As mice carrying mutations of the DNA mismatch repair genes MSH2 and MSH6 often develop lymphoid neoplasms, we addressed the prevalence of the replication error (RER+) phenotype, a manifestation of an underlying defect of DNA mismatch repair genes, in human lymphoid tumors. We compared microsatellite instability (MSI) at 10 loci in 37 lymphoid tumors, including 16 acute lymphoid leukemias (ALL) and 21 non-Hodgkin’s lymphomas (NHL), and in 29 acute myeloid leukemias (AML). Significant differences in MSI prevalence between AMLs and ALLs emerged, and MSI occurrence was more frequent in the NHLs versus AMLs. Indeed, only 3 of 29 (10%) AMLs exhibited MSI, thus confirming its paucity in myeloid tumors, while 10 of 37 (27%) lymphoid tumors, 6 ALLs and 4 NHLs, disclosed an RER+phenotype. In 1 ALL patient, the same molecular alterations were observed in correspondence with a relapse, but were not detected during remission over a 14-month follow-up; in another ALL patient, findings correlated with impending clinical relapse. These results suggest that the study of MSI in lymphoid tumors might provide a useful molecular tool to monitor disease progression in a subset of ALLs. To correlate MSI with other known genetic abnormalities, we investigated the status of the proto-oncogene, bcl-2, in the lymphoma patients and found that 4 of 4 NHL patients with MSI carried bcl-2 rearrangements, thus linking genomic instability to enhanced cell survival in NHL; moreover, no p53 mutations were found in these patients. Finally, we addressed the putative cause of MSI in hematopoietic tumors by searching for both mutations and deletions affecting DNA repair genes. A limited genetic analysis did not show any tumor-specific mutation in MLH1 exons 9 and 16 and in MSH2 exons 5 and 13. However, loss of heterozygosity (LOH) of markers closely linked to mismatch repair genes MLH1, MSH2, and PMS2 was demonstrated in 4 of 6 ALLs and 1 of 3 AMLs with MSI. These observations indicate that chromosomal deletions might represent a mechanism of inactivation of DNA repair genes in acute leukemia.
MICROSATELLITES are highly polymorphic genetic markers dispersed in the human genome and comprise di-, tri-, and tetranucleotide repeats. Microsatellite instability (MSI) was first observed in subjects with hereditary nonpolyposis colorectal cancer (HNPCC) as new alleles generated through errors of DNA replication.1,2 Later work showed that MSI may be attributed to an underlying defect of the DNA mismatch repair genes, including MLH1, MSH2, PMS1, PMS2, and MLH6.3-6 The overall findings of several studies that addressed MSI occurrence in hematologic neoplasms indicate that MSI is uncommon in acute myeloid leukemia (AML), as it is generally detected in less than 10% of patients.7-12 This is not surprising because the predominant type of genetic damage in hematopoietic tumors consists of specific translocations juxtaposing genes that are normally distant in the genome, thereby activating proto-oncogenes and creating new fusion proteins.13 Nevertheless, alterations in DNA repair genes might play a role in defined subsets of hematoproliferative disorders, and in particular, lymphoid tumors. Indeed, studies in MSH2- and MSH6-deficient mice clearly establish a link between mismatch repair gene defects and development of lymphoid tumors.14-16Consistent with this, MSI has been detected in lymphomas from human immunodeficiency virus (HIV)+ subjects,17 and MSH2 mutations have been reported in adult lymphoblastic lymphoma,18 thus suggesting that a similar connection might also exist in man. Our study explores this hypothesis by investigating microsatellite alterations in human lymphoid neoplasms, and addressing their relationship to rearrangements of the proto-oncogene, bcl-2, often involved in the pathogenesis of lymphoid neoplasms as an inhibitor of apoptosis (reviewed by Chao and Korsmeyer19).
In hereditary and sporadic solid tumors, MSI is generally linked to point mutations of DNA mismatch repair genes, and gene inactivation by deletions is uncommon.3-6 This relationship, however, has been rarely if at all addressed in hematopoietic tumors. As chromosomal aberrations are commonly found in leukemia and lymphoma, we investigated their contribution to the inactivation of MLH1, MSH2, PMS1, and PMS2 genes; we report that loss of heterozygosity (LOH) involving microsatellite markers closely associated with mismatch repair genes occurs in some hematopoietic tumors with MSI.
MATERIALS AND METHODS
Patients and DNA isolation.
We analyzed 66 samples of tumor DNA obtained from the Third Medical Clinic of the University of Heidelberg (Heidelberg, Germany) and from the Divisione di Ematologia of the University of Verona (Verona, Italy). These samples corresponded to 45 cases of leukemia (29 AML, 16 acute lymphoid leukemia [ALL]), and 21 of non-Hodgkin’s lymphoma (NHL), as detailed in Table 1. Tumor DNA was extracted from bone marrow biopsies or peripheral blood in the case of leukemias and from bone marrow or lymph nodes in the case of NHL. Control constitutional DNAs were extracted either from the leukocytes or bone marrow of the same patients during remission or from lymphoma-unaffected bone marrow samples of some NHL patients. The AMLs were classified morphologically according to the French-American-British classification,20 and the NHLs according to the Revised European-American Lymphoma (REAL) classification.21 ALLs were classified according to their immunophenotype.22 High-molecular-weight DNA was isolated by standard procedures.23 Cytogenetic analysis was performed on short-term cultures of bone marrow or peripheral blood cells. Chromosomes were analyzed using a modified GAG-banding technique, as described elsewhere.24
Study Population
| AML . | No. . | ALL . | No. . | NHL . | No. . |
|---|---|---|---|---|---|
| M0 | 1 | pre-B ALL | 9 | B-cell chronic lymphocytic leukemia | 2 |
| M1 | 3 | B ALL | 3 | Small lymphocytic lymphoma | 2 |
| M2 | 5 | T ALL | 4 | Immunocytoma | 1 |
| M3 | 8 | Plasmacytoma | 1 | ||
| M4 | 7 | Follicle center lymphoma | 7 | ||
| M5 | 4 | Mantle cell lymphoma | 3 | ||
| Hybrid | 1 | Diffuse large B-cell lymphoma | 5 | ||
| Total | 29 | total | 16 | total | 21 |
| AML . | No. . | ALL . | No. . | NHL . | No. . |
|---|---|---|---|---|---|
| M0 | 1 | pre-B ALL | 9 | B-cell chronic lymphocytic leukemia | 2 |
| M1 | 3 | B ALL | 3 | Small lymphocytic lymphoma | 2 |
| M2 | 5 | T ALL | 4 | Immunocytoma | 1 |
| M3 | 8 | Plasmacytoma | 1 | ||
| M4 | 7 | Follicle center lymphoma | 7 | ||
| M5 | 4 | Mantle cell lymphoma | 3 | ||
| Hybrid | 1 | Diffuse large B-cell lymphoma | 5 | ||
| Total | 29 | total | 16 | total | 21 |
Microsatellite analysis.
DNA was amplified by polymerase chain reaction (PCR), as previously described,8 in a volume of 25 μL containing 0.1 μg of genomic DNA as template, 50 mmol/L KCl, 10 mmol/L Tris-HCl (pH 8.3), 1.5 mmol/L MgCl2, 0.2 mmol/L of each deoxynucleoside triphosphate, 0.4 μmol/L of each sense and antisense primer, and 1 U of Taq Polymerase (Perkin Elmer, Norwalk, CT). The primer pairs listed in Table 2 were used. Forward primers were end-labeled with [γ33P] adenosine triphosphate (ATP) for 1 hour at 37°C using T4 polynucleotide kinase (Amersham, Little Chalfont, UK). Reaction conditions consisted of 1 minute at 94°C, 30 seconds at 55°C, and 30 seconds at 72°C for 30 cycles. PCR products were electrophoresed on denaturing 6% polyacrylamide formamide-containing gels. After electrophoresis, the gel was dried and exposed to x-ray film at −70°C for 24 to 72 hours.
Primer Sequences, Chromosomal Location, and Size of Expected PCR Products
| Primer Pair . | Sequence . | Locus . | Size . |
|---|---|---|---|
| D2S123-for | 5′-CCT TTC TGA CTT GGA TAC CAT CTA TCT ATC TA-3′ | 2 | 157-187 |
| D2S123-rev | 5′-CAG GAT GCC TGC CTT TAA CAG TG-3′ | ||
| D9S126-for | 5′-ATT GAA ACT CTG CTG AAT TTT CTG-3′ | 9p21 | 238-248 |
| D9S126-rev | 5′-CAA CTC CTC TTG GGA ACT GC-3′ | ||
| D10S197-for | 5′-AGC TGA GAT CGC ACC ACT GCA CTT CAG-3′ | 10 | 161-173 |
| D10S197-rev | 5′-AGG GTA GCC TTT CCT ATC CTC CCA TTC-3′ | ||
| VW-for | 5′-CCC TAG TGG ATG ATA AGA ATA ATC-3′ | 12p12 | 138-162 |
| VW-rev | 5′-GGA CAG ATG ATA AAT ACA TAG GAT GGA ATG G-3′ | ||
| D15S212-for | 5′-CAG ATT TCA GAT GTG CCT AGT CCA C-3′ | 15 | 203-217 |
| D15S212-rev | 5′-CCT TAG CCA GTG AGT GCA TGT G-3′ | ||
| D15S161-for | 5′-TCTGTGATTTTGCCATTATGAG-3′ | 15 | 277-291 |
| D15S161-rev | 5′-TAAACTGGAATTTTTGACTATGAGC-3′ | ||
| D18S61-for | 5′-ATT TCT AAG AGG ACT CCC AAA CT-3′ | 18 | 157-183 |
| D18S61-rev | 5′-ATA TTT TGA AAC TCA GGA GCA T-3′ | ||
| D18S65-for | 5′-AAT AAG TTT GGA AGC AGG TGG AG-3′ | 18 | 168-178 |
| D18S65-rev | 5′-AGA GGA GAG GCT GGT CTT ACT AT-3′ | ||
| AR-for | 5′-TCC GCG AAG TGA TCC AGA AC-3′ | Xq11-12 | 213-225 |
| AR-rev | 5′-CTT GGG GAG AAC CAT CCT CA-3′ | ||
| DxS1217-for | 5′-TCC AGT AAG TTT GGT CTA TAT GAC G-3′ | X | 231-243 |
| DxS1217-rev | 5′-ATG AAG TAT CGT ATC TGA ATC CCG-3′ |
| Primer Pair . | Sequence . | Locus . | Size . |
|---|---|---|---|
| D2S123-for | 5′-CCT TTC TGA CTT GGA TAC CAT CTA TCT ATC TA-3′ | 2 | 157-187 |
| D2S123-rev | 5′-CAG GAT GCC TGC CTT TAA CAG TG-3′ | ||
| D9S126-for | 5′-ATT GAA ACT CTG CTG AAT TTT CTG-3′ | 9p21 | 238-248 |
| D9S126-rev | 5′-CAA CTC CTC TTG GGA ACT GC-3′ | ||
| D10S197-for | 5′-AGC TGA GAT CGC ACC ACT GCA CTT CAG-3′ | 10 | 161-173 |
| D10S197-rev | 5′-AGG GTA GCC TTT CCT ATC CTC CCA TTC-3′ | ||
| VW-for | 5′-CCC TAG TGG ATG ATA AGA ATA ATC-3′ | 12p12 | 138-162 |
| VW-rev | 5′-GGA CAG ATG ATA AAT ACA TAG GAT GGA ATG G-3′ | ||
| D15S212-for | 5′-CAG ATT TCA GAT GTG CCT AGT CCA C-3′ | 15 | 203-217 |
| D15S212-rev | 5′-CCT TAG CCA GTG AGT GCA TGT G-3′ | ||
| D15S161-for | 5′-TCTGTGATTTTGCCATTATGAG-3′ | 15 | 277-291 |
| D15S161-rev | 5′-TAAACTGGAATTTTTGACTATGAGC-3′ | ||
| D18S61-for | 5′-ATT TCT AAG AGG ACT CCC AAA CT-3′ | 18 | 157-183 |
| D18S61-rev | 5′-ATA TTT TGA AAC TCA GGA GCA T-3′ | ||
| D18S65-for | 5′-AAT AAG TTT GGA AGC AGG TGG AG-3′ | 18 | 168-178 |
| D18S65-rev | 5′-AGA GGA GAG GCT GGT CTT ACT AT-3′ | ||
| AR-for | 5′-TCC GCG AAG TGA TCC AGA AC-3′ | Xq11-12 | 213-225 |
| AR-rev | 5′-CTT GGG GAG AAC CAT CCT CA-3′ | ||
| DxS1217-for | 5′-TCC AGT AAG TTT GGT CTA TAT GAC G-3′ | X | 231-243 |
| DxS1217-rev | 5′-ATG AAG TAT CGT ATC TGA ATC CCG-3′ |
Molecular analysis of bcl-2 rearrangements.
Ten micrograms of high-molecular-weight genomic DNA was digested independently with 2 restriction endonucleases (EcoRI andHindIII) under conditions recommended by the supplier (Boehringer Mannheim, Mannheim, Germany). After electrophoresis on 1% agarose gels, DNA was transferred to nylon membranes (Bio Rad, Munich, Germany) according to the manufacturer’s instructions. For the hybridization of Southern blots, denatured 32P-labeled DNA probes were independently used for the major (MBR) and the minor (mcr) breakpoint cluster regions, PFL1 and PFL2, respectively,25 26 both kindly supplied by Dr M.L. Cleary (Stanford University School of Medicine, Stanford, CA). The labeling of the DNA probes by random priming was performed under conditions recommended by the supplier (Amersham). Autoradiography was performed at −70°C using x-ray film with double high-speed intensifying screens.
Mismatch repair genes analysis.
We performed LOH studies on the MLH1, MSH2, PMS1, and PMS2 DNA repair genes.3-5 Briefly, LOH was investigated by the same PCR assay described above, using the D3S1611, CA21, D2S117, and D7S517 microsatellite markers to study the MLH1, MSH2, PMS1, and PMS2 genes, respectively. The sequence of these primers has been reported elsewhere.27,28 The intensity of the bands corresponding to the 2 alleles of a given marker was quantified by analysis with Instantimager (Packard, Grove Hills, IL). In a set of experiments, the microsatellite markers D2S119, D2S136, D2S288, D2S378, D3S1561, D3S1298, D7S531, D7S481, and D7S503 were used28 to characterize the LOH involving the MLH1, MSH2, and PMS2 genes.
For single-strand conformation polymorphism (SSCP) analysis, exons 9 and 16 of MLH1 and exons 5 and 13 of MSH2 were amplified by PCR according to previously reported procedures,29 using exon-specific oligonucleotide primers reported by others.30 31 Briefly, 25 PCR cycles were run in 50 μL standard reaction mixture containing 0.2 μg of DNA, 0.4 μmol/L of each primer, and 2 U Taq polymerase (Perkin Elmer) as follows: 1 minute denaturation at 94°C, 30 seconds annealing at 55°C, and 30 seconds extension at 72°C in a DNA thermal-cycler (Perkin Elmer). A total of 10 μL of the amplified products was mixed with 0.1 μCi α-33P-dATP and 0.1 U Taq polymerase, and 5 additional cycles were run. To detect DNA mutations, PCR product samples were heated at 95°C for 5 minutes and then electrophoresed through a 6% acrylamide gel containing 5% glycerol; the gel was then dried, and exposed to x-ray film at −70°C for 24 to 72 hours.
P53 gene mutation analysis.
P53 gene mutations were detected by PCR-SSCP analysis of exons 5 to 8 with specific primers as described above.32
RESULTS
Microsatellite alterations in leukemias and NHL.
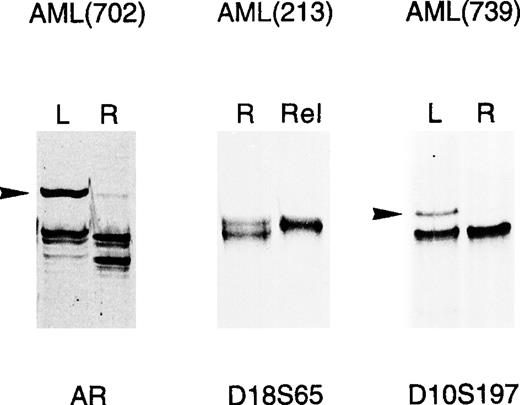
We looked for microsatellite alterations in 10 microsatellite markers scattered on 7 different chromosomes, as detailed in Table 2 and in Materials and Methods. MSI was defined as a gain of new alleles at a given locus; LOH was a significant attenuation (>50%), or loss of 1 normal allele in the tumor, compared with constitutional DNA; some representative alterations found in leukemias are shown in Fig 1. The bands indicating MSI, in some cases, were not equimolar to the germ line bands, likely due to contaminating normal cells, as also suggested by cytological analysis (data not shown), or heterogeneity within the leukemic poulation. PCR analysis disclosed molecular changes in only 3 of the 29 AML patients analyzed (10%), thus confirming the scarcity of MSI in adult myeloid leukemia.8-10 Interestingly, however, these alterations were detected in 10 of 37 lymphoid tumors (27%), including 6 of 16 ALL patients (37%) and 4 of 21 NHL patients (19%). Thus, in our study population, MSI was more frequently observed in ALL than AML patients (P < .05, Fisher’s exact test). Molecular alterations, listed in Table 3, were found on 1 to 3 loci in 9 of 13 cases; on the other hand, patient 213 (AML), and patients 32, 509, and VR5 (all with ALL) presented widespread genetic instability involving most of the loci analyzed and consisting of both MSI and LOH. Overall, MSI was more frequently detected, accounting for 33 of 44 molecular alterations found, while the remaining 11 were LOH at defined loci (Table 3). MSI was found in tumor DNA obtained at diagnosis in 6 leukemia patients (702, 739, 619, 32, 509, and VR5), and 2 NHL patients (11, 23), thus indirectly suggesting that alterations in mismatch repair genes might already occur in the early steps of the malignancy (Table 4). In the remaining 5 patients, alterations were detected in tumor DNA obtained at relapse, after chemotherapy. Overall, MSI in leukemia was detected in 3 of 9 patients studied at relapse (33%) and in 6 of 36 patients studied at diagnosis (16%) (Tables 4 and 5). However, because DNA samples at diagnosis were not available, it could not be established whether the same alterations were already present at disease onset or had developed during tumor progression.
Microsatellite alterations in adult acute leukemias. PCR amplification of the indicated microsatellites was performed as described in Materials and Methods on genomic DNA of AML patients at diagnosis (L) or relapse (Rel), and the resulting banding patterns were compared with those obtained at remission (R). Tumor-specific alterations consisted either of gain of new-length alleles (MSI), indicated by arrows, or loss of bands detected in constitutional DNA (LOH).
Microsatellite alterations in adult acute leukemias. PCR amplification of the indicated microsatellites was performed as described in Materials and Methods on genomic DNA of AML patients at diagnosis (L) or relapse (Rel), and the resulting banding patterns were compared with those obtained at remission (R). Tumor-specific alterations consisted either of gain of new-length alleles (MSI), indicated by arrows, or loss of bands detected in constitutional DNA (LOH).
Microsatellite Alterations in Leukemia and Lymphoma
| Patient No. . | Disease . | Microsatellite Marker . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| D2s123 . | D9s126 . | D10s197 . | VWA . | D15s212 . | D15s161 . | D18s61 . | D18s65 . | AR . | DXs1217 . | ||
| 213 | AML M1 | MSI | LOH | MSI | MSI | MSI | LOH | MSI | MSI | ||
| 702 | AML M2 | MSI | LOH | ||||||||
| 739 | AML M3 | MSI | |||||||||
| 32 | Pre-B ALL | MSI | MSI | MSI | MSI | LOH | MSI | LOH | |||
| 63 | Pre-B ALL | MSI | MSI | ||||||||
| 74 | B ALL | LOH | LOH | MSI | |||||||
| 509 | T ALL | MSI | MSI | MSI | MSI | MSI | MSI | ||||
| 619 | B ALL | MSI | MSI | ||||||||
| VR5 | T ALL | MSI | MSI | MSI | LOH | LOH | LOH | LOH | |||
| 11 | NHL | MSI | MSI | ||||||||
| 52 | NHL | MSI | |||||||||
| 23 | NHL | MSI | MSI | ||||||||
| 298 | NHL | MSI | |||||||||
| Patient No. . | Disease . | Microsatellite Marker . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| D2s123 . | D9s126 . | D10s197 . | VWA . | D15s212 . | D15s161 . | D18s61 . | D18s65 . | AR . | DXs1217 . | ||
| 213 | AML M1 | MSI | LOH | MSI | MSI | MSI | LOH | MSI | MSI | ||
| 702 | AML M2 | MSI | LOH | ||||||||
| 739 | AML M3 | MSI | |||||||||
| 32 | Pre-B ALL | MSI | MSI | MSI | MSI | LOH | MSI | LOH | |||
| 63 | Pre-B ALL | MSI | MSI | ||||||||
| 74 | B ALL | LOH | LOH | MSI | |||||||
| 509 | T ALL | MSI | MSI | MSI | MSI | MSI | MSI | ||||
| 619 | B ALL | MSI | MSI | ||||||||
| VR5 | T ALL | MSI | MSI | MSI | LOH | LOH | LOH | LOH | |||
| 11 | NHL | MSI | MSI | ||||||||
| 52 | NHL | MSI | |||||||||
| 23 | NHL | MSI | MSI | ||||||||
| 298 | NHL | MSI | |||||||||
The leukemia patients showing either MSI or LOH at the indicated loci are listed. Overall, 29 patients with AML and 37 patients with lymphoid tumors (16 ALL and 21 NHL) were studied for microsatellite alterations.
Clinical Characteristics and Genetic Markers of Patients With MSI
| Patient No. . | Clinical Stage . | Sex/Age (yr) . | Diagnosis . | Cytogenetics and Molecular Genetics . | Treatment . |
|---|---|---|---|---|---|
| 213 | Relapse | M/34 | AML M1 | ND | CT |
| 702 | Diagnosis | F/55 | AML M2 | t(8;21); −X | NT |
| 739 | Diagnosis | M/35 | AML M3 | t(15;17) | NT |
| 32 | Diagnosis | F/44 | Pre-B ALL | 46, XX | NT |
| 63 | Relapse | M/38 | Pre-B ALL | 46, XY | CT |
| 74 | Relapse | M/52 | B ALL | ND | CT |
| 509 | Diagnosis | M/21 | T ALL | t (10; 14) (q24; q11); t (3; 9) (p13-14; p22-24) | NT |
| 619 | Diagnosis | F/43 | B ALL | ND | NT |
| VR5 | Diagnosis | M/41 | T ALL | 46, XY; 47, XY, del(4)(p14) | NT |
| 11 | Diagnosis | M/68 | Diffuse large B-cell lymphoma | bcl-2 rearrangement | NT |
| 52 | Relapse | M/45 | Follicle center lymphoma | bcl-2 rearrangement | CT |
| 23 | Diagnosis | F/72 | Follicle center lymphoma | bcl-2 rearrangement | NT |
| 298 | Relapse | F/79 | Mantle cell lymphoma | bcl-2 rearrangement | CT |
| Patient No. . | Clinical Stage . | Sex/Age (yr) . | Diagnosis . | Cytogenetics and Molecular Genetics . | Treatment . |
|---|---|---|---|---|---|
| 213 | Relapse | M/34 | AML M1 | ND | CT |
| 702 | Diagnosis | F/55 | AML M2 | t(8;21); −X | NT |
| 739 | Diagnosis | M/35 | AML M3 | t(15;17) | NT |
| 32 | Diagnosis | F/44 | Pre-B ALL | 46, XX | NT |
| 63 | Relapse | M/38 | Pre-B ALL | 46, XY | CT |
| 74 | Relapse | M/52 | B ALL | ND | CT |
| 509 | Diagnosis | M/21 | T ALL | t (10; 14) (q24; q11); t (3; 9) (p13-14; p22-24) | NT |
| 619 | Diagnosis | F/43 | B ALL | ND | NT |
| VR5 | Diagnosis | M/41 | T ALL | 46, XY; 47, XY, del(4)(p14) | NT |
| 11 | Diagnosis | M/68 | Diffuse large B-cell lymphoma | bcl-2 rearrangement | NT |
| 52 | Relapse | M/45 | Follicle center lymphoma | bcl-2 rearrangement | CT |
| 23 | Diagnosis | F/72 | Follicle center lymphoma | bcl-2 rearrangement | NT |
| 298 | Relapse | F/79 | Mantle cell lymphoma | bcl-2 rearrangement | CT |
Abbreviations: M, male; F, female; AML, acute myeloid leukemia; ALL, acute lymphoblastic leukemia; CT, chemotherapy; NT, no treatment; ND, not determined.
Clinical Characteristics and Genetic Markers of Leukemia Patients Without MSI
| Patient No. . | Clinical Stage . | Sex/Age (yr) . | Diagnosis . | Cytogenetics and Molecular Genetics . | Treatment . |
|---|---|---|---|---|---|
| 13 | Diagnosis | M/68 | AML M5 | ND | NT |
| 34 | Diagnosis | F/46 | AML M2 | ND | NT |
| 168 | Diagnosis | F/22 | AML M4 | t (9; 11) (p21/22; q23) | NT |
| 191 | Diagnosis | F/39 | AML M2 | 45, X, t (10; 11) (p12; q13.1), inv (11) (q13.1) | NT |
| 192 | Diagnosis | M/29 | AML M1 | 46, XY, t (10; 11) (p12; q21) | NT |
| 197 | Diagnosis | M/19 | AML M4 | 46, XY | NT |
| 211 | Relapse | F/34 | AML M1 | 46, XX | CT |
| 217 | Diagnosis | M/59 | AML M4 | t (8; 21) (q22; q22) | NT |
| 240 | Diagnosis | F/57 | AML M4 | inv (16) (p13q22) | NT |
| 335 | Diagnosis | M/65 | AML M2 | ND | NT |
| 396 | Diagnosis | F/70 | AML M5 | ND | NT |
| 423 | Diagnosis | M/41 | AML M0 | 46, XY | NT |
| 432 | Diagnosis | M/25 | AML M3 | 46, XY | NT |
| 441 | Diagnosis | M/55 | AML M3 | 46, XY, 15q+, i(17q−) | NT |
| 448 | Relapse | M/64 | AML M2 | 45, X | CT |
| 453 | Diagnosis | M/54 | AML M3 | del (17) (q21; q23) | NT |
| 454 | Diagnosis | F/39 | AML M4 | 45, X, t (8; 21) (q22; q22) | NT |
| 484 | Relapse | M/32 | AML M5 | 46, XY | CT |
| 601 | Diagnosis | M/40 | AML M3 | t (15; 17) (q22; q21) | NT |
| 637 | Diagnosis | F/33 | AML M4 | 46, XY | NT |
| 643 | Diagnosis | M/55 | AML M3 | t (15; 17) (q22; q21) | NT |
| 722 | Diagnosis | F/49 | AML M3 | t (15;17) (q22; q21) | NT |
| 738 | Relapse | M/34 | AML M5 | Complex aberrations with inv (16) (p13;q22) and 47, XY, +22 | CT |
| 743 | Diagnosis | M/36 | AML M4 | inv (16) (p13q22) | NT |
| 774 | Diagnosis | F/23 | AML M3 | ND | NT |
| 860 | Diagnosis | M/38 | AML M2 | 46, XY, del (7) (q22) | NT |
| 106 | Diagnosis | F/22 | T ALL | 46, XX; t(9; 14)(p22; q11) | NT |
| 154 | Diagnosis | M/30 | Pre-B ALL | t (9; 22) (q34; q11) | NT |
| 176 | Diagnosis | M/20 | Pre-B ALL | t(9; 22)(q32; q11) | NT |
| 528 | Diagnosis | M/39 | Pre-B ALL | t (9; 22) (q34; q11) | NT |
| 698 | Relapse | F/35 | Pre-B ALL | ND | CT |
| 699 | Relapse | M/28 | Pre-B ALL | t (2; 9) (p13; p24) | CT |
| 786 | Diagnosis | M/18 | Pre-B ALL | 46, XY | NT |
| VR1 | Diagnosis | M/17 | T ALL | Hyperdiploid karyotype | NT |
| VR3 | Diagnosis | M/23 | B ALL | t(8/14) | NT |
| VR7 | Diagnosis | F/13 | Pre-B ALL | ND | NT |
| Patient No. . | Clinical Stage . | Sex/Age (yr) . | Diagnosis . | Cytogenetics and Molecular Genetics . | Treatment . |
|---|---|---|---|---|---|
| 13 | Diagnosis | M/68 | AML M5 | ND | NT |
| 34 | Diagnosis | F/46 | AML M2 | ND | NT |
| 168 | Diagnosis | F/22 | AML M4 | t (9; 11) (p21/22; q23) | NT |
| 191 | Diagnosis | F/39 | AML M2 | 45, X, t (10; 11) (p12; q13.1), inv (11) (q13.1) | NT |
| 192 | Diagnosis | M/29 | AML M1 | 46, XY, t (10; 11) (p12; q21) | NT |
| 197 | Diagnosis | M/19 | AML M4 | 46, XY | NT |
| 211 | Relapse | F/34 | AML M1 | 46, XX | CT |
| 217 | Diagnosis | M/59 | AML M4 | t (8; 21) (q22; q22) | NT |
| 240 | Diagnosis | F/57 | AML M4 | inv (16) (p13q22) | NT |
| 335 | Diagnosis | M/65 | AML M2 | ND | NT |
| 396 | Diagnosis | F/70 | AML M5 | ND | NT |
| 423 | Diagnosis | M/41 | AML M0 | 46, XY | NT |
| 432 | Diagnosis | M/25 | AML M3 | 46, XY | NT |
| 441 | Diagnosis | M/55 | AML M3 | 46, XY, 15q+, i(17q−) | NT |
| 448 | Relapse | M/64 | AML M2 | 45, X | CT |
| 453 | Diagnosis | M/54 | AML M3 | del (17) (q21; q23) | NT |
| 454 | Diagnosis | F/39 | AML M4 | 45, X, t (8; 21) (q22; q22) | NT |
| 484 | Relapse | M/32 | AML M5 | 46, XY | CT |
| 601 | Diagnosis | M/40 | AML M3 | t (15; 17) (q22; q21) | NT |
| 637 | Diagnosis | F/33 | AML M4 | 46, XY | NT |
| 643 | Diagnosis | M/55 | AML M3 | t (15; 17) (q22; q21) | NT |
| 722 | Diagnosis | F/49 | AML M3 | t (15;17) (q22; q21) | NT |
| 738 | Relapse | M/34 | AML M5 | Complex aberrations with inv (16) (p13;q22) and 47, XY, +22 | CT |
| 743 | Diagnosis | M/36 | AML M4 | inv (16) (p13q22) | NT |
| 774 | Diagnosis | F/23 | AML M3 | ND | NT |
| 860 | Diagnosis | M/38 | AML M2 | 46, XY, del (7) (q22) | NT |
| 106 | Diagnosis | F/22 | T ALL | 46, XX; t(9; 14)(p22; q11) | NT |
| 154 | Diagnosis | M/30 | Pre-B ALL | t (9; 22) (q34; q11) | NT |
| 176 | Diagnosis | M/20 | Pre-B ALL | t(9; 22)(q32; q11) | NT |
| 528 | Diagnosis | M/39 | Pre-B ALL | t (9; 22) (q34; q11) | NT |
| 698 | Relapse | F/35 | Pre-B ALL | ND | CT |
| 699 | Relapse | M/28 | Pre-B ALL | t (2; 9) (p13; p24) | CT |
| 786 | Diagnosis | M/18 | Pre-B ALL | 46, XY | NT |
| VR1 | Diagnosis | M/17 | T ALL | Hyperdiploid karyotype | NT |
| VR3 | Diagnosis | M/23 | B ALL | t(8/14) | NT |
| VR7 | Diagnosis | F/13 | Pre-B ALL | ND | NT |
Abbreviations: M, male; F, female; AML, acute myeloid leukemia; ALL, acute lymphoblastic leukemia; CT, chemotherapy; NT, no treatment; ND, not determined.
Microsatellite alterations as a disease marker in leukemia.
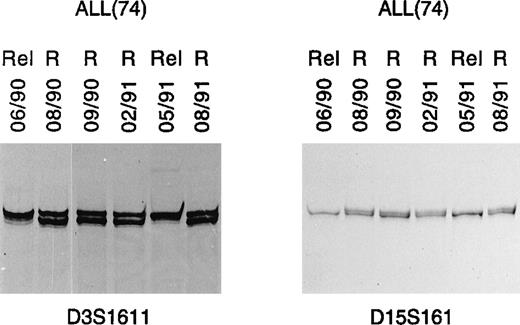
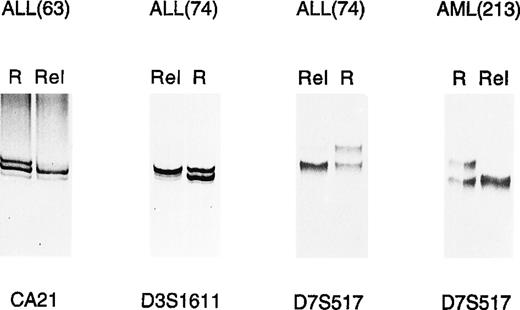
Given the relatively frequent occurrence of microsatellite alterations in ALL, it would be important to establish whether they might be useful as molecular markers of disease. To address this point, serial DNAs collected from a B-ALL patient (patient 74) over a 14-month follow-up were analyzed by PCR. Alterations in marker D15S161, consisting of LOH, were first shown in DNA collected during a relapse that occurred in June 1990 (Fig 2). As expected, LOH disappeared when this patient achieved remission (August 1990), and was no longer detected at different time points when the patient was still in remission (Fig 2). However, this same molecular alteration was again detected during a second relapse, which occurred about 1 year later and then disappeared after effective therapy and achievement of remission (Fig 2). Marker D3S1611, which is closely associated with mismatch repair gene MLH1, showed the same behavior (Fig 2).
LOH as clonal marker in ALL. PCR analysis in 1 B-ALL case (74) detected identical LOH patterns in leukemic cell DNA in 2 consecutive relapses (lanes 06/90 and 05/91, respectively). This was observed with 2 different microsatellite markers, respectively, on chromosome 3 (D3S1611) and 15 (D15S161). R, remission; Rel, relapse.
LOH as clonal marker in ALL. PCR analysis in 1 B-ALL case (74) detected identical LOH patterns in leukemic cell DNA in 2 consecutive relapses (lanes 06/90 and 05/91, respectively). This was observed with 2 different microsatellite markers, respectively, on chromosome 3 (D3S1611) and 15 (D15S161). R, remission; Rel, relapse.
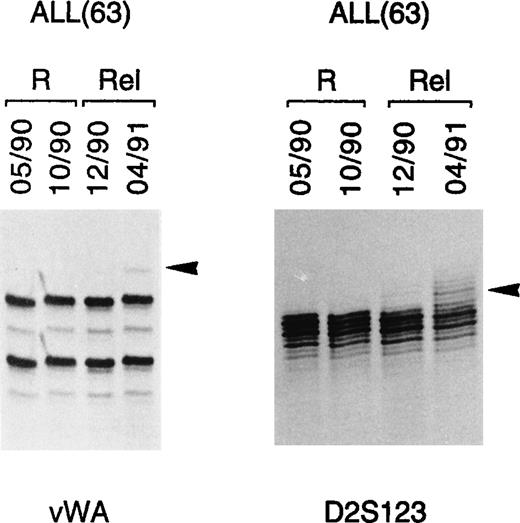
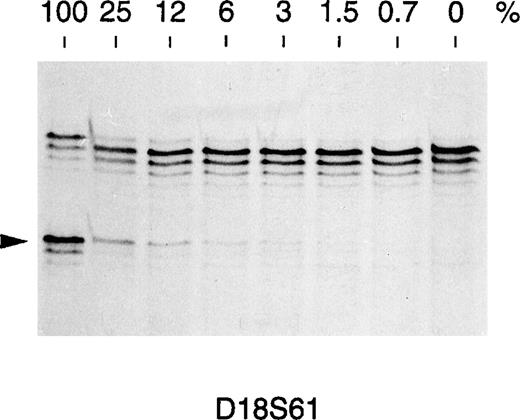
To determine whether microsatellite alterations might be a marker of impending relapse, 4 consecutive DNA samples from another ALL patient (patient 63) were analyzed for MSI involving markers vWA and D2S123 (Fig 3). MSI could be shown in both markers during impending and full-blown relapse and consisted of faint bands of increased size, compared with the alleles detected at remission (Fig3). Conventional bone marrow cytology of this patient showed hypercellularity with 50% blasts at initial relapse and 90% blasts at full relapse. In view of these findings, we attempted to measure the sensitivity of the PCR assay used to detect MSI. To this end, genomic DNAs from 2 healthy individuals showing a different allele for marker D18S61 were mixed in different proportions, amplified with D18S61-specific primers, and analyzed on acrylamide gel. As shown in Fig 4, the B-specific allele of 1 individual could be detected by this method when his DNA represented at least 3% of the total DNA used as template. According to our estimate, therefore, 3% leukemic cells in the bone marrow sample represent our current limit of detection in disease affected tissues. Even though these observations are limited to 2 cases, they suggest that the assay might potentially be applied to monitor minimal residual disease and disease progression in those leukemia patients who present with microsatellite alterations at diagnosis.
MSI as impending relapse marker in ALL. In 1 ALL case (63), MSI was observed by PCR analysis in 2 different loci (vWA, D2s123) at both early (lane 12/90) and full (lane 04/91) relapse. Tumor-specific alterations are indicated by arrows. R, remission; Rel, relapse.
MSI as impending relapse marker in ALL. In 1 ALL case (63), MSI was observed by PCR analysis in 2 different loci (vWA, D2s123) at both early (lane 12/90) and full (lane 04/91) relapse. Tumor-specific alterations are indicated by arrows. R, remission; Rel, relapse.
Sensitivity of the PCR-based assay in detecting microsatellite alterations. Two DNAs from healthy donors (A and B) showing a different allele for the D18S61 marker (arrowed) were mixed at the indicated percent to obtain a constant final amount of 100 ng DNA as a PCR template. A content of 3% of the donor B DNA showing the lower allele is required for a faint band to appear in the autoradiography. This corresponds to a sensitivity of about 1.5% novel allele DNA in a given sample under these PCR conditions.
Sensitivity of the PCR-based assay in detecting microsatellite alterations. Two DNAs from healthy donors (A and B) showing a different allele for the D18S61 marker (arrowed) were mixed at the indicated percent to obtain a constant final amount of 100 ng DNA as a PCR template. A content of 3% of the donor B DNA showing the lower allele is required for a faint band to appear in the autoradiography. This corresponds to a sensitivity of about 1.5% novel allele DNA in a given sample under these PCR conditions.
Association between MSI, bcl-2 rearrangements, and p53 mutations in NHL patients.
In an attempt to correlate MSI with genetic alterations of known oncogenes, the DNA from all 21 NHL patients was analyzed by Southern blotting, as detailed in Materials and Methods, for rearrangements of the bcl-2 gene, whose involvement in lymphomagenesis is well established.25,26,33 Strikingly, bcl-2 rearrangements were detected in 4 of 4 patients with MSI, but only in 3 of 17 patients without MSI (Fig 5 and Table 6). This difference, which is significant (P < .05, Yate’s corrected χ2test) cannot be accounted for by differences in lymphoma histotype between MSI+ and MSI− patients (Table 6) and suggests an association between MSI and bcl-2 rearrangements in NHL. Finally, in view of the possibility that lymphoma cells might carry multiple genetic alterations, we also addressed the presence of mutations involving the p53 gene. As previous reports showed that most p53 mutations in NHL cluster in exons 5 to 8,34 35 these exons were analyzed by reverse transcriptase (RT)-PCR in those NHL patients with MSI; no mutations were found in these patients: genomic DNA from colon cancer patients, with known mutations for the exons considered, was also analyzed and served as a positive control for the SSCP assay. In our patients, therefore, the presence of MSI and bcl-2 rearrangements did not correlate with p53 abnormalities.
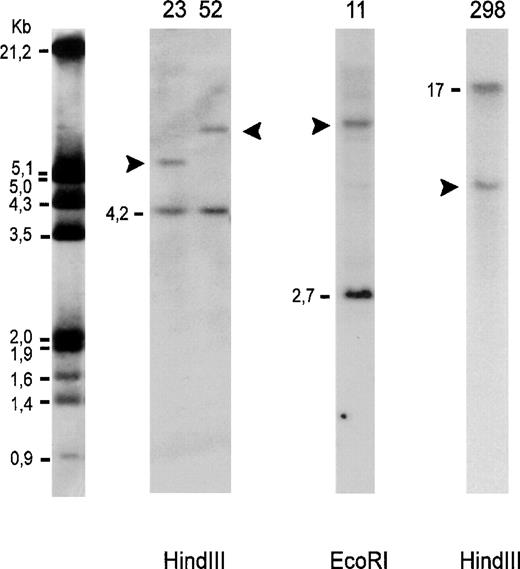
Southern blot analysis of genomic DNA. Autoradiography after hybridization with 32P-labeled DNA probes for detection of bcl-2 gene rearrangements in major (lanes 23, 52, 11) and minor breakpoint region (lane 298). Lanes 23, 52: follicle center lymphomas; lane 11: diffuse large B-cell lymphoma; lane 298: mantle cell lymphoma. Dashes indicate the germline bands and arrowheads the rearranged fragments. The molecular-weight marker is indicated on the left.
Southern blot analysis of genomic DNA. Autoradiography after hybridization with 32P-labeled DNA probes for detection of bcl-2 gene rearrangements in major (lanes 23, 52, 11) and minor breakpoint region (lane 298). Lanes 23, 52: follicle center lymphomas; lane 11: diffuse large B-cell lymphoma; lane 298: mantle cell lymphoma. Dashes indicate the germline bands and arrowheads the rearranged fragments. The molecular-weight marker is indicated on the left.
Histotype, bcl-2 Rearrangements, and MSI in NHL Patients
| Patient No. . | Diagnosis . | bcl-2 Status . | MSI . |
|---|---|---|---|
| 30 VK | B-cell chronic lymphocytic leukemia | N | − |
| 325 NR | N | − | |
| 83 BH | Small lymphocytic lymphoma | N | − |
| 185 WD | N | − | |
| 10 RS | Immunocytoma | N | − |
| 308 SW | Plasmacytoma | N | − |
| 170 HW | Follicle center lymphoma | N | − |
| 201 GE | N | − | |
| 52 LG | R | + | |
| 224 WM | N | − | |
| 237 MD | N | − | |
| 23 SM | R | + | |
| 85 MA | R | − | |
| 7 LI | Mantle cell lymphoma | R | − |
| 298 SH | R | + | |
| 398 AR | N | − | |
| 11 BR | Diffuse large B-cell lymphoma | R | + |
| 64 RA | N | − | |
| 261 StH | R | − | |
| 449 WE | N | − | |
| 155 LL | N | − |
| Patient No. . | Diagnosis . | bcl-2 Status . | MSI . |
|---|---|---|---|
| 30 VK | B-cell chronic lymphocytic leukemia | N | − |
| 325 NR | N | − | |
| 83 BH | Small lymphocytic lymphoma | N | − |
| 185 WD | N | − | |
| 10 RS | Immunocytoma | N | − |
| 308 SW | Plasmacytoma | N | − |
| 170 HW | Follicle center lymphoma | N | − |
| 201 GE | N | − | |
| 52 LG | R | + | |
| 224 WM | N | − | |
| 237 MD | N | − | |
| 23 SM | R | + | |
| 85 MA | R | − | |
| 7 LI | Mantle cell lymphoma | R | − |
| 298 SH | R | + | |
| 398 AR | N | − | |
| 11 BR | Diffuse large B-cell lymphoma | R | + |
| 64 RA | N | − | |
| 261 StH | R | − | |
| 449 WE | N | − | |
| 155 LL | N | − |
Abbreviations: N, normal; R, rearranged.
Detection of LOH involving mismatch repair genes in some leukemia patients showing MSI.
Because MSI in solid tumors is linked to the functional inactivation of mismatch repair genes, including MLH1, MSH2, PMS1, PMS2, and, recently, MSH6,3-6 we investigated whether this might also be true for MSI in leukemias and lymphomas. We looked for mutations affecting the MLH1 and MSH2 genes; given the complexity of these genes, we focused on exons 9 and 16 of MLH1, and exons 5 and 13 of MSH2, all of which have been reported to be mutated in some solid and also lymphoid tumors.18,36 We examined samples from all the patients who were positive for MSI and were unable to demonstrate tumor-specific mutations by SSCP analysis. Genomic DNA from a healthy donor, containing a known mutation in the second domain of the CD4 gene,29 was also simultaneously analyzed by SSCP and served as a control for our SSCP experimental conditions (data not shown).
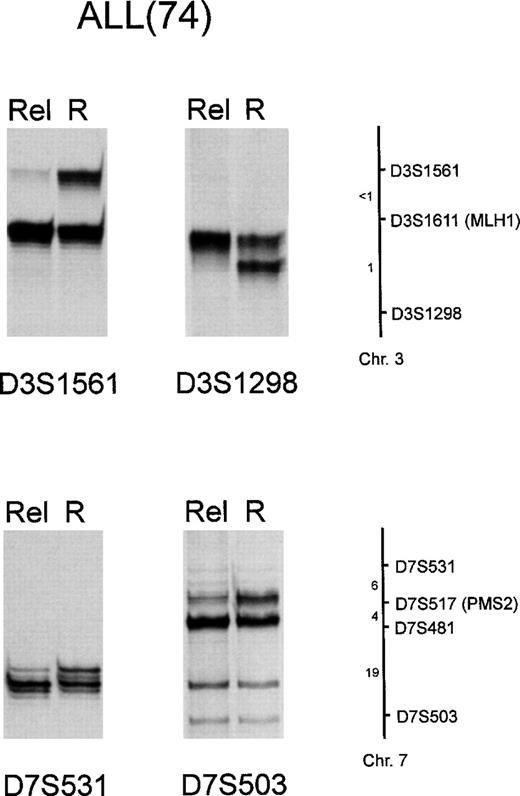
As chromosomal rearrangements in hematologic neoplasms are a major cause of gene disruption,37 we advanced that deletions affecting chromosomes 2, 3, and 7 carrying the MSH2 and PMS1, MLH1, and PMS2 genes, respectively, might occur, and lead to their inactivation in some patients. To test this hypothesis, genomic DNA from patients showing MSI was amplified with primer pairs for D3S1611, CA21, D2S117, and D7S517, which are microsatellite markers closely linked to MLH1, MSH2, PMS1, and PMS2, respectively.27 All mutated NHL and leukemia patients except patients 213 and VR5 were informative for D3S1611 and CA21; all patients, except VR5, were informative for D2S117, and all except patients 619, 52, and 298 for D7S517. LOH of markers closely linked to mismatch repair genes MLH1, MSH2, and PMS2 was demonstrated in 4 of 6 ALLs and 1 of 3 AMLs with microsatellite alterations. In particular, among the ALL patients, patient 32 presented LOH at the D3S1611 site (MLH1), patient 63 at the CA21 site (MSH2), patient 74 at the D3S1611 and D7S517 sites (MLH1 and PMS2), and patient 509 at the D2S117 (PMS1) site; AML patient 213 presented LOH at the D7S517 site (PMS2) (Fig 6). On the other hand, neither 6 RER− ALLs, nor 6 RER− AMLs and 4 RER+ NHL presented LOH at the same chromosomal loci (data not shown). To evaluate whether the observed LOH was confined to a particular marker or was indicative of larger choromosomal deletions, LOH at mismatch repair gene loci was analyzed in further detail in 3 patients (63, 74, and 213). To this end, microsatellite markers mapping close to D3S1611, CA21, and D7S517 were chosen for amplification, as reported in Materials and Methods. In the case of patient 74, this analysis disclosed that LOH extended to markers located both centromeric (D3S1561 and D7S531) and telomeric (D3S1298 and D7S503) to D3S1611 and D7S512 (Fig 7); marker D7S481 was not informative in patient 74. On the other hand, in the case of patients 63 and 213, no additional LOH was demonstrated at these loci. These findings suggest that in some leukemia patients MSI is linked to inactivation of DNA repair genes by chromosomal deletions.
LOH involving mismatch repair genes loci; some representative alterations are shown. Genomic DNA from ALL patients 63, 74, and AML patient 213 was PCR-amplified for microsatellites D3S1611, CA21, and D7S517, 3 genetic markers respectively associated with the MLH1, MSH2, and PMS2 genes, as reported in Materials and Methods. R, remission; Rel, relapse.
LOH involving mismatch repair genes loci; some representative alterations are shown. Genomic DNA from ALL patients 63, 74, and AML patient 213 was PCR-amplified for microsatellites D3S1611, CA21, and D7S517, 3 genetic markers respectively associated with the MLH1, MSH2, and PMS2 genes, as reported in Materials and Methods. R, remission; Rel, relapse.
LOH involving loci flanking D3S1611(MLH1) and D7S517 (PMS2) in ALL patient 74. Genomic DNA was PCR-amplified for microsatellites D3S1561, D3S1298, D7S531, and D7S503, as reported in Materials and Methods. R, remission; Rel, relapse. A schematic chromosome map indicating the location of the markers used and their approximate distances in centimorgans is also shown.28 Marker D7S481 was not informative in patient 74 (data not shown).
LOH involving loci flanking D3S1611(MLH1) and D7S517 (PMS2) in ALL patient 74. Genomic DNA was PCR-amplified for microsatellites D3S1561, D3S1298, D7S531, and D7S503, as reported in Materials and Methods. R, remission; Rel, relapse. A schematic chromosome map indicating the location of the markers used and their approximate distances in centimorgans is also shown.28 Marker D7S481 was not informative in patient 74 (data not shown).
DISCUSSION
The last few years have witnessed major advances in the decoding of the molecular basis of acute leukemias; indeed, in many cases (up to 50%) a specific chromosomal translocation, often involving genes that encode transcription factors, is involved in the pathogenesis of the disease.13,37 In the other cases, however, translocations are random or do not occur at all, thus leaving the origin of the malignancy unexplained. Therefore, other types of genetic damage, for example those affecting tumor suppressor genes and DNA repair genes, might occur and participate in the leukemic transformation. In view of this likelihood, a number of recent studies addressed the occurrence of microsatellite alterations in hematologic malignancies as indirect evidence of disruption of DNA repair genes.7-12,38 The overall conclusion reached by most of these studies was that, unlike findings in some hereditary and sporadic solid tumors, MSI is uncommon in human leukemia and is detected only in a minority of cases, with the possible exception of therapy-related leukemia.39Remarkably, AML was the predominant type of leukemia investigated in these studies; less is known about MSI occurrence in lymphoid tumors.
We compared the prevalence of microsatellite alterations in lymphoid versus myeloid tumors and found a higher prevalence of MSI in ALL than in AML. Although this finding was expected on the basis of previous observations, an explanation for it is presently not forthcoming. Indeed, it is known that knockout mice lacking the murine equivalent of mismatch repair genes MSH2 and MSH6 develop lymphoid malignancies resembling human lymphoblastic lymphoma with high frequency.14-16,18 Furthermore, a screening of human cell lines established from leukemia/lymphoma for MSI disclosed genetic instability exclusively in lymphoid cell lines.36 Finally, it is also known that HNPCC kindreds develop an excess of lymphoid tumors.40 41 Overall, these observations and our findings suggest a connection between inactivation of mismatch repair genes and subsequent MSI on 1 side and lymphoid tumorigenesis on the other.
In contrast to the results of Pabst et al,11 we detected MSI much more frequently than LOH; this discrepancy might probably be explained, at least partially, by differences in the microsatellite markers used, as well as the patient population. Interestingly, 2 of 6 ALLs with MSI belong to the rare immunophenotypic group of mature B-cell ALL, which constitute about 4% of ALLs: this might indicate a nonrandom association between the 2. Clearly, only investigation of MSI prevalence in this rare subset of ALLs and in the closely related B-lymphoblastic lymphoma will definitively resolve this issue. In our patients, MSI seemed to be an adverse prognostic factor. Two of the 6 leukemic patients with MSI detected at diagnosis (patients 702 and 619) relapsed within 15 months, and the relapse was refractory to chemotherapy. In patient 702, the leukemic cells harbored a translocation t(8;21) that normally associates with a favorable outcome.42 Furthermore, the 4 NHL patients with MSI also had an overall poor response to chemotherapy. This contrasts with findings in colorectal cancer, where MSI seems to be a good prognostic indicator2; 1 possible explanation for this discrepancy might be that MSI in leukemia is correlated with resistance to chemotherapy, which usually is the first-line therapy for hematologic neoplasms. Intriguingly, human cell lines with mutator phenotype carrying MLH1 mutations are tolerant to some DNA-alkylating agents, thus suggesting that a functioning mismatch repair system might favor the cytotoxic effects of these drugs.43 A future area of investigation will try to address whether MSI detected at diagnosis might represent a negative prognostic indicator for the outcome of chemotherapy.
Of particular relevance is the finding that in NHL patients MSI is associated with rearrangements of bcl-2, a proto-oncogene whose product is involved in inhibition of apoptosis.44 Chromosomal translocations resulting in high-level bcl-2 expression have been detected in the majority of follicular neoplasms, in about one third of diffuse large B-cell lymphomas and less commonly in other NHL.33 In agreement with this, we did not detect MSI in those lymphoma types, which are not commonly associated with bcl-2 rearrangements (Table 6). In view of the pathogenic relevance of bcl-2, it is tempting to speculate that an increased cell survival conferred by bcl-2 overexpression and genomic instability due to DNA repair gene-defects might cooperate in facilitating the inclusion of new mutations in the genome, possibly increasing the malignancy of the tumor. We also looked for other associated genetic abnormalities and could not detect p53 mutations in the RER+ NHL patients. This finding, however, was not completely unexpected, as a negative association between MSI and p53 gene mutation in colorectal cancer was previously reported.45
Given the relative frequency of MSI in ALL, we believe that microsatellite analysis might be useful in monitoring the disease course in some leukemia patients. Leukemias constitute a unique opportunity to study tumor progression because the neoplastic cells are easily accessible, and the bone marrow is routinely sampled for conventional cytologic evaluation. Although observed in only a few patients, our study shows that the same microsatellite alterations can be consistently detected during follow-up and might be predictive of impending relapse. The advantage of an early detection of leukemia relapse by microsatellite analysis would be to begin chemotherapy at an early stage, when the tumor burden is still low. However, the limited sensitivity of this technology is a critical factor; some investigators suggest that it might be feasible to increase it to detect 1 neoplastic cell among 500 normal cells.46 Compared with some other tumor-specific molecular markers, such as K-ras mutations (detected in 1 of 10,000 cells), this is still low, but might be sufficient in many situations. While specific genetic changes are very sensitive tumor markers, including the BCR-ABL, MLL-AF4, E2A-PBX1, and TEL-AML1 fusion genes in ALL, and AML1-ETO, CBFβ-MYH11, and PML-RARα in AML,13 unfortunately they are not always observed. Microsatellite analysis is easy to perform and also reproducible by nonradioactive methods47; these features should facilitate its application in future studies aimed at assessing the relevance of microsatellite alterations as a tumor marker in lymphoid malignancies.
Although many studies describe MSI in hematologic neoplasms, its relationship to genetic alterations of DNA repair genes has only been marginally explored. To address this issue, we looked for point mutations in a limited number of exons of MLH1 and MSH2, but found none. Admittedly, this was not surprising given the complexity of these genes and the reported absence of clusters of mutations in RER+ tumors.30,31 To fully characterize mutations of DNA repair genes in human hematologic malignancies, it would be necessary to analyze all of the exons of MLH1 and MSH2 and perhaps extend these investigations to the PMS1 and PMS2 genes. On the other hand, our analysis of the chromosomal loci of these genes using closely linked microsatellite markers showed LOH in 4 of 6 RER+ ALLs, and 1 of 3 RER+ AMLs, thus showing that chromosomal deletions might contribute to inactivate mismatch repair genes, at least in some leukemia cases. As defects in mismatch repair genes are apparent only in completely knocked-out cells,27 48 it could be speculated that the remaining allele in leukemias positive for LOH carries disabling mutations. We advance that chromosomal deletions might also contribute to leukemogenesis by targeting and inactivating genes involved in DNA repair.
ACKNOWLEDGMENT
We are grateful to Dr R. Zamarchi for statistical analysis, P. Gallo for artwork, and P. Segato for precious help in the preparation of the manuscript. We also thank Dr Fabrizio Vinante and the Divisione di Ematologia of the University of Verona, Italy, for bone marrow samples from ALL patients.
Supported by grants from Associazione Italiana Ricerca sul Cancro (AIRC) and Fondazione Italiana Ricerca sul Cancro (FIRC).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Stefano Indraccolo, MD, Department of Oncology and Surgical Sciences, via Gattamelata, 64, 35128-Padova, Italy; e-mail: indra@ux1.unipd.it.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal