo, and had engrafted after transplantation.
Evaluation of a cytotoxic T-cell response to mCD24.
In several gene-therapy protocols, development of a CTL response to foreign genes has resulted in a reduction of cells expressing foreign genes. Because the mCD24 protein expressed at the cell surface as a result of transduction mediated gene marking in our studies was of murine origin, we evaluated all 4 animals for a CTL response to mCD24 and the initial 2 animals for a CTL response to htNGFR. Using the sensitive in vitro stimulation protocol outlined in Materials and Methods, we were unable to detect a cellular immune response to either mCD24 or htNGFR in 3 animals for the duration of the study (6 months). In 1 animal (no. J546), we detected a CTL response to mCD24 at 15 weeks after transplantation, and this response persisted in measurements made at 16 and 23 weeks posttransplantation (Fig8). This CTL response was major histocompatability complex restricted, because allogeneic cells expressing mCD24 antigen (transducedhp B-LCL derived from a control animal, no. 4664, used as a target of CTL responses by no. J546 stimulated mononuclear cells) were not lysed (Fig 8). It should be noted that the level of mCD24 expression in animal J546 granulocytes did not change substantially at the point that a CTL response was first detected, but declined thereafter (see Figs 4 and 6). Additionally, it is of note that PCR detection of mCD24 sequence in leukocytes from animal no. J546 was below the level of the standard curve (<0.5%) after 16 weeks (see Table 5).
CTL lysis of the 51Chromium-labeled targethp B-LCL indicated in the legend. Effector cells were mononuclear cells derived from animal no. J546 at the times indicated on the horizontal axis after transplantation with autologous CD34+ PBSC transduced with MFGS-mCD24. This animal was not transplanted with any CD34+ PBSC transduced with MFGS-htNGFR. Controls included target autologous hp B-LCL transduced to express htNGFR and allogeneic hp B-LCL (from control animal no. 4664) transduced and selected to express mCD24 or htNGFR. All effector cells were stimulated with autologous hpB-LCL transduced with either mCD24 or htNGFR retrovirus vectors. CTL assays were performed using E:T ratios of 80:1 to 10:1, although the representative data shown in this figure were performed using an E:T ratio of 40:1. The percent lysis of targets indicated on the vertical axis was calculated by the formula given in Materials and Methods.
CTL lysis of the 51Chromium-labeled targethp B-LCL indicated in the legend. Effector cells were mononuclear cells derived from animal no. J546 at the times indicated on the horizontal axis after transplantation with autologous CD34+ PBSC transduced with MFGS-mCD24. This animal was not transplanted with any CD34+ PBSC transduced with MFGS-htNGFR. Controls included target autologous hp B-LCL transduced to express htNGFR and allogeneic hp B-LCL (from control animal no. 4664) transduced and selected to express mCD24 or htNGFR. All effector cells were stimulated with autologous hpB-LCL transduced with either mCD24 or htNGFR retrovirus vectors. CTL assays were performed using E:T ratios of 80:1 to 10:1, although the representative data shown in this figure were performed using an E:T ratio of 40:1. The percent lysis of targets indicated on the vertical axis was calculated by the formula given in Materials and Methods.
DISCUSSION
In our current study, a high level of in vivo durable gene transfer with prolonged mCD24 marker protein expression was achieved using a protocol of ex vivo transduction of PBSCs together with sublethal irradiation to enhance engraftment. To facilitate assessment of expression of the protein product of the transferred gene in different blood lineages, the retrovirus vectors used in this study encoded either mCD24 or htNGFR, both of which are expressed at the cell surface, allowing fluorescent antibody analysis using flow cytometry. Best results were achieved with the mCD24 vector, which was of higher titer, and most of the discussion will focus on the results with this marker. Highest levels of marking occurred during the first 3 months after transplantation, although there was persistence of mCD24 expression on peripheral blood cells at modest levels beyond 6 months of follow-up.
Theoretically, gene therapy targeting hematopoietic stem cells has the therapeutic potential to correct inherited errors of blood cell function or to correct other errors of metabolism. Such gene therapy might also provide a means to protect blood cells from the toxic effects of chemotherapeutic agents or provide protection against infectious agents such as the human immunodeficiency virus. In general, these goals require efficientgene transfer into long-lived stem cells, efficient engraftment with persistence of marked stem cells in vivo, the replacement or displacement of unaltered resident stem cells by the gene altered stem cells, and prolonged expression of the transferred gene. In general, these goals of stem cell gene transfer have not been realized or only partly realized in therapeutic or marking trials in humans or in marking studies in nonhuman primates. Until very recently, levels of in vivo marking of blood cells in published studies has been very low (often <1%, even in the setting of ablative marrow conditioning) and the duration of expression of the protein product from a transferred gene has been limited to a few weeks or months.
Very recently during the period of preparation of the data from our study for publication, two important studies of high level retrovirus marking of blood cells in vivo in nonhuman primates were published.6,7 While the studies by Tisdale et al6 focused primarily on demonstrating that prolonged culture of hematopoietic stem cells was detrimental to engraftment of vector marked cells, the in vivo marking in rhesus monkeys seen with a culture period of 4 days (similar to our regimen) was significant and prolonged. Particularly impressive are the results by Kiem et al7 showing a very high level of prolonged in vivo blood cell marking (20% for more than 20 weeks) in baboons. They attributed their high level of prolonged marking to several factors, including the use of gibbon ape leukemia virus envelope pseudotyped vector, the use of recombinant fibronectin fragment-coated flasks (RetroNectin; Takara Shuzo, Otsu, Japan), and the inclusion of Flt3-L and megakarocyte growth and development factor in the culture medium for transduction. Our study shares in common the use of high-titer vector and the inclusion of Flt3-L in the culture transduction medium. In addition, our study complements these studies by showing that nonablative sublethal marrow conditioning is sufficient to achieve engraftment of transduced stem cells. Our studies also complement those two studies by our demonstration that in all 4 animals there was prolonged expression in vivo in peripheral blood cells of the heterologous cell-surface protein (mCD24) encoded by the retrovirus vector. Thus, in addition to DNA marking, there was continued expression from the transduced gene in vivo. In the study by Kiem et al one of the animals did receive stem cells marked with a vector encoding human placental alkaline phosphatase, which they could detect by flow cytometry at the surface of 10% and 5% of peripheral blood cells at 2 and 4 weeks after transplantation.
Although our current study did not achieve all of the idealized goals of gene transfer to hematopoietic stem cells, it does represent a significant improvement over the results of most previously published studies of this type. We attribute these improved results to a combination of factors relating to some novel features of the procedures and materials used in this study. These potentially contributing factors were the following: (1) mobilization with the combination of Flt3-L and G-CSF achieved high levels of circulating CD34+ cells and may have improved the transduction and engraftment properties of the PBSC; (2) the use of a serum-free culture and transduction medium may have eliminated potential inhibitory factors associated with FBS; (3) inclusion of Flt3-L as part of the growth factor cocktail used for ex vivo culture of PBSC may have preserved long-term engraftment potential; (4) use of a high-titer MFGS-mCD24 vector exceeding 106 infective particles per milliliter together with a modified spinocculation protocol achieved high levels of transduction of CD34+ cells in general and of LT-CIC in particular; (5) the mCD24 marker protein appeared to lack toxicity and was of low immunogenicity, facilitating long-term protein expression by marked blood cells; and (6) use of a sublethal, nonablative conditioning regimen of 320 to 400 cGy X-irradiation was sufficient to enhance greatly the engraftment of transduced stem cells without the high toxicity and potential damage to marrow stroma associated with lethal radiation. A more detailed discussion of some of these issues follows.
Hematopoietic growth factors can mobilize large quantities of PBSCs into the peripheral blood that are capable of contributing to the long-term hematopoietic reconstitution of myeloablated mice,25-29 canines,30 and nonhuman primates.31,32 A very effective combination for mobilizing PBSC capable of engrafting myeloablated animals has been G-CSF plus SCF.26,32-34 In addition to the enhanced quality of these mobilized PBSCs for engraftment, Bodine et al33 showed that these PBSCs are a better target for murine retrovirus gene transfer. The long-term gene marking efficiencies were equivalent to or better than that demonstrated for 5-fluorouracil (5-FU)–treated bone marrow–derived hematopoietic stem cells. When this mobilization and transduction protocol was further evaluated in rhesus monkeys, the G-CSF/SCF–mobilized CD34+ cells as well as the primed, marrow-derived CD34+ cells showed superior transduction efficiency relative to steady-state bone marrow.35 These results suggested that the growth factor–induced manipulation of hematopoietic stem cells in vivo before harvest and ex vivo transduction could increase their responsiveness or susceptibility to retrovirus gene transfer. Orlic et al36,37 have suggested that the growth factor–induced modification of the target stem cells include upregulation of amphotropic retrovirus receptor expression. Such regulation may play an important role in the increased transduction efficiency noted both in our current study and the studies by Tisdale et al6 and by Kiem et al.7
Recent efforts to provide more effective mobilization relative to quantity and quality of PBSCs have focused on the use of Flt3-L in combination with G-CSF or GM-CSF,29,38-40 and also on the mobilization properties of such chimeric growth factor receptor agonists as myelopoietin41 and progenipoietin.42 In our study, the combination of Flt3-L and G-CSF resulted in a synergistic mobilization of PBSCs following kinetics over the 5- to 7-day period characteristic for G-CSF alone.42 Our data herein confirm the marked mobilization of hematopoietic progenitor cells using the Flt3-L/G-CSF combination. A single large-volume leukapheresis after 7 days of Flt3-L/G-CSF administration provided sufficient PBSCs for ex vivo transduction and subsequent transfusion into the autologous hosts. Except for animal no. J546, where laboratory handling problems led to a lower final yield of cells, the number of transduced CD34+ cells infused in the other 3 animals averaged 16 × 106 cells/kg. In fact, despite the small number of animals in our study, it appeared that the level of engraftment of transduced cells as measured by marked cells appearing in the peripheral blood was related to the number of CD34+ cells reinfused.
The ex vivo culture and transduction was performed in the presence of growth factors (Flt3-L, PIXY321, and SCF) as higher levels of CD34+ cell transduction with amphotropic retrovirus vectors are achieved in the presence of growth factors.8 These growth factors are included to induce cell cycling, a necessary requirement for the integration of DNA introduced via murine retrovirus vectors.43,44 There are, however, data to suggest that both the choice and dose of growth factors used for ex vivo transduction may impact negatively45-49 or positively8,9,50,51on the engraftment capacity of the transduced cells. In general, transduction conditions that are frequently used to enhance proliferation of hematopoietic cells generally lead to a loss of pluripotency and engraftment potential, particularly if the culture period is prolonged.6 Matsunaga et al47 and Hirayama et al49 noted that transduction of murine bone marrow cells in the presence of growth factors inhibited B-cell differentiation capacity of these stem cells. We have observed that high levels of IL-3, IL-6, and SCF inhibit T-cell differentiation of transduced hematopoietic progenitor cells as determined using an in vitro T-cell differentiation assay (M.R., R.P.J., unpublished data, June 1997). Furthermore, studies from a number of laboratories have shown that retrovirus transduction of human CD34+ cells in the presence of bone marrow stroma greatly enhances gene marking of cells capable of long-term hematopoietic engraftment in immunodeficient mice, but this effect can be reproduced in part by inclusion of Flt3-L in the ex vivo growth factor cocktail.8,9,51,52 In our current nonhuman primate study, we used a combination of Flt3-L, PIXY321, and SCF. This choice of growth factors was informed by the studies of others discussed above, but also influenced in part by a requirement to pilot those growth factors that would be available to us for a human clinical trial of gene therapy for chronic granulomatous disease.53The ex vivo culture conditions also included use of serum-free medium to reduce both the potential for effects of inhibitory factors present in FCS and the potential for immune responses to FCS. These serum-free culture and transduction conditions have been piloted in our past and ongoing human clinical trials of gene therapy for chronic granulomatous disease.21 53
Using the cells and culture conditions noted above, we employed a transduction protocol that involved centrifugations (spinocculation) with 3 changes of retrovirus supernate on each of 4 successive days to achieve high levels of transduction. Even when specifically analyzing cells that were CD34+ after 4 days of transduction, the MFGS-mCD24 vector transduced 43% ± 13% of cells while the MFGS-htNGFR vector transduced 21% ± 2% of cells. When ex vivo transduction was assessed in CFU and LTC-IC assays, we achieved highest levels of transduction using the mCD24+ vector. Of particular concern was the fact that no htNGFR-positive LTC-IC could be generated posttransduction with the MFGS-htNGFR vector, and this was accompanied by a significant decrease in the overall number of LTC-IC that could be generated from the ex vivo–transduced cells exposed to this vector. As both the mCD24 and htNGFR constructs are in the same vector backbone and use the same producer cell lines, this suggests the possibility of potential toxicity of htNGFR to LTC-IC. Although these studies used the spinocculation technique to enhance transduction, it has been shown that coating the surface of the culture vessel with a recombinant C-terminal fragment of fibronectin (CH-296 [RetroNectin]; Takara Shuzo Co) can also enhance transduction of CD34+.54 Although not available to us at the time of the nonhuman primate studies described in this report, we subsequently have used this fibronectin fragment for this purpose and found it to be superior to the spinocculation technique for transduction using MFGS-based amphotropic retrovirus vectors. In such studies we achieved enhancement of ex vivo transduction routinely to levels of 70% of human CD34+ cells.53 In their recent report, Kiem et al7 used the CH-296 recombinant human fibronectin fragment in their ex vivo hematopoietic cell gene marking studies of baboons to achieve high rates of transduction that may have contributed to persistence of the high levels of in vivo marking that they reported. It is likely that transduction enhancement with CH-296 could improve further on our current results with nonablative conditioning regimens in any future nonhuman primate gene transfer studies.
In primates undergoing autologous bone marrow transplantation, it is not possible to determine the level of partial engraftment without gene marking. Furthermore, in the absence of any treatments to cytoreduce or otherwise condition the marrow of recipients of transduced autologous stem cells, the levels of gene marking/engraftment has been very low.21,53,55,56 In mice it is possible to overcome the apparent barrier to engraftment in the nonconditioned host by infusion of very large doses of bone marrow cells.57 However, with doses of stem cells that would be available for autologous gene transfer studies in humans or in outbred large animal models, preparative conditioning of resident bone marrow is believed to be essential to ensure high levels of engraftment of autologous stem cells.
Ablative marrow conditioning has been thought to enhance engraftment of transplanted stem cells by opening up niches in the marrow that were occupied by endogenous stem cells. In the setting of allogeneic transplantation, conditioning also serves an essential role in reducing the immunologic barriers to transplantation. Experience from the allogeneic transplant setting resulted in the general use of ablative conditioning regimens in most experimental animal studies of gene transfer using autologous or syngeneic stem cells. However, in the autologous setting there presumably are no immunologic barriers to transplantation, and it raises the possibility that more modest sublethal, nonablative conditioning regimens might serve to enhance engraftment in that setting.16,58,59 Previous studies in mice have shown that low-dose (nonmyeloablative) radiation from doses as low as 30 cGy can enhance engraftment of congenic stem cells in a radiation dose-dependent fashion as relative to nonconditioned controls, without the morbidity and mortality associated with myeloablative radiation protocols.16 Based on those studies, the nonhuman primates in the current study received a nonmyeloablative dose of radiation (320 to 400 cGy) to capitalize on these previous observations and thus attempt to improve engraftment of transduced stem cells without the morbidity usually associated with myeloablation therapies. An additional factor that may have contributed to the high level of engraftment we observed was the pretreatment of the animals with Flt3-L and G-CSF as a mobilizing regimen, as it has been reported that some growth factors used before irradiation and transplantation may further enhance engraftment.16 This pretreatment with growth factors may also provide some radioprotection.60-63 Our studies establish, in the primate model, the feasibility of using a modest level of nonablative conditioning to enhance the level and durability of engraftment of transduced hematopoietic stem cells. In our study, the combination of adequate mobilization, conditioning with nonmyeloablative radiation exposure, and a large dose of transduced stem cells may have all been factors contributing to our observations of improved hematopoietic engraftment of the marked cells. Further work needs to be done using a variety of nonablative conditioning regimens for gene transfer studies in large animal models to optimize this observation.
The use of mCD24 to identify transduced cells greatly facilitated fluorescence-activated cell sorting (FACS) analysis of the different marked blood cell lineages appearing after transplantation. As shown, there is generally a peak of gene marking early after transplant in granulocytes that may reflect increased early contribution of transplanted stem and progenitor cells to hematopoiesis compared with that of endogenous stem cells that survived sublethal irradiation. In addition, the conditioning regimen caused peripheral depletion of lymphocytes, so that with this more long-lived cell type as well, gene-marked cells derived from transplanted progenitors may have contributed to a larger fraction of rapidly turning over cells. The fact that the transplanted cells had been exposed to growth factors ex vivo may have induced a more rapid differentiation and maturation of transduced progeny, contributing to the overall kinetics of appearance of a peak of marked peripheral blood cells at earlier time points.
The patterns of engraftment observed in different hematopoietic lineages were varied, suggesting a heterogeneous pool of hematopoietic stem and progenitor cells had been transduced. In general, high levels of mCD24 were first detectable in granulocytes, followed by monocytes and T- and B-lymphocyte subsets. Importantly, engraftment was persistent at easily detectable levels (>5%) for at least 24 weeks. Persistence of labeled cells in the granulocyte and monocyte pools for this period indicates that very primitive cells and not only the more mature, lineage committed progenitors were labeled. This is consistent with mCD24 detection in both CD34+ and CD34+38− subsets of stem cells in the bone marrow to at least 24 weeks posttransplantation. Evaluation of mCD24 detection in T, B, and NK cells shows that this protocol and this particular vector do not impact negatively on lymphocyte ontogeny. These observations are of importance in consideration of the development of gene-therapy strategies to prevent or treat infection with human immunodeficiency virus. Detection of mCD24 in the bulk of naı̈ve (CD45RA+ CD62L+) CD4+or CD8+ subsets of CD3+ T cells at 4 to 10 weeks posttransplantation provides additional evidence that in the conditions used for our study immature transduced PBSCs can develop into marked T cells.
The decrease from very high levels of mCD24-labeled cells to a persistent level of approximately 5% to 10% for 24 weeks and at lower levels thereafter may be explained in part by a resumption of endogenous hematopoiesis subsequent to sublethal irradiation. It is also possible that those very long-term or permanently repopulating totipotent stem cells that contribute to hematopoiesis at times later than 6 months after transplantation may have been transduced at a much lower frequency. Such permanently repopulating gene-marked totipotent hematopoietic cells persisting after 6 months are likely not measured in the LTC-IC assays and perhaps not even adequately measured in nonobese diabetic-severe combined immunodeficient (NOD-SCID) mouse models that assess human stem cell engraftment potential over 3 to 6 months in this model system. At later time points in our study both labeled progenitors in the bone marrow and more mature cells in the periphery were present, showing that small numbers of mCD24-labeled PBSCs were still contributing in part to the makeup of the peripheral blood compartment, though at a much lower level than the first 6 months.
It was important to address if a cellular immune response to mCD24 (a mouse antigen) had contributed to the decrease in the frequency of labeled cells, because this mechanism has been identified in other protocols that have used retrovirus vectors encoding the neomycin resistance gene.64 Although the rhesus CD24 gene sequence has not been reported, the published human and murine coding sequences share limited homology (57%) overall, though there are short regions of identity, suggesting that this antigen could be immunogenic in rhesus macaques.13 Interestingly, using in vitro antigen-specific stimulation, a CTL response was consistently absent in the 3 animals that showed high levels of gene marking. However, in no. J546, a CTL response to mCD24 was detectable at 15 weeks until 24 weeks posttransplantation. Despite this finding, mCD24+ cells were still detectable by flow cytometric analysis at 24 weeks, albeit at a very low frequency. It is worth noting that in this animal at 16 weeks after transplantation, our PCR assay that had been designed to yield quantitative information about copy number in the range of 2 to 0.005 was negative. The fact that a CTL response was only detectable in this animal may be a consequence of a low dose of transduced stem cells being transplanted. This is consistent with the dose dependency of certain antigens to induce a CTL response, where it has been observed that high dose of antigens can act as tolerizing agents (particularly where those antigens are presented by hematopoietic progenitor cells). The mCD24 clearly has the potential to induce an immune response in primates, based on the CTL detected in no. J546, so the absence of a response in the other animals may be in part due to these factors. It is also possible that transduced cells may have migrated to the thymus of these animals where they were able to induce tolerance of T cells that developed after irradiation. One of the important potential therapeutic uses for gene therapy is the treatment of inherited protein-null metabolic deficiency diseases. In that clinical setting, the therapeutic gene product may be immunogenic in the host. Therefore, these issues regarding immunogenicity of marker genes in animal studies likely are relevant to human clinical studies where therapeutic genes may be perceived as neoantigens. Despite the fact that we did not detect CTL response to the mCD24 antigen in some nonhuman primates after gene transfer, in the clinical setting of gene therapy for protein-null disorders it may still be necessary to apply some form of immune suppression to achieve permanent genetic correction after gene therapy.
In conclusion, we have shown that efficient and durable engraftment of transduced hematopoietic cells is possible using PBSC mobilized with Flt3-L and G-CSF, using ex vivo transduction with an MFGS-mCD24 vector in the presence of growth factors which include Flt3-L, and with engraftment enhanced by nonablative, sublethal irradiation. A high dose of autologous, transduced CD34+ cells may have contributed to the success of this protocol as well as the absence of a CTL response to mCD24 in 3 of 4 animals studied.
ACKNOWLEDGMENT
We thank Dr Stewart Lyman and the Immunex Corporation for kindly providing the Flt3-L and PIXY321, and Dr M. Gately, Hoffman LaRoche, for kindly providing the IL-2. We also thank Dr Fei Li for his contributions to development of the colony labeling assays used in this study.
Supported in part by National Institutes of Health Grants No. RR00168 (M.R. and R.P.J.), A139423 (M.R.), and CA73473 (R.P.J.).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to H.L. Malech, MD, Laboratory of Host Defenses, NIAID, Bldg 10 Room 11N113, 10 Center Dr MSC 1886, Bethesda, MD 20892-1886; e-mail: hmalech@nih.gov.

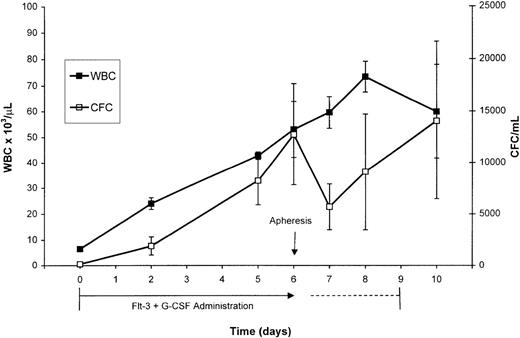
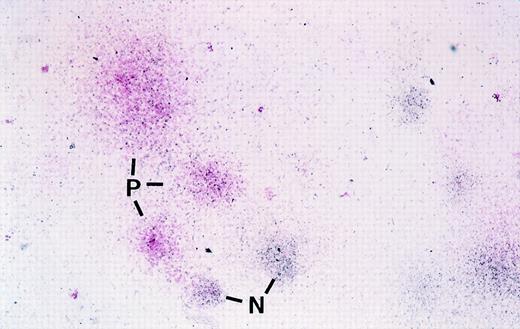
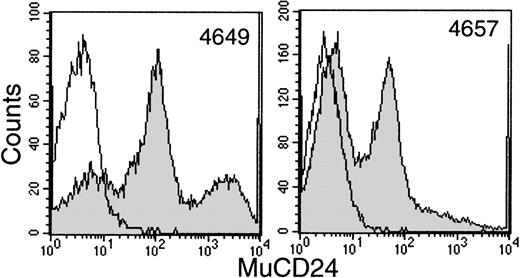


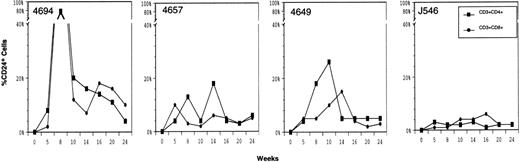
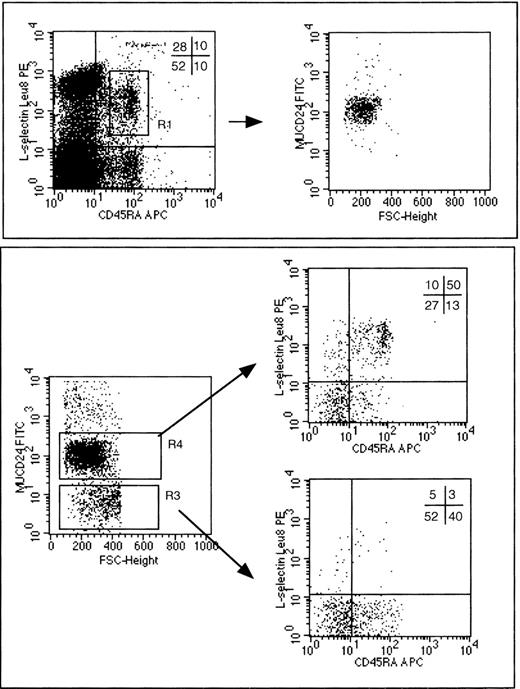
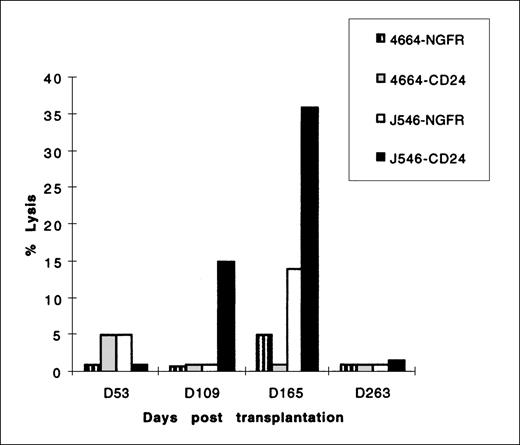
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal