The c-kit receptor and its ligand, steel factor (SLF), are critical for optimal hematopoiesis. We evaluated effects of transducing cord blood (CB) progenitor cells with a retrovirus encoding humanc-kit cDNA. CD34+ cells were sorted as a population or as 1 cell/well for cells expressing high levels of CD34+++ and different levels of c-kit (++, +, Lo/−), transduced and then cultured in the presence of granulocyte-macrophage colony-stimulating factor (GM-CSF), interleukin-3 (IL-3), IL-6, erythropoietin (Epo) +/− SLF in the absence of serum. At a single-cell level, transduction with c-kit, but not with control (neo only), virus significantly increased colony formation, especially by erythroid and multipotential progenitors. The enhancing effect of c-kit transduction was inversely correlated with expression of c-kit protein before transduction. The greatest enhancing effects were noted in CD34+++kitLo/− cells transduced with c-kit. The stimulating effect was apparent even in the absence of exogenously added SLF, but in the presence of GM-CSF, IL-3, IL-6, and Epo. Enzyme-linked immunosorbent assay (ELISA) of SLF protein, reverse transcriptase-polymerase chain reaction (RT-PCR) analysis of SLF mRNA expression in CD34+ cells, and use of neutralizing antibodies to SLF and/or c-kit suggested the presence of endogenous, although probably very low level, expression of SLF by these progenitor cells. Transduction of c-kit significantly decreased sensitivity of progenitor cells to the inhibitory effects of transforming growth factor-β1 and tumor necrosis factor-.c-kit–transduced cells had increased expression ofc-kit protein and decreased spontaneous or cytokine-induced apoptosis. Our results suggest that transduced c-kit into selected progenitor cells can enhance proliferation and decrease apoptosis and that endogenous SLF may mediate this effect.
HEMATOPOIETIC CELL proliferation, differentiation, migration, and apoptosis are mediated via the interaction of various hematopoietic stimulatory and inhibitory cytokines and their receptors. Among these, SLF and its receptor,c-kit, are critical for maintenance of steady-state hematopoiesis.1c-kit, a 145-kD transmembrane glycoprotein, is the normal cellular counterpart of the viral oncogene v-kit, and a member of the receptor tyrosine kinase subclass III family that includes receptors to platelet-derived growth factor (PDGF), macrophage colony-stimulating factor (M-CSF), and flt3 ligand.2-4 The c-kit gene product is expressed on a variety of murine5-7 and human8-10hematopoietic stem/progenitor cells, mast cells, and germ cells. The ligand of c-kit exists in both transmembrane and soluble forms and is known as mast cell growth factor (MGF), SLF, stem cell factor (SCF), and kit ligand (KL).11-15 We and others have shown that SLF is produced by human and murine hematopoietic stromal cells including fibroblasts, endothelial cells, and bone marrow (BM)-derived stromal cells.16 17
The dependence of normal hematopoiesis on an intact c-kit/SLF axis is demonstrated in mice that have specific mutations of either thec-kit (W) or SCF (SL) gene loci. These mice exhibit BM failure, as well as white spotting, sterility, and a profound decrease in tissue mast cells.1 Studies of variousW mutations in mice have suggested that the severity of the observed phenotype correlates with residual functional activity of thec-kit polypeptide and that there may be a threshold level ofc-kit kinase activity required for functional hematopoiesis.18 Additional evidence supporting the importance of c-kit in regulating hematopoiesis has been provided by in vivo or in vitro experiments in which c-kitprotein functional activity is blocked with neutralizing antibodies. Treatment of mice with an anti–c-kit monoclonal antibody that blocks SLF binding results in aplastic anemia.5 Several groups of investigators have demonstrated that treatment of murine or human long-term BM culture (LTBMC) with a c-kit neutralizing monoclonal antibody completely inhibits the production of differentiated hematopoietic progenitors and mature myeloid blood cells. After removal of the antibody, hematopoiesis recovers and the number of transplantable stem cells in antibody-treated cultures is not different than that in treated cultures. It has been suggested that functional c-kit is necessary for the generation of differentiated hematopoietic progenitors and mature myeloid blood cells, but may not be necessary for the survival of stem cells.19,20 In addition, treatment of LTBMC withc-kit antisense oligonucleotides causes suppression of the generation of burst-forming unit-erythroid (BFU-E) which continues for at least 3 weeks.21
Transforming growth factor-β1 (TGF-β1), a multifunctional cytokine, affects the proliferation and differentiation of a variety of tissue and cell types including cells of the hematopoietic system.22-25 TGF-β1 is produced by both hematopoietic cells and stromal cells of the hematopoietic microenvironment,22,26 and inhibits hematopoietic progenitor cells (HPC) in LTBMC.22,27 In LTBMC, addition of TGF-β1 to the medium blocks the recruitment of HPC into cell cycle, and addition of neutralizing antibody to TGF-β1 increases the percentage of primitive progenitors in cell cycle.22,27TGF-β1 has also been reported to inhibit the proliferation of progenitors in colony assays in vitro,23,28,29 and treatment of mice with TGF-β1 suppresses blood cell formation in vivo.30 TGF-β1 inhibits the expression of receptors on progenitor cells for hematopoietic growth factors including receptors for interleukin-1 (IL-1), IL-3, SLF, and granulocyte-macrophage colony-stimulating factor (GM-CSF),31,32 increases expression of cyclin-dependent kinase inhibitors (CDKI) such as p27kip1, p21cip1, and p15INK4B,33-36 and reduces phosphorylation of the Retinoblastoma protein (Rb).37
Tumor necrosis factor-α (TNF-α) is another bifunctional cytokine that is able to interact with many growth factors and their receptors.38-42 It has been reported to upregulate the expression of IL-3, IL-5, and GM-CSF receptors40 and downregulate receptors for GM-CSF and SLF.41,42 The mechanism by which TNF-α inhibits the proliferation of hematopoietic stem/progenitor cells remains largely unknown, but may involve induction of apoptosis and/or cell-cycle arrest. Some investigators have found that TNF-α decreases c-kit expression on both normal and leukemic CD34+ cells,42 whereas others have reported that TNF-α enhances the expression ofc-kit in acute myelogenous leukemia (AML) cells through posttranscriptional stabilization of the c-kittranscripts.43 TNF-α decreases expression ofc-kit on murine HPC in a manner analogous to TGF-β1.44
We have previously described a subpopulation of CD34+ cells expressing a high level of this antigen (CD34+++) from cord blood (CB), which are enriched for primitive HPCs and can be isolated and studied as single cells.45 These cells can be transduced with retroviral vectors at both a population and a single-cell level.10 46-49 In the present report, we examined if enforced expression of c-kit by retroviral gene transduction in CD34+++ CB cells could change the proliferation capacity of these cells to growth cytokines and evaluated their responsiveness to the inhibitory effect of TGF-β1 and TNF-α.
MATERIALS AND METHODS
Cells and cell separation.
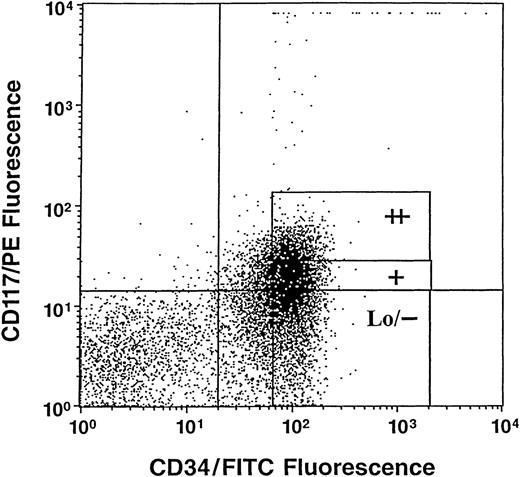
Cells were obtained from normal human umbilical CB scheduled for discard after delivery of the infant and after prior need for samples for clinical study had been satisfied. CD34+++ cells were obtained after sorting magnetic activated cell separation (MACS) beads (Miltenyi Biotec Inc, Auburn, CA) separated CD34+ cells (90% to 95% pure) on a flow cytometer (FacStarplus; Becton Dickinson, San Jose, CA) as previously described.45 These cells are enriched for stem and immature subsets of progenitor cells.45 The kit++, kit+, and kitLo/− subsets of cells were sorted from CD34+++ cells based on expression ofc-kit using a phycoerythrin-conjugated monoclonal antibody against human c-kit (CD117/PE) (PharMingen, San Diego, CA) as previously described.10 Sorting gates are shown in Fig 1. Cells sorted by CD34 andc-kit expression were either directly sorted by an autoclone device as 1 cell/well into single wells containing 0.1 mL semisolid culture medium (with a single cell per well verified as described previously), or cells were sorted into tubes as populations of cells and assayed for colony formation as previously described.46-49
Representative analysis of c-kit (CD117)/PE and CD34/fluorescein isothiocyanate (FITC) immunofluorescence on gated high densities of CD34+ (CD34+++) cells. Sorting windows shown as ++, +, and Lo/− are for CD34+++ kit++, kit+, and kitLo/− cells, respectively.
Representative analysis of c-kit (CD117)/PE and CD34/fluorescein isothiocyanate (FITC) immunofluorescence on gated high densities of CD34+ (CD34+++) cells. Sorting windows shown as ++, +, and Lo/− are for CD34+++ kit++, kit+, and kitLo/− cells, respectively.
Colony assay.
Cultures contained Iscove’s modified Dulbecco’s medium (IMDM; GIBCO-BRL, Gaithersburg, MD), 1% methylcellulose, 30% fetal calf serum (FCS; Hyclone Laboratory, Logan, UT), and 0.1 mmol/L hemin (Eastman Kodak Co, Rochester, NY). Serum-depleted medium contained bovine serum albumin (1 mg), iron-saturated transferrin (300 μg), cholesterol (7.8 μg), and CaCl2 (200 μg)/mL instead of serum as described.29 Recombinant human SLF, IL-3, and GM-CSF were kind gifts from Immunex Corporation (Seattle, WA) and were used per/mL, respectively, at 0 to 50 ng, 200 U, and 200 U. Recombinant human IL-6 was a gift from Genetics Institute (Cambridge, MA) and was used at 10 ng/mL. Recombinant human erythropoietin (Epo) was purchased from Amgen Corp (Thousand Oaks, CA) and was used at 1 U/mL. TGF-β1 and TNF-α were purchased from R&D Systems (Minneapolis, MN) and Genentech, Inc (San Francisco, CA), respectively, and added at 5 ng and 20 ng/mL, respectively. After gene transduction at a population level, TGF-β1 and TNF-α were added at the initiation of the colony assays. After gene transduction at the single-cell level, TGF-β1 and TNF-α were added 1 day after transduction to avoid the inhibitory effect on cell growth, which could have resulted in a decrease in the gene transduction efficiency. Cells were incubated under humidified conditions at 37°C, 5% O2 and 5% CO2 for 14 days.
Retroviral vectors and packaging cell line.
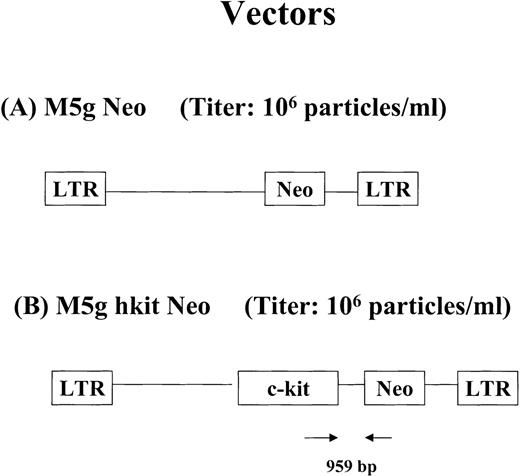
To construct the retroviral vector encoding human (h) c-kit, a polymerase chain reaction (PCR) was used with pCVN hc-kitplasmid as a template. This plasmid encodes a full-length humanc-kit cDNA that contains the 12-bp insert encoding an asparagine-rich insert in the extracellular domain (kit A+ isoform).50 The sense primer sequence was CTC GAG GCC GCCATG AGA GGC GCT CGC GGC GCC TGG. This primer contains a synthetic XhoI site and Kozak consensus sequence (underlined). The c-kit protein initiation codon is shown in bold. The antisense primer used had a sequence of GGA TCCTCAGAC ATC GTC GTG CAC AAG CAG and contains a synthetic BamHI site (underlined). Bases complementary to the c-kit stop codon are shown in bold. The product of the PCR reaction was cloned into the pCRII vector (Invitrogen, Carlsbad, CA) and the insert sequence was verified by automated DNA sequencing. The insert from this plasmid clone was cloned into the BamHI site of the M5g Neo vector to yield M5g hkit Neo. The M5g Neo vector was constructed by excision of the murine c-kit sequence from the plasmid M5gNeo-kitwt (a generous gift of Dr Hitoshi Kitayama, Osaka University Medical School, Osaka, Japan).50 The M5g Neo vector contains the long terminal repeat (LTR) of the murine myeloproliferative sarcoma virus (MPSV) and encodes the sequence for the bacterial neomycin phosphotransferase gene (Neo). This plasmid has been previously described.51 Two transcripts are derived from the M5g hkit Neo vector: a monocistronic message containing sequences for human c-kit coding sequence as well as Neo gene and an alternatively spliced message containing only the Neo sequences (Fig2).52 Vector lacking hkit cDNA (M5g Neo) was used as mock virus control. The retroviral packaging cell lines, Ψ-2 and PA317, were maintained in Dulbecco’s modified Eagle’s medium, supplemented with 10% FCS. High titer amphotropic PA317 packaging cells for M5g Neo or M5g hkit Neo were produced by standard techniques.47 The viral titers for both the PA317 M5g Neo and PA317 M5g hkit Neo packaging cells were in the range of 1 to 3 × 106G418 resistant (G418R) colony-forming unit (CFU)/mL on NIH3T3 cells. Viral supernatant was harvested and filtered through 0.45-μm and 0.22-μm filters and stored at −80°C until use.
Diagram of retroviral vectors encoding humanc-kit cDNA used in the studies. (A) M5g Neo vector. (B) M5g hkit Neo vector. The positions of the PCR primers and expected size of amplified products are shown.
Diagram of retroviral vectors encoding humanc-kit cDNA used in the studies. (A) M5g Neo vector. (B) M5g hkit Neo vector. The positions of the PCR primers and expected size of amplified products are shown.
Retroviral transduction protocol.
For population cell gene transduction, sorted CD34+++ cells and their subsets of kit++, kit+, or kitLo/− cells were prestimulated with IL-3 (200 U), GM-CSF (200 U), Epo (1 U), and IL-6 (10 ng)/mL for 2 days and transduced with viral supernatant as described.46-49 The cells were then washed twice and plated for colony formation at 200 or 300 cells/mL for serum-containing or 800 cells/mL for serum-depleted cultures in the presence of GM-CSF, IL-3, Epo, and IL-6, minus and plus varying concentrations of SLF (2, 10, and 50 ng/mL). An alternative protocol was used for single-cell gene transduction. Single cells were directly sorted into single wells containing 0.1 mL methylcellulose in the presence of GM-CSF, IL-3, Epo, IL-6 at the same concentrations as mentioned above, minus and plus varying concentrations of SLF (2, 10, and 50 ng/mL) in the serum-containing or serum-depleted cultures for 2 days. Viral supernatant was added once at 20 μL/well with polybrene (8 μg/mL) as described previously.46-48 The cells remained in the same growth factor medium throughout the study. Two days after addition of viral supernatant, fewer than 2% of the cells had formed doublets and the predicted short half-life of the virus preparation suggested that in more than 95% of the cases, single cells had been transduced.
Statistics.
The probability of significant differences between cells plated at 200 or 800/mL (expressed as mean ± 1 standard error of mean [SEM]) was determined by Student’s t-test, and that between groups in the single-cell experiments was determined by χ2 test.
PCR analysis.
Genomic DNA was isolated from individual colonies as previously described.46-48 Briefly, individual colonies were removed from methylcellulose culture medium and washed with 1 mL of phosphate-buffered saline (PBS). Cell pellets were resuspended in the small volume of remaining PBS to which 200 μL of a chelex 100 solution (200 to 400 mesh; BioRad, Richmond, CA) was added. Cells were mixed well with chelex and lysed by boiling for 5 minutes, chilled on ice for 5 minutes, and pelleted for 30 seconds at 2,800g. A total of 10 μL of supernatant from the lysate was used for PCR. The plasmid DNA of M5g hkit Neo was used as PCR positive controls. As a negative control, DNA was obtained from cells incubated with viral supernatant collected from packaging cell lines that had been transfected with the retroviral vector lacking the hkit cDNA insert, M5g Neo (mock control). A pair of primers was used at 1 μmol/L in the PCR reaction (see Fig 2 for PCR primer positions). The following primers were used: 5′ TCT GCT TGT GCA CGA CGA TGT CT 3′ (residues: 2928-2950 sense strand from human c-kit cDNA sequences according to the numbering convention of Yarden et al2) and 5′ CAT GCG AAA CGA TCC TCA TCC 3′ (residues: 5464-5484 antisense strand from M5g Neo sequences). Each sample was amplified with AmpliTaq (Perkin-Elmer/Cetus, Roche Molecular Systems, Inc, Branchburg, NJ) using a DNA thermocycler (Perkin-Elmer/Cetus) for 35 cycles (94°C for 30 seconds to denature the DNA, 55°C for 30 seconds for primer annealing, and 72°C for 1 minute for primer extension) resulting in a 959-bp product. Ten microliters of reaction mixture was electrophoresed on a 1% agarose gel. To ensure that amplified products were the correct sequences, Southern blot analysis was performed by standard methods. A 3.4-kb fragment of human c-kit was isolated from pM5g hkit Neo plasmid digested with BamHI and XhoI and used as probe.
Reverse transcriptase (RT)-PCR analysis.
RNA was extracted from CD34+ cells or individual colonies as described previously.46-48 Briefly, colonies were removed, cell pellets were resuspended in 20 μL double distilled water treated with dimethyl pyrocarbonate (DEPC), 100 U RNasin, and 1 μg tRNA, cooled on ice for 30 minutes, and pelleted for 30 seconds. A 9-μL supernatant of the resulting RNA solution was used in the reverse transcription reaction. To eliminate contaminating DNA, the RNA extracts were treated with 1 U DNase I for 15 minutes at room temperature. The DNase was inactivated by adding 1 μL of 20 mmol/L EDTA solution to the reaction mixture and heating at 65°C for 10 minutes before reverse transcription. A total of 20 μL of the RNA lysate in RT buffer (RT buffer: 25 mmol/L Tri-HCL [pH 8.3], 37.5 mmol/L KCL, 1.5 mmol/L MgCl, 5 mmol/L dithiotreitol [DTT], 10 mmol/L deoxyribonucleoside triphosphate [dNTP] mixture), 0.5 μg oligo (dT)15primer, 26 U RNasin, and 200 U murine Molony leukemia virus reverse transcriptase (Promega Corp, Madison, WI) were used for reverse transcription. After cDNA synthesis, 10 μL of the DNA solution was used for the RT-PCR reaction. The primers and PCR conditions for the RT-PCR reaction were the same as for PCR. To detect mRNA of SLF on CD34+++ cells and their subsets, RNA was isolated from cells and reverse transcription was performed. The following primers were used: 5′ TGG ATA AGC GAG ATG GTA GT 3′ (residues: 388-407 sense strand from human SLF cDNA) and 5′ TGG GTA GCA AGA ACA GAT AAA (residues: 1124-1144 antisense strand from hyman SLF cDNA) for PCR analysis. Each sample was amplified for 35 cycles (94°C for 1 minute to denature the DNA, 60°C for 1 minute for primer annealing, and 72°C for 2 minutes for primer extension) resulting in 757-bp and 673-bp products, respectively, representing soluble and membrane-bound forms of SLF as described previously.16 A 1.0-kb fragment of human SLF was isolated from pBluescript SK-plasmid digested with BamHI and HindIII and used as probe.16 To detect mRNA c-kit expression on CD34+ cells and their subsets, RNA was isolated and reverse transcription was performed as above. The following primers were used: 5′ CGT TGA CTA TCA GTT CAG CGA G 3′ (residues: 843-864 sense strand from human c-kit cDNA) and 5′ CTG GGA ATG TGT AAG TGC CTC C (residues: 1180-1201 antisense strand from humanc-kit cDNA) for PCR analysis. Each sample was amplified for 35 cycles (94°C for 30 seconds to denature the DNA, 55°C for 30 seconds for primer annealing, and 72°C for 2 minutes for primer extension) resulting in 360-bp products as described.53 Human stromal cells (NFF-6)16 were used as positive control for RT-PCR reaction for SLF and c-kit, and PCR reaction reagents were used as negative control for PCR analysis. β-Actin was used as internal control. The primers were as follows: 5′ATC TGG CAC CAC ACC TTC TAC AAT GAG CTG CG 3′ (sense) and 5′ CGT CAT ACT CCT GCT TGC TGA TCC ACA TCT GC 3′ (antisense). The β-actin primers were used to amplify for 35 cycles (94°C for 45 seconds to denature the DNA, 60°C for 45 seconds for primer annealing, and 72°C for 2 minutes for primer extension) resulting in an 838-bp fragment.
Terminal deoxynucleotidyl transferase (TdT)-mediated deoxyuridine triphosphate (dUTP) nick end-labeling (TUNEL) assay.
Apoptosis was determined using the In Situ Cell Death Detection Kit (Boehringer Mannheim, Indianapolis, IN) as described by the manufacturer to detect the incorporation of labeled nucleotides to DNA strand breaks by TUNEL assay. Briefly, cells were washed with PBS containing 1% bovine serum albumin (BSA) and fixed by 2% paraformaldehyde solution (Sigma, St Louis, MO) for 30 minutes at room temperature and permeabilization was performed with 0.1% Triton-X-100 (Bio-Rad Laboratories, Richmond, CA) in 0.1% sodium citrate (Sigma) at 4°C for 2 minutes. The cells were then washed and labeled with TUNEL reaction mixture at 37°C for 60 minutes in the dark. After washing, cells were resuspended in cold PBS and analyzed by a FACScan flow cytometer (Becton Dickinson).54
Enzyme-linked immunosorbent assay (ELISA).
To measure the soluble form of SLF endogenously produced and released into the cultures, ELISA assay was applied using polyclonal antibody against SLF with an ELISA kit (R&D Systems) as described.52To measure soluble SLF, medium conditioned by CD34+, sorted CD34+++kit+, and CD34+++kitLo/− cells at 106cells/mL or no cells were cultured in IMDM containing 10% FCS or serum-depleted medium in the presence of IL-3, GM-CSF, IL-6, and Epo at 200 U, 200 U, 1 U, and 10 ng/mL for 72 hours at 37°C, 20% CO2 and 5% O2. Medium was collected as conditioned medium (CM) by centrifugation at 3,000 rpm for 10 minutes. To measure cell-associated SLF, MACS-separated CD34+ cells were washed extensively and whole cell lysates were prepared using 1 mL of lysis buffer (20 mmol/L TRIS [pH 7.4], 1 mmol/L EDTA, 0.1% sodium dodecyl sulfate [SDS], 1% Triton X-100, 0.02% sodium azide, 1 mol/L NaCl). A postnuclear fraction of the whole cell lysate was obtained by pelleting the nuclei in a microcentrifuge and then collecting the supernatant. Soluble and cell-associated SLF was measured using a commercially available ELISA kit (R&D Systems) according to the manufacturer and read by a microplate reader (Titetek Multiskan Plus) at optical density (OD) 450 nm. Standards for soluble SLF were prepared using culture medium diluent, whereas standards for cell-associated SLF were prepared using lysis buffer as a diluent. The minimum detectable level of SLF is about 9 pg/mL.
RESULTS
Influence of c-kit gene transduction on colony formation by CD34+++ cells.
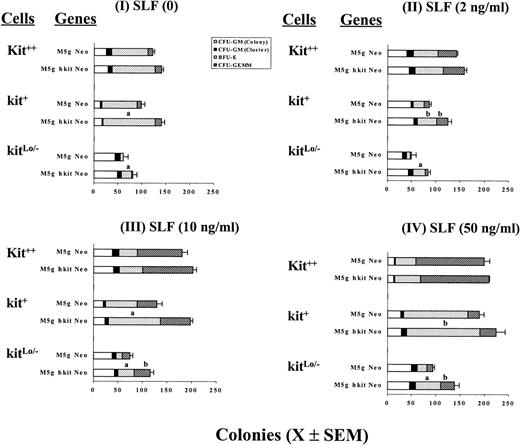
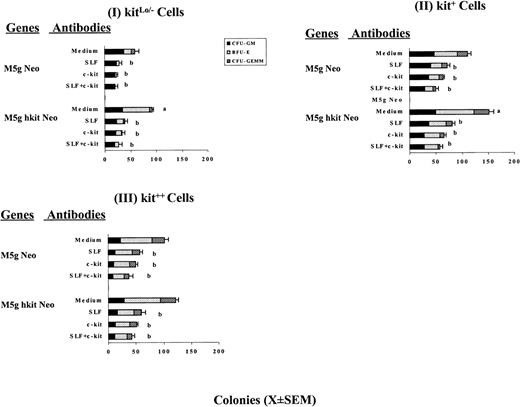
CD34+++ cells were separated into 3 subsets denoted as kit++, kit+, and kitLo/− cells based on cell surface expression of c-kit protein (Fig 1). Cells were transduced with c-kit or mock control virus using either bulk population or single-cell gene transduction methods.46-49 Bulk population transduced cells were plated in the presence of Epo (1 U/mL), IL-3 (200 U/mL), IL-6 (10 ng/mL), GM-CSF (200 U/mL) without or with SLF at 2, 10, or 50 ng/mL in the presence of FCS or in serum-depleted cultures. Figure 3 shows the results from a representative of 3 separate experiments using a serum-depleted culture assay and cells plated at 800 cells/mL. Transduction of these cells with c-kit, but not mock (neo only), retrovirus significantly increased colony formation by BFU-E and sometimes CFU-granulocyte, erythrocyte, macrophage, megakaryocyte (GEMM) in the presence of GM-CSF, IL-3, IL-6, Epo with or without addition of SLF. Transduction with c-kit cDNA had little or no effect on numbers of colonies formed by CFU-GM. There was no difference between Neo only virus and medium without virus (data not shown). The enhancing effect of transduction inversely correlated with c-kit protein expression before transduction; the greatest effects were noted in kitLo/− cells transduced with c-kit, either without or with addition of SLF (2 to 50 ng/mL). For example, transduction of kitLo/− cells with c-kit cDNA followed by culture in the presence of GM-CSF + IL-3 + IL-6 + Epo increased BFU-E and CFU-GEMM colonies up to 256% and 300%, respectively, compared with the mock virus-transduced cells. In contrast, transduction of c-kit cDNA and culturing in the same cytokine combination resulted in an increase of 48% for BFU-E in kit+ and 17% in kit++ cells and an increase of 54% for CFU-GEMM in kit+ and 38% in kit++cells, respectively. Less enhancing effects were obtained when SLF was added at 50 ng/mL. Similar results were observed in the presence of serum (data not shown). It has been reported that the action of SLF is particularly critical when levels of other growth factors are limited.55 We thus tested to see if the effects would be greater when one fourth concentrations of GM-CSF (50 U), IL-3 (50 U), IL-6 (2.5 ng), and Epo (0.25 U)/mL were used. Similar, but not greater, increases were noted in the absence or presence of different concentrations of SLF (data not shown).
Influence of cytokines on colony formation by CD34+++kit++, kit+, and kitLo/− cells transduced with c-kit cDNA. Sorted cells were prestimulated with full dosages of IL-3 (200 U), GM-CSF (200 U), Epo (1 U), and IL-6 (10 ng/mL) and then transduced withc-kit cDNA (M5g hkit Neo) or mock (M5g Neo) viruses as described in Materials and Methods. After washing, cells were assayed at 800 cells/mL for colony formation in semisolid culture in the presence of IL-3, GM-CSF, and Epo with SLF at 0 (I), 2 (II), 10 (III), and 50 (IV) ng per mL in the serum-depleted cultures as described in Materials and Methods. Results are expressed as mean ± SEM from 1 representative of 3 separate experiments. Significant differences from mock virus control are: aP < .01;bP < .05.
Influence of cytokines on colony formation by CD34+++kit++, kit+, and kitLo/− cells transduced with c-kit cDNA. Sorted cells were prestimulated with full dosages of IL-3 (200 U), GM-CSF (200 U), Epo (1 U), and IL-6 (10 ng/mL) and then transduced withc-kit cDNA (M5g hkit Neo) or mock (M5g Neo) viruses as described in Materials and Methods. After washing, cells were assayed at 800 cells/mL for colony formation in semisolid culture in the presence of IL-3, GM-CSF, and Epo with SLF at 0 (I), 2 (II), 10 (III), and 50 (IV) ng per mL in the serum-depleted cultures as described in Materials and Methods. Results are expressed as mean ± SEM from 1 representative of 3 separate experiments. Significant differences from mock virus control are: aP < .01;bP < .05.
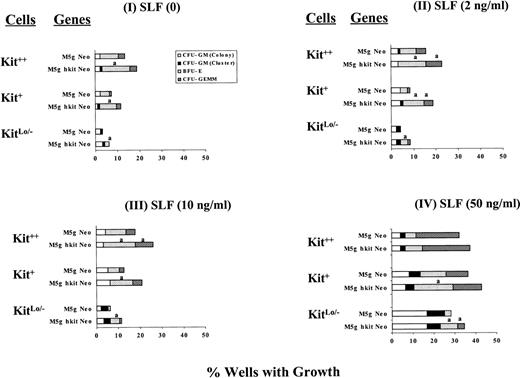
We previously reported that transduction and culture of single CD34+++ cells with retroviruses encoding EpoR and/or IL-9R increased Epo-dependent erythroid colony formation compared with mock transduced cells.46-49 We therefore tested if transduction of single cells with c-kit cDNA resulted in similar effects as seen after transduction and culture of the bulk populations of cells. The 3 subsets of c-kit expressing cells were directly deposited at a frequency of 1 cell/well into wells containing full dosages of GM-CSF, IL-3, IL-6, and Epo without or with SLF (2 to 50 ng/mL) in the presence or absence of serum for prestimulation. Single-cell gene transduction was performed as described previously.46-48 The results from a total of up to 192 wells per point for 3 separate experiments using serum-depleted cultures are shown in Fig 4. The increase in colony numbers of progenitor cells transduced with c-kit cDNA was confirmed at the single-cell level in the absence of serum. These effects were most notable for BFU-E. This increase was more apparent in kitLo/− cells, especially in the absence of exogenously added SLF or the low concentration of SLF (2 ng/mL). Similar results were obtained using serum-containing cultures (data not shown). Our results suggest that c-kit transduction has a direct growth enhancing effect on progenitors.
Influence of cytokines on colony formation by single sorted CD34+++ kit++, kit+, and kitLo/− cells transduced at the single-cell level with c-kit cDNA or mock viruses. Single cells were directly sorted into wells as 1 cell/well containing 0.1 mL of semisolid culture in the presence of full dosages of IL-3 (200 U), GM-CSF (200 U), Epo (1 U), IL-6 (10 ng) with SLF at 0 (I), 2 (II), 10 (III), and 50 (IV) ng per mL in the serum-depleted culture as described in Materials and Methods. Results are expressed as percent wells with growth for a total of 192 wells per point from 3 separate experiments. Significant differences from mock virus control are: aP < .05.
Influence of cytokines on colony formation by single sorted CD34+++ kit++, kit+, and kitLo/− cells transduced at the single-cell level with c-kit cDNA or mock viruses. Single cells were directly sorted into wells as 1 cell/well containing 0.1 mL of semisolid culture in the presence of full dosages of IL-3 (200 U), GM-CSF (200 U), Epo (1 U), IL-6 (10 ng) with SLF at 0 (I), 2 (II), 10 (III), and 50 (IV) ng per mL in the serum-depleted culture as described in Materials and Methods. Results are expressed as percent wells with growth for a total of 192 wells per point from 3 separate experiments. Significant differences from mock virus control are: aP < .05.
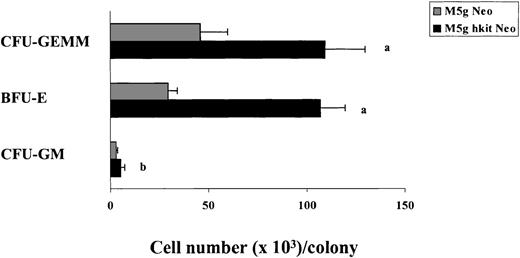
c-kit transduction not only increased clonogenic efficiency of progenitors, but also increased the size of individual BFU-E and CFU-GEMM colonies (Fig 5). The number of cells per BFU-E (106 × 103) and CFU-GEMM (109 × 103) colony from c-kit transduced cells was greater than that of the mock-transduced cells (29 × 103 and 46 × 103). Interestingly, the number of cells per CFU-GM colony was significantly increased as well in c-kittransduced cells (5.2 × 103) compared with that of mock-transduced cells (2.9 × 103).
Influence of c-kit cDNA transduction on the size of colonies derived from CD34+++ cells in the presence of full dosages of IL-3, GM-CSF, Epo, and SLF (10 ng) per mL. The 30 largest CFU-GM, BFU-E, and CFU-GEMM colonies fromc-kit–cDNA or mock-transduced cells were removed from a total of 2 experiments and the cell numbers per colony were counted. Significant differences from mock virus control are:aP < .0001; bP < .05.
Influence of c-kit cDNA transduction on the size of colonies derived from CD34+++ cells in the presence of full dosages of IL-3, GM-CSF, Epo, and SLF (10 ng) per mL. The 30 largest CFU-GM, BFU-E, and CFU-GEMM colonies fromc-kit–cDNA or mock-transduced cells were removed from a total of 2 experiments and the cell numbers per colony were counted. Significant differences from mock virus control are:aP < .0001; bP < .05.
Influence of c-kit transduction on responsiveness of kit++, kit+, and kitLo/− cells to the inhibitory effects of TGF-β1 and TNF-α.
One of the known effects of TGF-β1 and TNF-α on progenitor cells is to suppress c-kit protein.52 56 Therefore, we assessed if enforced expression of c-kit in progenitors could influence the sensitivity of c-kit transduced cells to these inhibitory cytokines. We tested the inhibitory effects of TGF-β1 and TNF-α on colony formation using dosages ranging from 10 pg to 5 ng/mL and 62.5 U to 1,000 U/mL, respectively, added to cultures containing full dosages of SLF, IL-3, GM-CSF, and Epo using 3 subsets of CD34+++ cells (kit++, kit+, and kitLo/−), transduced in bulk cell populations withc-kit cDNA or mock virus. We found a dose-dependent inhibitory effect of TGF-β1 and TNF-α on colony formation in the kit++, kit+, and kitLo/−populations of cells transduced with the mock control (M5gNeo) virus and grown in the presence of serum (data not shown). Significant inhibition was detected at concentrations as low as 50 to 100 pg/mL of TGF-β1 and 125 U/mL of TNF-α in kit++, kit+, and kitLo/− cells. However, the inhibitory effect was significantly reduced by 50% to 75% in cells transduced with c-kit compared with the mock virus transduced cells at the same dosages of TGF-β1 and TNF-α (data not shown). The effects of c-kit transduction in reducing the sensitivity of progenitor cells to TGF-β1 was similar between the subsets of kit++, kit+, and kitLo/−cells.
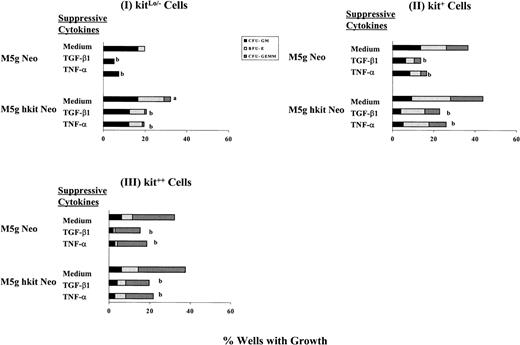
These results suggested that transduction with c-kit cDNA reduced the sensitivity of progenitors to the growth inhibitory effects of TGF-β1 and TNF-α. To further test this hypothesis, we transduced single cells with c-kit cDNA and cultured them under serum-depleted conditions. The results summarized from 3 separate experiments are shown in Fig 6. Using kitLo/− cells in the presence of full dosages of GM-CSF, IL-3, IL-6, Epo, and SLF, transduction of c-kit reduced the inhibitory effect of TGF-β1 from 74% in mock virus-transduced cells to 45% in c-kit–transduced cells (Fig 6I). Using kit+ cells, similar, but slightly less, reduced inhibitory effects were detected in c-kit transduced cells (48% decrease) compared with mock-transduced cells (63% decrease) (Fig 6II). In contrast, no significant reduction in inhibitory effects by TGF-β1 was noticed in Kit++ cells transduced with eitherc-kit (47% decrease) or mock (51% decrease) virus (Fig 6III). Similar results were obtained when lower dosages of SLF (10 ng/mL) were used (data not shown), and the same pattern was noticed in the presence of serum (data not shown).
Inhibitory effects of TGF-β1 and TNF- on colony formation from single sorted CD34+++kitLo/− (I), kit+(II), and kit++ (III) cells transduced withc-kit cDNA or mock control and growth in the presence of full dosages of IL-3, GM-CSF, Epo, and SLF with or without TGF-β1 (5 ng) or TNF- (20 ng) per mL in the serum-depleted cultures. Results are expressed as percent wells with growth from a total of 96 wells per point from 3 separate experiments. Significant differences for cells transduced with c-kit cDNA from that with mock control;aP < .05; significant differences for cells incubated with TGF-β1 or TNF- from medium control;bP < .05.
Inhibitory effects of TGF-β1 and TNF- on colony formation from single sorted CD34+++kitLo/− (I), kit+(II), and kit++ (III) cells transduced withc-kit cDNA or mock control and growth in the presence of full dosages of IL-3, GM-CSF, Epo, and SLF with or without TGF-β1 (5 ng) or TNF- (20 ng) per mL in the serum-depleted cultures. Results are expressed as percent wells with growth from a total of 96 wells per point from 3 separate experiments. Significant differences for cells transduced with c-kit cDNA from that with mock control;aP < .05; significant differences for cells incubated with TGF-β1 or TNF- from medium control;bP < .05.
Using kitLo/− cells in the presence of full dosages of GM-CSF, IL-3, IL-6, Epo, and SLF and in the absence of serum, transduction of c-kit reduced the inhibitory effect by TNF-α from 63% in mock-transduced cells to 48% in c-kit–transduced cells (Fig 6I). Using kit+ cells, similar, but slightly less, reduced inhibitory effects were detected inc-kit–transduced cells (41% decrease) compared with mock virus transduced cells (54% decrease) (Fig 6II). However, no significant reduction in inhibitory effects by TNF-α was obtained in Kit++ cells transduced with c-kit (42% decrease) compared with that of mock virus-transduced cells (42% decrease) (Fig6III). Similar results were obtained when SLF was used at 10 ng/mL (data not shown), and the same pattern was noticed in the presence of serum (data not shown).
Proviral integration and expression of c-kit cDNA by PCR and RT-PCR analysis.
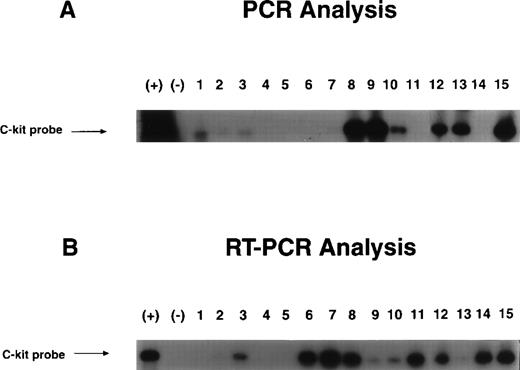
Proviral integration and gene expression of c-kit cDNA were determined from individual colonies by PCR and RT-PCR analysis using primers that contained both c-kit cDNA and viral vector sequences as shown in Fig 2. Figure 7 shows a sample of DNA integration and mRNA expression of the transducedc-kit gene in cells from individual colonies derived from single sorted kit++ and kit+ cells transduced with c-kit cDNA and growth in the presence of full dosages of GM-CSF, IL-3, IL-6, Epo, and SLF. DNA and RNA were extracted from individual colonies derived from single sorted kit++ and kit+ cells. Integration and expression of retroviralc-kit was found in 47.6% and 50.5%, respectively, from a total of 105 and 97 colonies analyzed. There was no difference in integration or expression for kit++ and kit+cells.
Examples of integration and expression of transducedc-kit cDNA by PCR (A) and RT-PCR (B) analysis. Sorted single CD34+++kit++ and kit+cells were prestimulated, transduced with c-kit cDNA or mock control viruses, and growth in the presence of cytokine combination as described in Materials and Methods. Individual BFU-E colonies were removed and DNA and RNA were extracted for PCR and RT-PCR analysis. The PCR generates a 959-bp fragment.
Examples of integration and expression of transducedc-kit cDNA by PCR (A) and RT-PCR (B) analysis. Sorted single CD34+++kit++ and kit+cells were prestimulated, transduced with c-kit cDNA or mock control viruses, and growth in the presence of cytokine combination as described in Materials and Methods. Individual BFU-E colonies were removed and DNA and RNA were extracted for PCR and RT-PCR analysis. The PCR generates a 959-bp fragment.
Detection of endogenous hSLF in cultures.
We noted that c-kit–transduced cells had enhanced colony formation even in the absence of exogenously added SLF, suggesting that endogenous SLF might be present in the cultures. Neutralizing antibodies against human SLF and c-kit (R&D Systems) were assessed for their capacity to neutralize endogenous SLF activity potentially present in our cultures. Sorted kit++, kit+, and kitLo/− subsets of CD34++ cells transduced with c-kit cDNA were incubated with neutralizing antibodies against SLF (1 μg/mL) and/orc-kit (100 ng/mL) in the presence and absence of serum with full dosages of GM-CSF, IL-3, IL-6, and Epo/mL at room temperature for 1.5 hours. No exogenous SLF was added. The cells were then assayed at 300 cells/mL for serum-containing or 800 cells/mL for serum-depleted cultures in the presence of the same cytokine combination and antibodies. Results from 1 of 2 representative serum-depleted experiments are shown in Fig 8. Either anti-SLF or anti-c-kit partially blocked colony formation stimulated by the cytokine combination in both c-kitor mock transduced cells, suggesting the presence of endogenous SLF in the cultures. Slightly greater, but not complete, neutralizing activity was noted using anti–c-kit than anti-SLF. No further inhibition was seen using both anti-SLF and anti–c-kitantibodies. Similar results were obtained in 2 experiments for serum-containing cultures (data not shown). These results indicate that endogenous SLF might contribute to the enhancing effects ofc-kit transduction in these cultures even in the absence of the addition of SLF, but in the presence of GM-CSF + IL-3 + IL-6 + Epo.
Effects of neutralizing antibodies against human SLF and/or c-kit on colony formation by CD34+++kitLo/− (I), kit+ (II), and kit++ (III) cells transduced with c-kit cDNA in the serum-depleted cultures. Transduced cells were treated with or without neutralizing antibodies at room temperature for 1.5 hours and assayed for colony formation at 800 cells/mL in the presence of full dosages of IL-3, GM-CSF, IL-6, and Epo. Results are expressed as mean ± SEM from 1 of 2 representative experiments. Significant differences for cells transduced with c-kit cDNA from that with mock virus control,aP < .05. Significant differences for cells treated with neutralizing antibodies from that of medium without added antibodies as control, bP < .05.
Effects of neutralizing antibodies against human SLF and/or c-kit on colony formation by CD34+++kitLo/− (I), kit+ (II), and kit++ (III) cells transduced with c-kit cDNA in the serum-depleted cultures. Transduced cells were treated with or without neutralizing antibodies at room temperature for 1.5 hours and assayed for colony formation at 800 cells/mL in the presence of full dosages of IL-3, GM-CSF, IL-6, and Epo. Results are expressed as mean ± SEM from 1 of 2 representative experiments. Significant differences for cells transduced with c-kit cDNA from that with mock virus control,aP < .05. Significant differences for cells treated with neutralizing antibodies from that of medium without added antibodies as control, bP < .05.
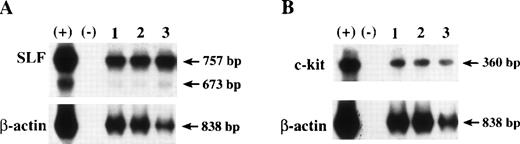
To determine whether the cells were releasing soluble SLF protein, conditioned medium (CM) was collected from 106MACS-separated CD34+ cells or no cells in the presence or absence of serum and presence of GM-CSF, IL-3, IL-6, and Epo and assayed for SLF using an ELISA method. In 3 separate experiments, SLF levels were below the level of assay sensitivity with or without serum or cells. We then sought to determine if cell-associated SLF was expressed by these cells. Whole cell lysates were prepared from MACS-separated CD34+ cells and measured by ELISA. An average of 60.4 ± 14.4 pg (n = 4) of SLF was detected in the lysates from 3 × 105 cells per sample. The results suggest that cell-associated SLF may contribute to the enhancing effects of tranduction of c-kit. Because only small numbers of cells were available, we were unable to compare the level of SLF in the 3 subsets of CD34+++ cells. Therefore, RT-PCR was used to assess SLF expression in CD34+++ cells and their subsets. Results from 4 experiments show that SLF expression was detected in all 3 subsets. Figure 9A shows a sample of SLF expression by RT-PCR in the 3 subsets of cells. Interestingly, it seems that the expression level of SLF was higher in kitLo/− cells (lane 3) than in the other 2 subsets (lanes 1 and 2). No obvious differences were noted in the level of SLF in these 3 populations before and after c-kit gene transduction (data not shown). We were unable to detect SLF expression on the cell surface using a flow cytometric-based assay system.52
Examples of expression of SLF (A) and c-kit (B) by RT-PCR in c-kit subsets of CD34+++ cells in 1 representative of 4 experiments. RNA was extracted from cells and reverse transcription was performed. PCR for SLF was performed using a pair of primers as described in Materials and Methods, which generate 757-bp and 673-bp products, respectively, representing soluble and membrane-bound SLF (A). PCR analysis for c-kit was performed using a pair of primers as described in Materials and Methods, which generates a 360-bp product (B). β-Actin, used as internal control, was performed using a pair of primers as described in Materials and Methods, and generates an 838-bp product. RNA extracts from human stromal cells (NFF) were used as positive control (+), and PCR reaction reagents were used as negative control (−). Lanes 1 through 3 are samples from CD34+++kit++, kit+, and kitLo/−cells, respectively.
Examples of expression of SLF (A) and c-kit (B) by RT-PCR in c-kit subsets of CD34+++ cells in 1 representative of 4 experiments. RNA was extracted from cells and reverse transcription was performed. PCR for SLF was performed using a pair of primers as described in Materials and Methods, which generate 757-bp and 673-bp products, respectively, representing soluble and membrane-bound SLF (A). PCR analysis for c-kit was performed using a pair of primers as described in Materials and Methods, which generates a 360-bp product (B). β-Actin, used as internal control, was performed using a pair of primers as described in Materials and Methods, and generates an 838-bp product. RNA extracts from human stromal cells (NFF) were used as positive control (+), and PCR reaction reagents were used as negative control (−). Lanes 1 through 3 are samples from CD34+++kit++, kit+, and kitLo/−cells, respectively.
Influence of transduction of c-kit on expression of c-kit protein on CD34+ cells and subsets by flow cytometer analysis.
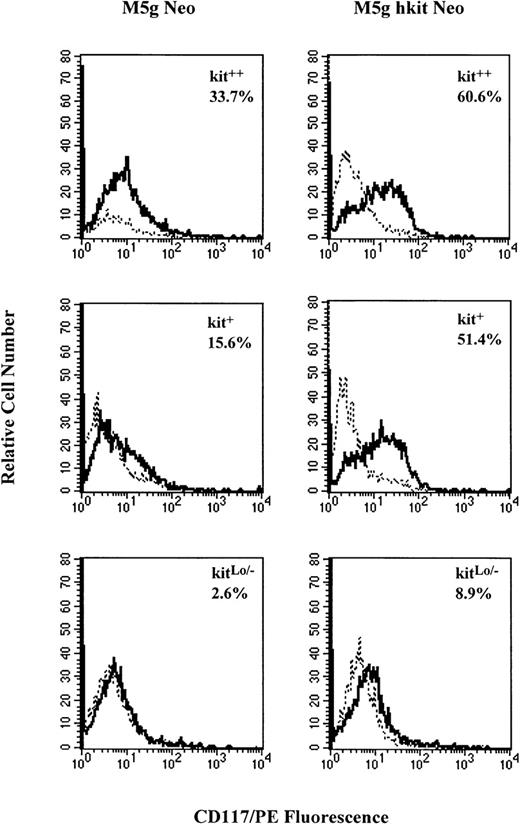
The number of CD34+++ cells available was too small to allow serial measurement of c-kit expression over time. Therefore, MACS-separated CD34+ cells (90% to 95% purity and with a 40% cloning efficiency for progenitors under optimal cytokine combinations) were used for flow cytometry analysis with monoclonal antibody against c-kit. Results from 5 separate experiments showed that CD34+ cells expressed c-kiton 64.2% ± 10% of the cells and this percent expression declined to 40.2% ± 7% after 2 days of prestimulation with full dosages of GM-CSF, IL-3, IL-6, and Epo, but without the addition of SLF. After gene transduction, the cells were incubated with the same cytokines for 24, 48, and 72 hours. The percent of c-kit expressing cells transduced withc-kit (80.2% ± 8%) was significantly greater than the mock virus-transduced cells (50.2% ± 7%) at 48 hours posttransduction. No significant decreases in percent of c-kitexpressing cells were seen by TGF-β1 or TNF-α in cells transduced with either c-kit or control virus (data not shown). To compare the influence of c-kit gene transduction on the c-kitprotein expression in subsets of CD34+++ cells, kit++, kit+, and kitLo/−cells were prestimulated and transduced with c-kit cDNA.c-kit expression on kitLo/− cells could also be detected by RT-PCR analysis (Fig 9B). After transduction, the cells were incubated with the same cytokine combination (without SLF) for 48 hours, and c-kit expression was analyzed by flow cytometry. Results from 4 separate experiments are summarized in Table 1. The percent of c-kitexpressing cells was increased in all 3 subsets of cells transduced with c-kit cDNA compared with the mock virus-transduced cells. The greater increase was seen in transduced kit+ (118%) and kitLo/− (101%) cells as compared with the kit++ cells (17%). Figure 10shows a representative experiment of c-kit expressing cells in the 3 c-kit subsets of CD34+++ cells transduced withc-kit cDNA or mock virus as analyzed by flow cytometer. Not only was the percent of c-kit expressing cells increased in the 3 subsets, but the fluorescence intensity was increased as well. The mean peak fluorescence channel was increased by 24% ± 11%, 37% ± 22%, and 34% ± 10%, respectively, inc-kit–transduced kit++, kit+, and kitLo/− cells compared with mock virus-transduced cells. The results indicate that transduction of c-kitincreased c-kit protein expression on the cell surface and increased c-kit fluorescence intensity as well.
Effect of Transduced c-kit on c-kitExpression on Subsets of CD34+++ CB Cells Analyzed by Flow Cytometry
| Subsets of CD34+++ Cells . | Percent c-kit Expressing Cells . | ||
|---|---|---|---|
| M5g Neo . | M5g hkit Neo . | Percent Change . | |
| Kit++ | 45.2 ± 14 | 53.1 ± 15 | +17 |
| Kit+ | 21.1 ± 4 | 46.0 ± 17 | +118 |
| KitLo/− | 7.2 ± 3 | 14.5 ± 4 | +101 |
| Subsets of CD34+++ Cells . | Percent c-kit Expressing Cells . | ||
|---|---|---|---|
| M5g Neo . | M5g hkit Neo . | Percent Change . | |
| Kit++ | 45.2 ± 14 | 53.1 ± 15 | +17 |
| Kit+ | 21.1 ± 4 | 46.0 ± 17 | +118 |
| KitLo/− | 7.2 ± 3 | 14.5 ± 4 | +101 |
Sorted CD34+++ kit++, kit+, and kitLo/− cells were prestimulated with GM-CSF (200 U), IL-3 (200 U), Epo (1 U), and IL-6 (10 ng) per mL without SLF and transduced with c-kit (M5g hkit Neo) or mock (M5g neo) viruses as described in Materials and Methods. The cells were washed and incubated with the same cytokine combination for 48 hours. Cells were washed and stained with CD117/PE or isotope control and analyzed by flow cytometer. Results are expressed as mean percent of c-kitexpressing cells ± SEM from an average of 4 separate experiments.
Representative analysis of c-kit protein expression on CD34+++ kit++, kit+, and kitLo/− cells transduced withc-kit cDNA by flow cytometer. Sorted cells were prestimulated with 10% FCS and full dosage of IL-3, GM-CSF, IL-6, and Epo and transduced with c-kit cDNA (M5g hkit Neo) or mock (M5g Neo) control viruses as described in Materials and Methods. The cells were washed and incubated with the same cytokine combination for 48 hours and then stained with monoclonal CD117/PE (solid line) or mouse isotope control (dashed line) and analyzed by flow cytometer.
Representative analysis of c-kit protein expression on CD34+++ kit++, kit+, and kitLo/− cells transduced withc-kit cDNA by flow cytometer. Sorted cells were prestimulated with 10% FCS and full dosage of IL-3, GM-CSF, IL-6, and Epo and transduced with c-kit cDNA (M5g hkit Neo) or mock (M5g Neo) control viruses as described in Materials and Methods. The cells were washed and incubated with the same cytokine combination for 48 hours and then stained with monoclonal CD117/PE (solid line) or mouse isotope control (dashed line) and analyzed by flow cytometer.
Influence of transduction of c-kit on apoptosis in MACS-separated CD34+ cells as assessed by TUNEL assay.
One reported function of SLF is to inhibit apoptosis.57 We tested whether enforced expression of c-kit altered cellular susceptibility to apoptosis. Summarized results from 4 experiments are shown in Table 2. In mock virus-transduced cells, TGF-β1 and TNF-α significantly increased the percent of apoptotic cells from 28.6% ± 6% to 44.1% ± 4% and 36.2% ± 5%, respectively, in the absence of SLF, but in the presence of full doses of GM-CSF, IL-3, and Epo. Addition of SLF at 50 ng/mL resulted in reduction of apoptotic cells to 14.8% ± 4% in mock virus-transduced cells. Addition of TNF-α, but not TGF-β1, to cultures of mock virus-transduced cells cultured with SLF slightly, but significantly, increased apoptosis. Transduction of c-kitinduced a marked reduction of apoptotic cells (4.4% ± 3%) in the absence of exogenously added SLF, and no further reduction was seen with addition of SLF at 50 ng/mL to cultures ofc-kit–transduced cells. These results suggest that transducedc-kit, even without exogenous addition of SLF, plays an important role in preventing apoptosis, possibly due to the small amount of endogenous cell-associated SLF expressed by these cells.
Effect of Transduced c-kit on Apoptotic CD34+ CB Cells Analyzed by TUNEL Assay and Flow Cytometry
| . | M5g Neo . | M5g hkit Neo . | ||
|---|---|---|---|---|
| Apoptotic Cells (%) . | No. of Experiments . | Apoptotic Cells (%) . | No. of Experiments . | |
| (I) No SLF | ||||
| Medium | 28.6 ± 6 | 4 | 4.4 ± 3† | 3 |
| TNF-α | 44.1 ± 4* | 4 | 8.5 ± 3† | 3 |
| TGF-β1 | 36.2 ± 5* | 4 | 9.5 ± 4† | 3 |
| (II) SLF (50 ng/mL) | ||||
| Medium | 14.8 ± 4 | 7 | 9.3 ± 3† | 3 |
| TNF-α | 20.1 ± 5* | 7 | 7.8 ± 2† | 3 |
| TGF-β1 | 17.2 ± 3* | 7 | 9.4 ± 3† | 3 |
| . | M5g Neo . | M5g hkit Neo . | ||
|---|---|---|---|---|
| Apoptotic Cells (%) . | No. of Experiments . | Apoptotic Cells (%) . | No. of Experiments . | |
| (I) No SLF | ||||
| Medium | 28.6 ± 6 | 4 | 4.4 ± 3† | 3 |
| TNF-α | 44.1 ± 4* | 4 | 8.5 ± 3† | 3 |
| TGF-β1 | 36.2 ± 5* | 4 | 9.5 ± 4† | 3 |
| (II) SLF (50 ng/mL) | ||||
| Medium | 14.8 ± 4 | 7 | 9.3 ± 3† | 3 |
| TNF-α | 20.1 ± 5* | 7 | 7.8 ± 2† | 3 |
| TGF-β1 | 17.2 ± 3* | 7 | 9.4 ± 3† | 3 |
MACS-separated CD34+ cells were prestimulated with GM-CSF (200 U), IL-3 (200 U), Epo (1 U), and IL-6 (10 ng)/mL and transduced with c-kit or mock viruses as described in Materials and Methods. The cells were washed and incubated with GM-CSF, IL-3, and Epo in the absence or presence of SLF at 50 ng/mL for 48 hours. Cells were washed and labeled with TUNEL reaction mixture and analyzed by flow cytometer. Results are expressed as percent apoptotic cells from an average of 3 to 7 separate experiments.
Significant enhancement in apoptosis with TNF-α or TGF-β1,P < .05.
Significant decrease in apoptosis of c-kit–transduced compared with mock virus-transduced cells, P < .05.
DISCUSSION
Studies of mice with partial or complete inactivating mutations of thec-kit (W locus) or SLF (Sl locus) genes have highlighted the essential role of an intact SLF/c-kit axis in maintaining steady state hematopoiesis. Mice with a complete deficiency of wild-type c-kit protein product are not viable due to severe anemia. Mice with partial function of c-kit protein are viable, but have varying degrees of macrocytic anemia and tissue mast cell deficiency.18 These studies of c-kit function in murine hematopoiesis suggest that under certain conditions the amount of functional c-kit protein is limiting to stem/progenitor cell proliferation.
We tested the hypothesis that increasing c-kit expression on CD34+++ cells and their subsets with a human c-kitencoding retrovirus would alter proliferation and/or differentiation. Transduction of CD34+++kit++ cells with ac-kit encoding retrovirus had only a small effect on the total number of colonies formed compared with mock virus-transduced cells, while transduction of CD34+++kit+ or CD34+++kitLo/− cells with thec-kit encoding retrovirus resulted in substantial increases in total colony number when cells were plated in the presence of a combination of cytokines in the presence and absence of serum. The increase in colony numbers was due to a marked increase in BFU-E– and a smaller increase in CFU-GEMM–derived colonies. Transduction ofc-kit did not significantly increase CFU-GM proliferation even in the starting population of CD34+++kitLo/− cells. In the absence of exogenously added SLF, but in the presence of added GM-CSF, IL-3, and Epo, the enhancing effects of transduced c-kit were more significant in all 3 subsets of the cells than in the presence of added GM-CSF, IL-3, Epo, and SLF. Moreover, the enhancing effect of transduced c-kit on colony formation was confirmed at the single-cell level and in the absence of serum. The largest effects of the transduced c-kit were seen in CD34+++kitLo/− cells grown in the presence of GM-CSF, IL-3, and Epo, but in the absence of exogenously added SLF under serum-depleted culture conditions.
The exact mechanism for the increase of erythroid and multipotential progenitor cell proliferation in c-kit–transduced cells is not clear. However, it is highly likely that this is directly due to the increased c-kit protein expression by c-kit–transduced cells. The percent of c-kit expressing cells was significantly increased in the 3 subsets of CD34+++ cells transduced withc-kit cDNA. Among the 3 subsets, the increase of c-kitexpressing cells was greater in the lower density of c-kit(kit+ and kitLo/−) expressing cells before c-kit transduction. Not only was the percent of c-kit expressing cells increased, but the fluorescence intensity was increased as well. Likewise, the enhancing effect of c-kittransduction on colony formation was greater inc-kit–transduced kit+ and kitLo/− cells than in kit++ cells.
We hypothesize that optimal hematopoiesis might require a certain minimal level of c-kit function and that by increasingc-kit expression in kit+ and kitLo/− cells, a threshold level of expression is reached thereby recruiting additional progenitors into a proliferative state. However, there may be an upper limit to c-kit expression above which no further biologic response is elicited due to maximal stimulation of downstream signal transduction pathways. Alternatively, heterogeneity of c-kit expression in CD34+++ cells may also reflect differences in differentiation status. We have reported previously that CD34+++kit++ cells are enriched for CFU-GEMM and BFU-E, CD34+++kit+cells are enriched for more primitive high proliferative potential–colony-forming cell (HPP-CFC), while CD34+++kitLo/− cells are enriched for CFU-GM.10 Thus, transduction of c-kit into different classes of HPC may result in disparate effects on differentiation.
Transduction of progenitors with c-kit may increase colony formation at least in part by inhibition of apoptosis.57Our data clearly show that apoptosis in the c-kit–transduced cell population (4.4% ± 2%) was lower than in the mock virus-transduced cell population (28.6% ± 6%), even in the absence of exogenously added SLF. Our data suggest that SLF has different dose response effects on apoptosis versus proliferation. Inc-kit–transduced cells, endogenous SLF is sufficient to maximally inhibit apoptosis even in the presence of the inhibitory cytokines TNF-α and TGF-β1. However, maximal colony growth is obtained only with the addition of exogenous SLF.
Our colony formation experiments and apoptosis assays indicated a biologic effect of c-kit transduction even in the absence of exogenous SLF. The enhancing effects of c-kit transduction on colony formation were blocked by neutralizing anti-SLF and/or anti–c-kit antibodies indicating that the observed effect was likely due to interaction of SLF with its receptor. Similar results were obtained in serum-depleted cultures indicating that the progenitor cells rather than the serum was the source of endogenous SLF in our system. Indeed, any SLF that might be present in our serum-containing culture medium was below the level of detectability, nor could we detect soluble SLF in conditioned medium from progenitor cells. However, endogenous cell-associated SLF was detected in MACS-separated CD34+ cells by ELISA assay. The results suggest that cell-associated SLF is the major source of SLF in our culture system and likely contributes to the enhancing effects of transduced c-kit gene seen in the absence of exogenous SLF. Because of the small number of subsets of CD34+ cells, we were unable to compare the level of cell-associated SLF between the 3 subsets of cells using ELISA assay. However, using RT-PCR analysis, transcripts for both the soluble and membrane-bound isoforms of SLF were found in CD34+ cells and all 3 subsets of CD34+++ (kit++, kit+, and kitLo/−) cells.
Taken together, our antibody neutralization studies, ELISA, and RT-PCR experiments strongly suggest that endogenous SLF expression by CD34+ cells may play a role in regulating progenitor proliferation and/or apoptosis. Our results are in agreement with those of Ratajczak et al58 who previously reported that CD34+ cells transcribe SLF mRNA and that treatment of such cells with antisense oligonucleotides complementary to SLF mRNA reduced BFU-E and CFU-Mix, but not CFU-GM, colony formation.
TGF-β1 appears to be a critical negative regulator of hematopoiesis. One reported effect of TGF-β1 on HPC is to repress expression ofc-kit protein by accelerating the degradation of c-kittranscripts.9,52,56 Because the effects of c-kit on CDK1 and Rb phosphorylation are potentially antagonistic to the effects of TGF-β1 on cell-cycle progression, we reasoned that the effect of TGF-β1 on c-kit expression might be critical to the overall inhibitory effect of TGF-β1 on HPC proliferation/differentiation.35-37,59 60 Our data provide evidence that enforced expression of c-kit antagonizes the antiproliferative effects of TGF-β1.
TNF-α is another complex regulator of hematopoiesis. The actions of TNF-α on HPC are biphasic and can be either stimulatory or inhibitory depending on the target cell type. Additional complexity arises because TNF-α is a potent inducer of hematopoietic growth factor release by accessory cells.61 A physiological role in regulating primitive HPC proliferation is suggested by the report of significantly increased numbers of Lin− Sca1+kit+ HPC in the bone marrow of mice with deficiency of TNF-receptor p55 protein.62 The actions of TNF-α on HPC are poorly understood, but might be mediated via cell-cycle arrest or increased apoptosis, as reported by several groups.63Another reported action of TNF-α is to accelerate the degradation ofc-kit transcripts.43 44 Signaling through thec-kit receptor is associated with increased cell-cycle progression and is antiapoptotic, especially for erythroid progenitors. Our data provide evidence that enforced expression of c-kitantagonizes the antiproliferative and/or antiapoptotic effects of TNF-α.
In summary, our experiments have demonstrated that transduction of CD34+++ cells and their subsets with a c-kitencoding retrovirus increases growth factor-dependent erythroid and multipotential progenitor cell proliferation. These data are consistent with other data concerning the in vitro and in vivo biological effects of c-kit and further support the notion that optimal growth of HPC may require c-kit receptor. Transduction of progenitors with c-kit cDNA was associated with increased expression ofc-kit protein, decreased spontaneous or cytokine-induced apoptosis, and expression of endogenous cell-associated SLF. In addition, c-kit transduction of CD34+++ cells and their subsets decreased the sensitivity of these cells to the inhibitory effects of TGF-β1 and TNF-α.
ACKNOWLEDGMENT
We thank Rebecca Miller and Linda Cheung for secretarial assistance and Drs Jie He, Yung-xing Li, and Hong-Jun Liu for technical assistance.
L.L. and M.C.H. contributed equally to this work.
Supported by Public Health Service Grants No. RO1 HL 56416, RO1 HL 54037, RO1 DK 53674 and a project in RO1 HL-53586 from the National Institutes of Health (to H.E.B.), grants from the Phi Beta Psi Sorority and Genetics Institute (to L.L.), and a VA Merit Review Grant from the Department of Veterans Affairs (to M.C.H.).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Li Lu, MD, the Walther Oncology Center, Indiana University School of Medicine, 1044 W Walnut St, Room 302, Indianapolis, IN 46202-5254.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal