γ-Glutamylcysteine synthetase catalyzes the first step in glutathione synthesis. The enzyme consists of 2 subunits, heavy and light, with the heavy subunit serving as the catalytic subunit. A patient with hemolytic anemia and low red blood cell glutathione levels was found to have a deficiency of γ-glutamylcysteine synthetase activity. Examination of cDNA from the patient and her mother showed that she was homozygous and that her mother was heterozygous for a A→T transversion at nt1109 producing a deduced amino acid change of His370Leu. The partial genomic structure of the catalytic subunit of γ-glutamylcysteine synthetase (GLCLC) was determined, providing some intron/exon boundaries to make it possible to sequence an affected part of the coding region from genomic DNA. The 1109A→T mutation was not present in the DNA of 38 normal subjects. In the course of these studies we found a diallelic polymorphism in nt +206 of an intron and another polymorphism that consisted of a duplication of a CAGC at cDNA nt1972-1975 in the 3′ untranslated region. The 2 polymorphisms were found to be only in partial linkage disequilibrium.
REDUCED GLUTATHIONE (GSH) is the major sulfhydryl compound of erythrocytes. It is transported out of cells1 and resynthesized, giving a turnover half-time of approximately 3 days.2 Abnormally low levels of this tripeptide are encountered in glucose-6-phosphate dehydrogenase deficiency3 and in deficiencies of the 2 enzymes of GSH synthesis, γ-glutamylcysteine synthetase4 (γ-GCS) and glutathione synthetase.5 Each of these states is associated with hemolytic anemia.
Numerous cases of glutathione synthetase deficiency have been documented, and the molecular lesion in some cases has been identified.6,7 In contrast, γ-GCS deficiency is rare. Only 5 patients from 4 unrelated families have been documented previously,4 8-10 and the mutation has not been reported in any of these cases.
γ-Glutamylcysteine synthetase consists of 2 subunits, heavy and light, with the heavy subunit serving as the catalytic subunit, designated γ-GCSh, and the light subunit, designated γ-GCSl, with regulatory functions. These subunits are encoded by 2 genes, GLCLC and GLCLR. We now report a sixth patient with this enzyme deficiency and show that the patient is homozygous for a mutation at cDNA nt 1109 in GLCLC, in which we identify an A→T transversion that predicts a His→Leu substitution at amino acid 370 of the catalytic subunit. Previously, a polymorphic trinucleotide repeat was identified in the 5′ untranslated region (UTR).11 We now demonstrate 2 additional polymorphisms, a tetranucleotide insertion in the 3′ untranslated region of the gene and a new diallelic intronic polymorphism.
MATERIALS AND METHODS
Enzyme Assays
Assays of red blood cell enzymes and intermediates were performed essentially as described previously,12 13 but spectrophotometric assays of glycolytic enzymes were performed in an automated instrument (Thermomax; Molecular Devices, Sunnyvale, CA) at 30°C. RNA and DNA were prepared from peripheral blood leukocytes by standard methods.
Sequence Analysis
cDNA was prepared from total RNA extracted from a lymphocyte and monocyte fraction obtained after hypaque/ficoll sedimentation. Five micrograms of total RNA was incubated with 300 ng of random primers in a 38 μL system for 10 minutes at 65°C and allowed to cool slowly at room temperature. First-strand cDNA was synthesized in a buffer containing 50 mmol/L Tris-HCl, pH 8.3, 75 mmol/L KCl, 3 mmol/L MgCl2, 1 mmol/L dithiothreitol, 4 mmol/L dNTPs, 0.8 U/μL RNase Block Inhibitor, and 1 U/μL Moloney murine leukemia virus (MMLV)-reverse transcriptase to which the RNA mixture was added and incubated at 37°C for 1 hour in a final volume of 50 μL. The reaction was terminated with a 5 minute incubation at 90°C. Subsequent polymerase chain reaction (PCR) reactions contained 2 μL of the reverse-transcribed cDNA.
The human gene encoding the catalytic subunit of γ-glutamylcysteine synthetase (GLCLC) was amplified by PCR using the primers from the GenBank sequence accession no. M90656. Primers are shown in Table 1. For primers in the 5′ flanking region of the GLCLC gene containing the putative antioxidant responsive element and the CAAT and TATA signals, accession no. L39773 was used. All numbering was based on the adenine in the initiator ATG being designated 1. The cDNA was amplified in 4 fragments, nt −77 to 684, 535 to 1183, 1028 to 1570, and 1527 to 2044. Genomic DNA was used to amplify the 5′ flanking region. Nested primers were used for sequencing. The amplified PCR products were sequenced on an Applied Biosystems Inc (Foster City, CA) automatic sequencer using the dideoxy termination method. Some attempts at amplifying genomic DNA with the cDNA primers yielded products that were also sequenced and yielded some information on the genomic structure of the gene.
Oligonucleotide Primers Used in Sequencing theGLCLC Gene
| . | Oligo Sequence 5′ to 3′ . | Region Amplified by PCR . | Fragment Size (bp) . |
|---|---|---|---|
| Sense | TCATGTACTCACTCATTTAGC | Promoter putative antioxidant responsive element | 243 |
| Antisense | CTAGAGTTGGCCTTATCTCG | ||
| Sense | CGCGTTTGCGTAAAGCGAGG | Promoter TATA and CAAT boxes | 331 |
| Antisense | GGAGCATGCCCAGTCTTTGC | ||
| Sense | GCTGAGTGTCCGTCTCGCG* | cDNA −77 to 684 | 761 |
| Antisense | TGAAGCTTCATCATCCTCAG* | ||
| Sense | GCAATAAACAAGCACCCTCG* | cDNA 535 to 1183 | 649 |
| Antisense | GACTCATTAGCATCATCCAGG* | ||
| Sense | TATAATGACATCGACTTGACG* | cDNA 1028 to 1570 | 543 |
| Antisense | TTCCCATTGATGATGGTGTC* | ||
| Sense | AGGCCCAGAACAGCACGGAG* | cDNA 1527 to 2044 | 518 |
| Antisense | TCAATTTCTGGCTCACTGGC* | ||
| Sense | ACTGACTCATCCAACTAGAC | cDNA 1896 to 2044 for 3′ polymorphic insertion | 149 or 153 |
| Antisense | TCAATTTCTGGCTCACTGGC | ||
| Sense | CCTCCAAACTCAGACATTGG | Genomic fragment containing G/A intron polymorphism | 397 |
| Antisense | CAAACACCACATAGGCAGAG |
| . | Oligo Sequence 5′ to 3′ . | Region Amplified by PCR . | Fragment Size (bp) . |
|---|---|---|---|
| Sense | TCATGTACTCACTCATTTAGC | Promoter putative antioxidant responsive element | 243 |
| Antisense | CTAGAGTTGGCCTTATCTCG | ||
| Sense | CGCGTTTGCGTAAAGCGAGG | Promoter TATA and CAAT boxes | 331 |
| Antisense | GGAGCATGCCCAGTCTTTGC | ||
| Sense | GCTGAGTGTCCGTCTCGCG* | cDNA −77 to 684 | 761 |
| Antisense | TGAAGCTTCATCATCCTCAG* | ||
| Sense | GCAATAAACAAGCACCCTCG* | cDNA 535 to 1183 | 649 |
| Antisense | GACTCATTAGCATCATCCAGG* | ||
| Sense | TATAATGACATCGACTTGACG* | cDNA 1028 to 1570 | 543 |
| Antisense | TTCCCATTGATGATGGTGTC* | ||
| Sense | AGGCCCAGAACAGCACGGAG* | cDNA 1527 to 2044 | 518 |
| Antisense | TCAATTTCTGGCTCACTGGC* | ||
| Sense | ACTGACTCATCCAACTAGAC | cDNA 1896 to 2044 for 3′ polymorphic insertion | 149 or 153 |
| Antisense | TCAATTTCTGGCTCACTGGC | ||
| Sense | CCTCCAAACTCAGACATTGG | Genomic fragment containing G/A intron polymorphism | 397 |
| Antisense | CAAACACCACATAGGCAGAG |
Oligonucleotides used for amplification. Some additional nested oligonucleotides were used for sequencing.
The 1109A→T mutation creates an Alu I restriction site, and this was used to confirm the mutation. A genomic fragment 282 bp in length was amplified using a sense intronic primer and an antisense cDNA primer shown in Table 2. The normal and mutant patterns are also described in Table 2. Thirty-eight control DNA samples from white subjects were examined for the presence of the mutation.
PCR Characterization of the 1109A → T Mutation
| . | PCR Amplification Primers . | Fragment Size (bp) . | Alu I Digest Normal Pattern (bp) . | Alu I Digest Mutant Pattern . |
|---|---|---|---|---|
| Sense Antisense | 5′GACACTCCCGCAGTGTCCGC3′ 5′GACTCATTAGCATCATCCAGG3′ | 282 | 178,62,32,10 | 101,77,62,32,10 |
| . | PCR Amplification Primers . | Fragment Size (bp) . | Alu I Digest Normal Pattern (bp) . | Alu I Digest Mutant Pattern . |
|---|---|---|---|---|
| Sense Antisense | 5′GACACTCCCGCAGTGTCCGC3′ 5′GACTCATTAGCATCATCCAGG3′ | 282 | 178,62,32,10 | 101,77,62,32,10 |
Population Studies
The 3′ tetranucleotide insertion polymorphism was studied in 28 subjects with European ancestry. A PCR fragment from nt 1896 to 2044 was amplified (Table 1) from genomic DNA. Amplified products were either 149 bp without the insertion or 153 bp if the additional 4 bp were inserted, indicating that no intron was present. On electrophoresis, the presence of heteroduplexes could be seen in those samples heterozygous for the repeat. Sequencing of the PCR product confirmed presence or absence of the repeat.
Immunologic Studies
Preparation of recombinant fusion proteins.
Preparation of constructs, purification, and immunization by recombinant fusion proteins were performed as described.14Briefly, a 764-bp DNA fragment (865-1628 bp) of full-length γ-GCS heavy subunit (γ-GCSh) cDNA15 was obtained by digestion with Pst I. Once a 12-bp sequence of BamHI linker had been added to both sides of the 764-bp γ-GCShcDNA, the cDNA was digested with BamHI and the DNA fragment was subcloned into the corresponding sites of the pGEX-2T vector (Amersham Pharmacia, Uppsala, Sweden) for expression as a fusion protein of GST.
A 601-bp DNA fragment (nt 181-781) containing the complete coding sequence of the human γ-GCS light subunit (γ-GCSl) was generated by reverse transcriptase-PCR (RT-PCR) from total RNA of human glioblastoma T98G cells serving as a template for the γ-GCSl cDNA.16 The forward (5′-CTCGGATCCGAGGAGCTTCGAGACTGTATCC-3′) and reverse (5′-GTACCCGGGCCTGGGCTTCTTCAATGTCAGGGAT-3′) primers were designed to facilitate in-frame insertion into the pGEX-2T vector. The human γ-GCSl-coding sequence was amplified and digested with BamHI and Sma I and ligated into theBamHI/Sma I site of pGEX-2T. Recombinant clones were analyzed by DNA sequencing to verify that the coding sequence of γ-GCS was intact and in-frame with GST.17
The plasmid construct containing GST-human γ-GCS-subunits was transfected into Escherichia coli JM109 cells. Production of GST-human γ-GCS-subunit fusion proteins was induced with isopropyl thiogalactoside, and the fusion protein was batch-purified using glutathione-Sepharose 4B beads (Amersham Pharmacia). Polyclonal antibodies against GST-human γ-GCSh and GST-human γ-GCSl were prepared by immunization of these purified GST-fusion proteins in rabbits with complete Freund adjuvant twice monthly. The antibodies were specific to γ-GCSh or γ-GCSl and did not show any cross-reactivity.
Western blots.
Packed cells (0.3 mL) were lysed in a final volume of 1 mL of distilled water. The solution was applied to a diethyl aminoethyl (DEAE)-cellulose membrane and the membrane was washed with ice-cold 3 mmol/L sodium phosphate buffer, pH 7.0, until there was no absorbance at 280 nm. The membrane was then treated with 3 mL of 50 mmol/L Tris-HCl, pH 7.5, containing 0.5 mmol/L β-mercaptoethanol, 0.5 mmol/L MgCl2 (buffer A), and 0.3 mol/L NaCl. The eluant was dialyzed against 1 L of buffer A for 4 hours and then with the same volume of buffer A for 12 hours. The dialyzed sample was centrifuged at 18,500g for 10 minutes at 4°C, and the supernatant was concentrated to be 0.025 mL using a Speed Vac Concentrator (Savant, Hicksville, NY). Seventy-five micrograms of the sample was applied to a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 12.5%), transferred to a nitrocellulose membrane, and immunologically stained using the rabbit polyclonal antibody against GST-human γ-GCSh and GST-human γ-GCSl. The bound antibodies were visualized with an alkaline-phosphatase-coupled second antibody using a ProtoBlot Kit (Promega, Madison, WI). The protein concentration was determined according to Redinbaugh and Turley18 with bovine serum albumin as the standard.
CASE REPORT
N.S. is a 14-year-old white female who had a blood count performed for evaluation of menorrhagia. Her hemoglobin was found to be 12.2 g/dL, with a mean corpuscular volume (MCV) of 105 μm3. Her mother noted that she becomes jaundiced 2 or 3 times per month. She had been told that she has iron deficiency and had been placed on oral iron supplementation. More extensive investigation was performed, showing the hemoglobin to be 11.9 g/dL, the MCV to be 106 μm3, and the reticulocyte count to be 4.9%. Platelets were 263,000/μL and white blood cells were 7,800/μL. Her serum folate level was 10.8 ng/mL, the B12 level was 508 pg/mL, and the thyroid stimulating hormone (TSH) was 2.12 μIU/mL. The direct Coombs test was negative, haptoglobin was less than 8 mg/dL, and total bilirubin was 1.5 mg/dL.
In the newborn period, exchange transfusion was required with a total bilirubin having increased to 23 mg/dL. During childhood, there had been 2 episodes of head trauma, 1 of which had been followed by a seizure. However, results of a neurologic evaluation (including computerized tomography of the head), an electroencephalogram, and a sleep study were normal. The patient was considered to have a learning disability with dyslexia. She has difficulty in concentration, impaired memory, and an auditory processing delay.
The patient’s mother is of Pennsylvania Dutch/German/Swedish descent, while the father is of Pennsylvania Dutch/German/Swedish/Native American descent. The parents are believed to be half-siblings.
On physical examination, including neurologic appraisal, no abnormalities were found.
RESULTS
Erythrocyte Enzymes and Intermediate Values
The levels of erythrocyte enzymes and GSH are summarized in Table 3.
Erythrocyte Enzyme Levels of Patient N.S.
| Enzyme or Intermediate Level . | Normal Range . | Patient . | Mother . | Brother . |
|---|---|---|---|---|
| Hexokinase | 0.67 to 1.79 | 3.26 | ||
| Glucose phosphate isomerase | 25.45 to 51.60 | 40.50 | ||
| Phosphofructokinase (PFK) | 3.02 to 6.38 | 2.18 | ||
| Aldolase | 1.01 to 3.29 | 6.10 | ||
| Triose phosphate isomerase | 805 to 1971 | 1463 | ||
| Glyceraldehyde phosphate dehydrogenase | 119.5 to 258.4 | 190.6 | ||
| Diphosphoglycerate mutase | 1.21 to 5.27 | 5.29 | ||
| Phosphoglycerate kinase | 175.9 to 260.6 | 223.4 | ||
| Monophosphoglycerate mutase | 13.40 to 24.10 | 32.56 | ||
| Enolase | 2.91 to 6.00 | 5.49 | ||
| Pyruvate kinase (PK) | 7.32 to 16.0 | 22.7 | ||
| Lactate dehydrogenase | 60.4 to 214.0 | 170.9 | ||
| Glucose-6-phosphate dehydrogenase | 9.3 to 14.2 | 21.8 | ||
| 6-Phosphogluconate dehydrogenase | 6.75 to 10.70 | 15.85 | ||
| Glutathione reductase | 6.62 to 8.83 | 7.62 | ||
| Glutathione peroxidase | 21.40 to 40.30 | 25.77 | ||
| GOT | 2.19 to 5.20 | 15.71 | ||
| Adenylate kinase | 87.7 to 241.0 | 192.0 | ||
| GST | 1.45 to 4.15 | 0.83 | ||
| Pyrimidine 5′ nucleotidase (mU/g Hb) | 100.5 to 176.1 | 262.7 | ||
| Adenosine deaminase | 0.70 to 1.60 | 0.92 | ||
| GSH | 4.50 to 8.70 | 0.21 | 5.14 | 5.31 |
| γ-Glutamylcysteine synthetase | 0.88 to 1.98 | 0.14 | 0.42 | 0.70 |
| Glutathione synthetase | 0.24 to 0.53 | 0.40 |
| Enzyme or Intermediate Level . | Normal Range . | Patient . | Mother . | Brother . |
|---|---|---|---|---|
| Hexokinase | 0.67 to 1.79 | 3.26 | ||
| Glucose phosphate isomerase | 25.45 to 51.60 | 40.50 | ||
| Phosphofructokinase (PFK) | 3.02 to 6.38 | 2.18 | ||
| Aldolase | 1.01 to 3.29 | 6.10 | ||
| Triose phosphate isomerase | 805 to 1971 | 1463 | ||
| Glyceraldehyde phosphate dehydrogenase | 119.5 to 258.4 | 190.6 | ||
| Diphosphoglycerate mutase | 1.21 to 5.27 | 5.29 | ||
| Phosphoglycerate kinase | 175.9 to 260.6 | 223.4 | ||
| Monophosphoglycerate mutase | 13.40 to 24.10 | 32.56 | ||
| Enolase | 2.91 to 6.00 | 5.49 | ||
| Pyruvate kinase (PK) | 7.32 to 16.0 | 22.7 | ||
| Lactate dehydrogenase | 60.4 to 214.0 | 170.9 | ||
| Glucose-6-phosphate dehydrogenase | 9.3 to 14.2 | 21.8 | ||
| 6-Phosphogluconate dehydrogenase | 6.75 to 10.70 | 15.85 | ||
| Glutathione reductase | 6.62 to 8.83 | 7.62 | ||
| Glutathione peroxidase | 21.40 to 40.30 | 25.77 | ||
| GOT | 2.19 to 5.20 | 15.71 | ||
| Adenylate kinase | 87.7 to 241.0 | 192.0 | ||
| GST | 1.45 to 4.15 | 0.83 | ||
| Pyrimidine 5′ nucleotidase (mU/g Hb) | 100.5 to 176.1 | 262.7 | ||
| Adenosine deaminase | 0.70 to 1.60 | 0.92 | ||
| GSH | 4.50 to 8.70 | 0.21 | 5.14 | 5.31 |
| γ-Glutamylcysteine synthetase | 0.88 to 1.98 | 0.14 | 0.42 | 0.70 |
| Glutathione synthetase | 0.24 to 0.53 | 0.40 |
Activity of enzymes is presented as international units per gram of Hb. GSH levels are millimoles per liter.
cDNA Sequence
cDNA from the patient and her mother were sequenced from −58 to 2044 and only the 1109A→T mutation and the polymorphic insertion were found to differ from the published sequence. The patient was homozygous for 1109A→T and her mother was a heterozygote for this transversion. The deduced amino acid change is His370Leu. Genomic DNA from 76 control alleles was tested by restriction analysis, and none contained this mutation. The patient’s brother was heterozygous for the 1109A→T mutation.
A Polymorphism in the 3′ UTR
A 3′ UTR polymorphism, a duplication of the CAGC at cDNA nt1972-1975, 61 bp 3′ to the TAG stop codon, was found in the patient and her mother. Both were homozygous for this insertion. The DNA of 26 normal white subjects was examined for the presence of the CAGC insertion (Table 4).
Genomic Sequence and Structure
Genomic DNA from the patient and her mother were sequenced in the region of the putative antioxidant responsive element and the CAAT and TATA regions of the GLCLC promoter. No abnormalities were noted.
We have not determined the entire genomic structure of GLCLC, but in the course of defining the mutation in this patient, 3 introns were sequenced in their entirety. These sequences have been deposited in GenBank (accession no. AF118846), and the sequences flanking exons are shown in Fig 1. An intronic polymorphism was discovered at position +206 of the last intron shown in Fig 1. The propositus, her mother, and brother all had the G/G genotype. The DNA of 26 white subjects was examined for this polymorphism as well as for the insertional polymorphisms in the 3′ UTR (see above). The results of this analysis are shown in Table 4. The gene frequency of the G allele at nt +206 was 0.79; that of the A allele was 0.21. The insert in the 3′ UTR had a gene frequency of 0.62; the absence of the insert, then, had a gene frequency of 0.38. The G allele was significantly associated with the presence of the insert in the 3′ UTR (χ2 = 15; DF = 4; P = .004). However, as shown in Table 4, the intronic polymorphism was not in complete linkage disequilibrium with the 3′ UTR polymorphism. Crossovers had clearly occurred.
Partial sequences of three GLCLC introns. The complete 2,076-bp sequence has been deposited in GenBank (Accession no.AF118846). cDNA is indicated by upper case letters and introns are shown as lower case letters.
Partial sequences of three GLCLC introns. The complete 2,076-bp sequence has been deposited in GenBank (Accession no.AF118846). cDNA is indicated by upper case letters and introns are shown as lower case letters.
Comparison With Other Species
The amino acid sequence around the histidine 370 mutated in our patient was compared with the published sequences of mouse (Genbank accession no. P97494) and rat (P19468). Human, mouse, and rat had identical amino acid sequences in the entire region from amino acid 361 through 475. The sequence surrounding amino acid 370 is shown in Fig 2. Using the GAP program (Wisconsin Package Version 9.1, Genetics Computer Group [GCG], Madison, WI) with the default settings, comparisons were also made with all of the more distantly related species for which a sequence of a homolgous enzyme was known, vizSchizosaccharomyces pombe (S59234), Leishmania tarentolae (CAA71144), Trypanosoma brucei (AAC47195), Saccharomyces cerevisiae (S59234), Plasmodium falciparum(CAA07354), Onchocerca volvulus (AAB96970), andCaenorhabditis elegans (CAA90955). Astonishingly, in each case, a histidine was aligned with the histidine in the human sequence.
The human, mouse, and rat amino acid sequence surrounding the amino acid mutated in the patient, histidine 370. The mutated amino acid is shown in bold type. The 3 sequences are identical in this region and extending all the way to amino acid 475.
The human, mouse, and rat amino acid sequence surrounding the amino acid mutated in the patient, histidine 370. The mutated amino acid is shown in bold type. The 3 sequences are identical in this region and extending all the way to amino acid 475.
DISCUSSION
The synthesis of GSH in red blood cells occurs in 2 steps. First, in a reaction catalyzed by γ-glutamylcysteine synthetase, a peptide bond is formed between the γ-carboxyl group of glutamic acid and the amino group of cysteine, forming γ-glutamylcysteine. Next, glutathione synthetase catalyzes the formation of a peptide bond between the carboxyl group of the cysteine of γ-glutamylcysteine and glycine. In each of these reactions, a molecule of ATP is dephosphorylated to ADP and the rate of reaction can conveniently be measured by estimating the release of inorganic phosphate from β-γ-labeled ATP.13Genetically determined deficiencies of both enzymes of glutathione synthesis are known. Glutathione synthetase deficiency results in hemolytic anemia and, in some cases in 5-oxoprolinuria, with associated neurologic deficits.6 γ-Glutamylcysteine synthetase deficiency also is known to cause hemolytic anemia. The first reported patient also had spinocerebellar degeneration,4,19 and for a time it was believed that this was a part of the syndrome. The fact that subsequent cases, including the 1 reported here, have been free of these neurological findings casts doubt on this relationship. However, it must be admitted that, in the case of a number of genetic disorders of the red blood cell, variants with and without neurological stigmata occur, particularly in glutathione synthetase deficiency20and NADH-diaphorase deficiency.21 The patient reported here was somewhat mentally retarded, but the relationship to the enzyme deficiency is not clear. She had neonatal hyperbilirubinemia and repeated head trauma, and was the offspring of a consanguineous marriage, so that other autosomal recessive disorders might be expected to be present.
γ-Glutamylcysteine synthetase consists of 2 subunits, a heavy catalytic subunit and a light, regulatory subunit.11,22,23Its complete sequence has not been determined, and we document here the sequence of several introns, making it possible to amplify the portion of the gene in which this mutation occurred, enabling us to use genomic DNA to screen other samples. Several polymorphisms have been detected in the GLCLC gene. In addition to a previously documented trinucleotide repeat in the 5′ UTR,11 we found a tetranucleotide repeat in the 3′ UTR sequence of the gene and a diallelic substitution in an intron. The latter 2 polymorphisms were in partial linkage disequilibrium. The fact that some crossovers had occurred suggests that these closely spaced polymorphisms are quite ancient or that crossing over in this region occurs with an unusually high frequency.
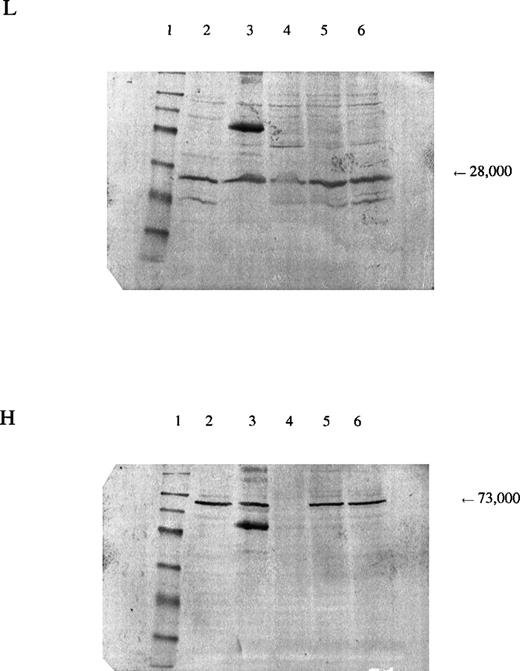
In the patient reported here, we sequenced the entire coding region of the catalytic subunit and found her to be homozygous for a mutation 1109A→T (H370L). The mutation resulted in a marked decrease in the amount of enzyme protein in the erythrocytes, as indicated by Western blots (Fig 3). Because we were able to isolate cDNA from a reticulocyte lysate from the patient and the amount produced was similar to that from normal lysates, the mutant allele is transcribed. Presumably, it is also translated, which suggests that the mutant subunit is unstable. The smaller amount of γ-GCSl subunit found on Western blot analysis suggests that the γ-GCSlmay be stabilized normally by the γ-GCSh subunit and that, in the absence of the latter, be proteolyzed.
Panel L is stained for the regulatory subunit (γ-GCSl), which has a molecular weight of 28,000 (arrow). Panel H is stained for the catalytic subunit (γ-GCSh), which has a molecular weight of 73,000 (arrow). The first lane is a molecular weight marker: 175,000, 83,000, 62,000, 47,500, 32,500, 25,000, 16,500, and 6,500, respectively, from the top. Lanes 2 and 3, hemolysate from a normal subject; lane 4, hemolysate from the patient; lanes 5 and 6, hemolysates from the mother and brother of the patient, respectively. A nonspecific band observed on the third lane of both sheets may represent an aggregation product formed during preparation of the samples.
Panel L is stained for the regulatory subunit (γ-GCSl), which has a molecular weight of 28,000 (arrow). Panel H is stained for the catalytic subunit (γ-GCSh), which has a molecular weight of 73,000 (arrow). The first lane is a molecular weight marker: 175,000, 83,000, 62,000, 47,500, 32,500, 25,000, 16,500, and 6,500, respectively, from the top. Lanes 2 and 3, hemolysate from a normal subject; lane 4, hemolysate from the patient; lanes 5 and 6, hemolysates from the mother and brother of the patient, respectively. A nonspecific band observed on the third lane of both sheets may represent an aggregation product formed during preparation of the samples.
The parents of the propositus were believed to be half-siblings, explaining the otherwise unexpected homozygosity for a rare mutant allele. The sequence abnormalities in the other 4 reported families have not been reported.
ACKNOWLEDGMENT
The authors gratefully acknowledge the assistance of Jennifer Bojanowski, MS, the genetic counselor, who was of great help in obtaining the family history and ensuring understanding of the defect; we acknowledge the cooperation of the family; and we thank Megumi Sakamoto for her excellent technical assistance.
Supported by National Institutes of Health Grants No. HL25552 and RR00833 and the Stein Endowment Fund. This is manuscript no. 12255-MEM.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Ernest Beutler, MD, Department of Molecular and Experimental Medicine, The Scripps Research Institute, 10550 N Torrey Pines Rd, La Jolla, CA 92037.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal