Abstract
To elucidate the molecular background for the heterogeneity in protein S plasma concentrations observed in protein S deficient individuals, the in vitro synthesis of recombinant protein S missense mutants was investigated. Six different naturally occurring mutations identified in the protein S gene (PROS1) of thrombosis patients were reproduced in protein S cDNA by site directed mutagenesis. Two mutants, G441C and Y444C (group A), were associated with low total plasma concentration of protein S. Modestly low protein S was found in families with R520G and P626L (group B) mutants. T57S and I518M (group C), which was associated with marginally low protein S, did not segregate with protein S deficiency in the respective families, raising doubts as to whether they were causative mutations or rare neutral variants. The 6 protein S mutants were transiently expressed in COS 1 cells. The Y444C mutant showed the lowest level of secretion (2.5%) followed by the G441C mutant (40%). Group B demonstrated around 50% reduction in secretion, whereas group C mutants showed normal secretion. Pulse-chase experiments demonstrated impaired protein S processing with intracellular degradation and decreased secretion into the culture media of group A and B mutants. Interestingly, there was a good correlation between in vitro secretion and the concentration of free protein S in the plasma of heterozygous carriers. These results demonstrate impaired protein S secretion to be an important mechanism underlying hereditary protein S deficiency and that variations in protein secretion is a major determinant of the phenotypic heterogeneity observed in protein S deficiency. (Blood. 2000;95:173-179)
Hereditary thrombophilia is a complex disorder in which different genetic factors predispose to the development of thromboembolic events.1-3 Among known genetic risk factors of thrombosis, many mutations in the genes of antithrombin, protein C and protein S have been described.4,5 In some cases, specific mutations or sets of mutations in a certain gene confer different risks of thrombosis. For instance, in antithrombin deficiency, mutations affecting the heparin-binding site associate with a lower prevalence of thrombosis than other types.6 In general, it is logical to assume that mutations leading to a complete null allele will be more deletereous than mutations leading to reduced expression of a functional protein.
Protein S is a plasma glycoprotein with anticoagulant properties, acting as cofactor to activated protein C (APC) in the degradation of factor Va and factor VIIIa.7 Protein S has also prothrombinase inhibitory properties because of its high affinity for negatively charged phospholipid membranes.8 In human plasma, protein S forms an equimolar complex with the complement regulatory protein C4b-binding protein (C4BP).9 The formation of this complex affects protein S function, as only the free protein S is active as APC cofactor.10 The protein S-C4BP interaction is of very high affinity, especially at physiological calcium concentration.11 In plasma, free protein S represents the molar excess of protein S concentration to C4BP binding sites,12 which are found in the beta chain of C4BP.13,14 Beta chain containing C4BP and protein S concentrations seem to be regulated coordinately to maintain a fairly constant concentration of free protein S in plasma.15,16Protein S is a multimodular protein composed of a γ-carboxyglutamic acid–containing module (Gla module), a thumb loop sensitive to thrombin (thrombin-sensitive region, TSR), 4 epidermal growth factor (EGF)-like modules, and a C-terminal region similar to the sex hormone binding globulin (SHBG-like region). The SHBG-like region consists of a spacer region followed by 2 G modules, a structural motive present in laminin A and other extracellular matrix proteins.17Structure-function studies have demonstrated that crucial APC interaction sites are located in the Gla, TSR, and first EGF-like modules of protein S.17-20 The binding site for C4BP is located in the SHBG-like region, which is also important for full anticoagulant activity.18 21
Congenital protein S deficiency is an autosomal dominant disease present in 2% to 6% of patients with thrombosis. The important anticoagulant role of protein S is dramatically illustrated by the severity of homozygous or compound heterozygous cases reported.22-24 Heterozygous carriers have an increased frequency of thrombosis close to 10-fold of that of their healthy relatives.25,26 In contrast, in population-based studies, the relative risk of thrombosis associated with low plasma protein S values is only 2-fold or less.27,28 Protein S deficiency is diagnosed using laboratory tests for both antigen and activity, and specific tests for the free-form of protein S have been developed.29 On the basis of these measurements, protein S deficiency is classified into 3 classes. Type I deficiency is characterized by a decrease in the total protein S antigen and, concomitantly, of free protein S. Type II or qualitative deficiency, is characterized by normal antigen levels and reduced protein S activity due to a dysfunctional protein S in plasma. Although a few type II deficiencies have been reported,30-33 they seem to be rare.34 Most of the cases initially reported were found to be caused by resistance to APC.35-37 Type III deficiency, in turn, is characterized by low free protein S levels, whereas the total plasma concentration of protein S is normal. The distinction between type I and type III could be of clinical importance to assess the risk of thrombophilia in a given individual, but its biologic basis and significance has been controversial.4,25,38-42 In fact, it has been proposed that the 2 types of deficiencies may be phenotypic variants of the same genetic disease.16 However, a causative mutation in the protein S gene (PROS1) is more frequently found in type I PS deficient patients/families than in type III.40 43
To clarify the causes underlying protein S deficiency and the heterogeneity observed in its manifestation, it is necessary to know more on the particular effects of naturally occurring PROS1mutations. To date, only 1 study has characterized a missense protein S mutation causative of type I deficiency, protein S Nagoya, where Arg 474 is changed to Cys.44 In the current study, to further analyze the molecular basis of protein S deficiency, we tested the particular effects of other naturally occurring missense mutations identified in thrombophilic protein S–deficient families. We have produced 6 naturally occurring protein S mutants and studied their expression in cultured mammalian cells. The results shed light on the mechanisms involved in the observed phenotypic heterogeneity of protein S deficiency.
Materials and methods
Mutations analyzed
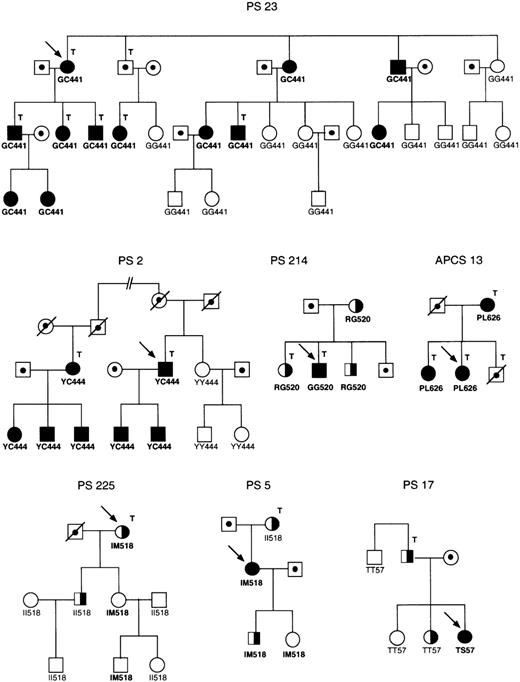
Six different naturally occurring PROS1 missense mutations were analyzed (Table 1). The mutations were previously identified in Spanish families referred for genetic analysis after diagnosis of different types of protein S deficiency associated with venous thrombotic disease (Figure1). Protein S determinations and other laboratory analyses in these individuals as well as PROS1sequencing analysis were performed as previously described.40,43 As shown in Table 1, the mutations were classified into 3 groups according to plasma concentrations of protein S, as well as cosegregation analysis in the family. Group A included 2 mutations (G441C and Y444C) associated with low levels of total protein S in heterozygotes, in most cases lower than 50% of normal reference values (only 2 of 15 had 52% and 61%, respectively). All carriers of these mutations were diagnosed as suffering from type I protein S deficiency.43 Group B included 2 mutations (R520G and P626L) identified in families in which heterozygotes present with moderately low levels of total protein S antigen, with values higher than 59% of normal reference plasma. Carriers of these mutations were diagnosed as having either type I or type III protein S deficiency. Interestingly, in kindred PS 214, the type I protein S deficient propositus was found to be homozygous for the R520G mutation, wheras their heterozygote relatives presented with type III protein S deficiency.43 Group C included 2 mutations (I518 mol/L and T57S) that did not cosegregate with protein S deficiency, raising the question as to whether they really were causative of the protein S deficient phenotype. The I518 mol/L mutation was identified in 2 families, a type III protein S deficient pedigree43 and a pedigree with some members presenting protein S values compatible with type I deficiency, whereas others had type III deficiency (type I-III). Segregation analysis in the pedigrees showed absence of cosegregation of this mutation with the type I or type III protein S deficient phenotype. The T57S mutation was identified in the only type I patient of a type I-III protein S deficient pedigree in which the type III protein S deficient relative is not carrier of the mutation. Apart from these amino acid substitutions, 8 other mutations were analyzed to study the effect of more conservative changes at the positions where the natural mutations studied occurred: G441A, Y444F, R520A, R520H, R520K, R520E, P626G, and P626X (amino acids are named by their 1 letter code, X = stop codon).
Classification and characteristics of the PROS1 missense mutations analysed
| . | Mutation . | Amino acid change . | Location . | Cosegregation* . | Deficiency type . | Total protein S† mean (range) . | Free protein S† mean (range) . |
|---|---|---|---|---|---|---|---|
| Group A | |||||||
| PS 23 | 1590G → T | G441C | Exon 12 | 12/24 (+) | I | 47 (35 − 61, n = 9) | 26 (5 − 38, n = 9) |
| PS 2 | 1600A → G | Y444C | Exon 12 | 7/10 (+) | I | 41 (21 − 52, n = 6) | 13 (9 − 26, n = 6) |
| Group B | |||||||
| PS 214‡ | 1827C → G | R520G | Exon 14 | 4/4 (+) | I-III | 82 (71 − 99, n = 3) | 26 (20 − 35, n = 3) |
| APCS 13 | 2147C → T | P626L | Exon 15 | 3/3 (+) | I | 62 (59 − 65, n = 3) | 38 (30 − 49, n = 3) |
| Group C | |||||||
| Ps 17 | 439C → G | T57S | Exon 4 | 1/4 (−) | I-III | 44 (n = 1) | 41 (n = 1) |
| PS 5 | 1823A → G | I518M | Exon 14 | 3/4 (−) | I-III | 65 (49 − 79, n = 3) | 45 (26 − 63, n = 3) |
| PS 225 | 1823A → G | I518M | Exon 14 | 3/8 (−) | III | 97 (89 − 112, n = 3) | 75 (53 − 104, n = 3) |
| . | Mutation . | Amino acid change . | Location . | Cosegregation* . | Deficiency type . | Total protein S† mean (range) . | Free protein S† mean (range) . |
|---|---|---|---|---|---|---|---|
| Group A | |||||||
| PS 23 | 1590G → T | G441C | Exon 12 | 12/24 (+) | I | 47 (35 − 61, n = 9) | 26 (5 − 38, n = 9) |
| PS 2 | 1600A → G | Y444C | Exon 12 | 7/10 (+) | I | 41 (21 − 52, n = 6) | 13 (9 − 26, n = 6) |
| Group B | |||||||
| PS 214‡ | 1827C → G | R520G | Exon 14 | 4/4 (+) | I-III | 82 (71 − 99, n = 3) | 26 (20 − 35, n = 3) |
| APCS 13 | 2147C → T | P626L | Exon 15 | 3/3 (+) | I | 62 (59 − 65, n = 3) | 38 (30 − 49, n = 3) |
| Group C | |||||||
| Ps 17 | 439C → G | T57S | Exon 4 | 1/4 (−) | I-III | 44 (n = 1) | 41 (n = 1) |
| PS 5 | 1823A → G | I518M | Exon 14 | 3/4 (−) | I-III | 65 (49 − 79, n = 3) | 45 (26 − 63, n = 3) |
| PS 225 | 1823A → G | I518M | Exon 14 | 3/8 (−) | III | 97 (89 − 112, n = 3) | 75 (53 − 104, n = 3) |
Cosegregation indicates the number of individuals carrying the identified mutation over the total genotyped; presence (+) or absence (−) of cosegregation between protein S deficiency and the presence of the mutation is indicated.
Total and free protein S is given as percentage of a standard plasma pool; the PS values shown correspond to the mean and range for the heterozygous carriers of the mutations.
Protein S mean levels of total and free protein S for the R520G homozygote are not included in the table. The homozygote is the only individual diagnosed as type I in this family.
Segregation of the mutations analyzed in families PS 23, PS 2, PS 214, APCS 13, PS 225, PS 5, and PS 17.
Filled and half filled symbols, type I and type III protein S deficiency, respectively; dotted symbols, individuals not tested; arrow, propositus; T, thrombotic disease. The genotypes for the G441C, Y444C, R520G, P626L, I518 mol/L, and T57S mutations are indicated below each symbol. The pedigrees of families PS 214 and PS 225 are taken from Figures 1 and 3 of reference 43.
Segregation of the mutations analyzed in families PS 23, PS 2, PS 214, APCS 13, PS 225, PS 5, and PS 17.
Filled and half filled symbols, type I and type III protein S deficiency, respectively; dotted symbols, individuals not tested; arrow, propositus; T, thrombotic disease. The genotypes for the G441C, Y444C, R520G, P626L, I518 mol/L, and T57S mutations are indicated below each symbol. The pedigrees of families PS 214 and PS 225 are taken from Figures 1 and 3 of reference 43.
Site-directed mutagenesis and construction of expression vectors
A full-length human protein S cDNA was subcloned into theBamHI site of the expression vector pcDNA3 (Invitrogen, San Diego, CA). Mutants were generated by PCR using a set of synthetic oligonucleotides (available from the authors on request) and the Quickchange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The presence of the expected mutations was confirmed by direct DNA sequencing using the ABIprism Taq polymerase–based sequencing kit with fluorescent dye terminators and an Applied Biosystems 373A DNA sequencer (Perkin-Elmer Applied Biosystems, Foster City, CA).
Transient expression of recombinant protein S
Monkey kidney COS 1 cells were cultured in Dulbecco's modified Eagle's medium (GIBCO-BRL, Gaithersburg, MD) supplemented with 10% fetal calf serum, 3.5 mmol/L glutamin, 44 IU penicillin, 44 μM streptomycin, and 10 mg/L vitamin K1. Six-centimeter dishes containing nearly confluent COS 1 cells were transiently transfected with 4 μg of pcDNA3 containing wild type or mutant protein S cDNA, using the DEAE-dextran-chloroquine method previously described.45 A mock transfection was carried out in parallel by transfecting the plasmid pcDNA3 without any insert. Cell media were changed after 40 hours, to serum-free medium (Optimem Glutamax, GIBCO-BRL) supplemented with 44 IU penicillin, 44 μM streptomycin, and 10 mg/L vitamin K1. After 24 hours, protein S containing media were harvested for further analysis.
Electrophoretic and blotting techniques
Media of transiently transfected cells were concentrated 10-fold by filtration with CENTRICON-50 (Amicon, Beverly, MA). The proteins were separated on 7.5% SDS-PAGE gels and transferred to nylon membranes. Protein S was detected by immunoblotting with a rabbit polyclonal antibody and detected colorimetrically with a goat antirabbit IgG antibody conjugated with alkaline phosphatase (DAKO, Glostrup, Denmark).
Quantification of protein S secretion
Protein S antigen concentration in the conditioned media of transfected cells was measured by an enzyme-linked immunosorbent assay (ELISA), essentially following a described method.19Culture medium diluted in 50 mmol/L Tris-HCl, 150 mmol/L NaCl, and 2 mmol/L CaCl2; pH 7.5 with 0.1% (w/v) BSA was added to microtiter wells containing immobilized antihuman protein S rabbit polyclonal antibodies. After 2 hours incubation at room temperature (RT), the plates were washed and bound protein S was detected using the HPS67 antihuman protein S monoclonal antibody,19 followed by horseradish peroxidase-coupled antimouse IgG antibodies (DAKO). The plates were developed with 0.65 mg/ mL 1,2-phenylenediamine (DAKO) and 0.012% H2O2 (v/v) diluted in 0.1 mol/L citrate buffer, pH 5.0. After 5 minutes, the reaction was stopped by the addition of 1 mol/L H2SO4 and the absorbance measured at 490 nm. A calibration curve was constructed using purified recombinant wild type human protein S of known concentration diluted in expression media.
Pulse-chase experiments
Pulse-chase experiments of recombinant protein S by radioactive labeling, immunoprecipitation, and electrophoresis were performed essentially as previously described.46 Eight micrograms of the expression vectors were transiently transfected into COS 1 cells by a Lipofectin (GIBCO-BRL)-mediated method on 10-cm dishes, then divided into several 6-cm dishes on the next day. On the third day after transfection, the cells were incubated with 1 mL of Met/Cys free DMEM, supplemented with glutamine and vitamin K, for 30 minutes to deplete intracellular Met and Cys, then radiolabeled with 800 μL of 100 μCi/mL [35S] Met and [35S] Cys for 30 minutes disolved in the previous medium. After washing twice with 2 mL of phosphate-buffered saline (PBS), the cells were chased for the indicated times with 1 mL of Optimem medium supplemented with 2 mmol/L cold Met and 2 mmol/L cold Cys, and vitamin K. Culture media were harvested and centrifuged at 10 000 rpm for 3 minutes at room temperature to remove cell debris. Labeled cells were washed once with 2 mL of PBS, then lysed on ice with 1 mL of 10 mmol/L Tris pH 7.5, 150 mmol/L NaCl, 2 mmol/L EDTA, 1% NP-40 (v/v), 0.1% SDS (w,v), and 1 mmol/L PMSF for 15 minutes. The supernatants of the cell lysates were collected by centrifugation at 14 000 rpm for 5 minutes at RT. Culture media and supernatants of the cell lysates were precleared with 50 μL of 10% suspension of Pansorbin (Calbiochem, La Jolla, CA) for 2 hours with rocking at RT. After centrifugation at 14 000 rpm for 1 minute at RT, the supernatants were incubated with 3 μg of antiprotein S polyclonal antibody (DAKO) for 16 hours at 4°C. The immune complexes were precipitated by incubation with 50 μL of a 10% suspension of Pansorbin for 1 hour with shaking at RT, then centrifuged at 14 000 rpm for 1 minute. Precipitates were washed 5 times with 1 mL of 10 mmol/L Tris pH 7.5, 150 mmol/L NaCl, 2 mmol/L EDTA, 0.5% NP-40 (v/v), and 0.1% SDS (w/v) to remove nonspecifically bound proteins. Bound proteins were eluted by boiling for 5 minutes in 25 μL of 2 × SDS sample buffer containing 2% mercaptoethanol, then centrifuged at 14 000 rpm for 1 minute. Supernatants were separated by 7.5% SDS-PAGE. Gels were treated with 10% acetic acid and 30% methanol for 15 minutes and dried for 1 hour at 80°C, and analyzed with a Phosphor Imager (Molecular Dynamics, Sunnyvalle, CA) to quantify the radioactivity of the bands on the gels. The total radioactivity immunoprecipitated at time zero was not significantly different among the different mutants, with the exception of Y444C that demonstrated a level of approximately 50% of that observed for wild type protein S.
Results
Transient expression of protein S mutants in COS 1 cells
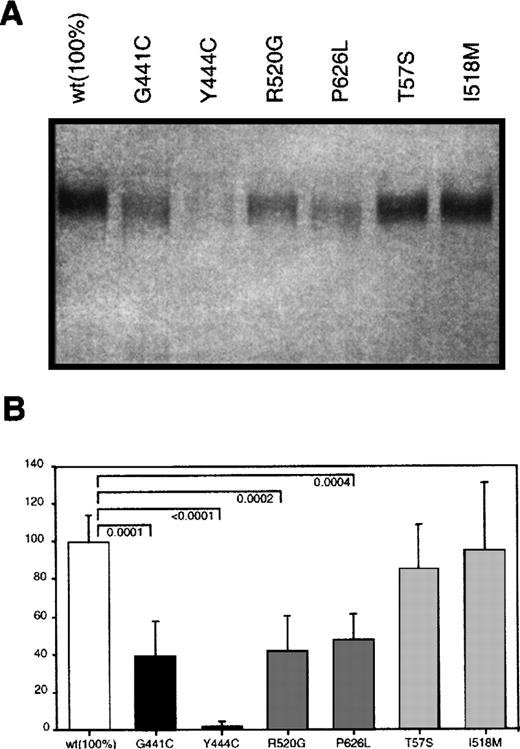
Eukaryotic expression plasmids carrying either wild type protein S cDNA or the protein S variants found in the different kindreds described in Table 1 were constructed as indicated in the methods section. COS 1 cells were transiently transfected with the different plasmids and their conditioned media analyzed for protein S content. In Western blot analysis under nonreducing conditions, protein S was detected in all media as a single band with the expected molecular weight (Figure 2). However, the amount of protein detected was clearly different between the various conditioned media. To accurately quantify the level of protein S expression of the different mutants, we standarized an ELISA-based protocol for determining the concentration of protein S in media from 8 different transfections. Analyses were carried out in parallel using 2 cell plates per transfected protein S variant. Wild type protein S was expressed at a concentration around 100 ng/mL, as determined by using purified protein S as standard. The concentration of protein S in the media (Figure 2) was determined by using sequential dilutions of conditioned media containing known protein S concentration. Group A mutants showed the lowest concentrations, although the level of secretion observed was very different between both mutants. Although the concentration of G441C mutant in conditioned media was 40% of the wild type, the effect of the Y444C mutation was much more severe, giving the lowest levels of expression detected, 2.5%. In group B mutants, the levels of expression were approximately half of the wild type, being 42% in the case of R520G and 48% in the case of P626L. All these values were significantly different from the concentration of wild type protein S. In contrast, group C mutants did not produce a significant reduction on the protein expressed, T57S and I518 mol/L had a concentration in the media that was 97% and 96% of the concentration of wild type protein S, respectively.
Transient expression of wild type and mutant recombinant protein S in COS 1 cells.
(A) Western blot analysis of 10 μL of conditioned media was performed according to Materials and Methods section. (B) ELISA determination of the concentrations of the different mutants. Bars represent the mean ± SD of 16 different protein S determinations corresponding to duplicates from 8 transfection experiments per mutant. An exception is P626L, for which 4 transfections were performed. A value of 100% was arbitrarily given to the mean value of the wild type expression. Comparison between the mutant and the wild type expression levels was performed using an unpaired t test. OnlyP-values considered significant (≤ .05) are shown.
Transient expression of wild type and mutant recombinant protein S in COS 1 cells.
(A) Western blot analysis of 10 μL of conditioned media was performed according to Materials and Methods section. (B) ELISA determination of the concentrations of the different mutants. Bars represent the mean ± SD of 16 different protein S determinations corresponding to duplicates from 8 transfection experiments per mutant. An exception is P626L, for which 4 transfections were performed. A value of 100% was arbitrarily given to the mean value of the wild type expression. Comparison between the mutant and the wild type expression levels was performed using an unpaired t test. OnlyP-values considered significant (≤ .05) are shown.
Pulse-chase experiments
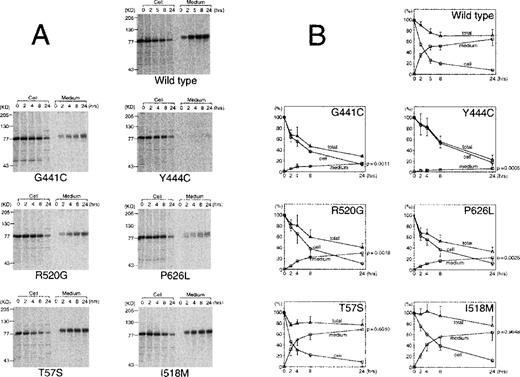
To analyze the mechanism of the observed reduction in the concentration of protein S in the media, transiently transfected cells were pulse-labeled and the incorporation of radioactive label into protein S was followed for up to 24 hours both in the media and inside the cells (Figure 3). The same effect in the reduction of protein S secreted in the media observed in the previous experiments was confirmed in the pulse-chase experiment. Group A mutants had the lowest secretion levels and group B mutants had an intermediate effect. These 4 mutants were clearly degraded during the experiment, as their total radioactivity at 24 hours was between 23% (Y444C) and 41% (R520G) of the initial radioactivity. In comparison, wild type protein S (72%) and group C mutants (78% for T57S and 77% for I518 mol/L) all had higher levels. The secretion efficiency, measured as the level of protein S in the media at 24 hours and expressed as a percentage of the initial value, was higher in wild type (65%) and group C mutants (69% for T57S and 64% for I518 mol/L), whereas it was decreased to < 30% in the other mutants. Among group A mutations, the difference observed between G441C and Y444C in the transient expression was confirmed. The G441C secretion efficiency was 2.5-fold higher than that of Y444C (15± 3 vs 6 ± 3,P = .14). Y444C had also the longest intracellular half life of all proteins tested, around 8 hours. In the pulse-chase experiment, the results are not affected by the transfection, transcription, and translation efficiency, as they refer at each time point to the protein S detected at the time of the pulse. Thus, the pulse-chase results confirm that the differences shown in Figure 2 are due to the effect of each mutation in the folding and/or secretion of the protein.
Quantitative analysis of pulse-chase experiments using transient expression in COS 1 cells.
Radiolabeled media and cell lysates were immunoprecipitated and electrophoresed on SDS-PAGE. (A) A representative experiment of wild type protein S and each mutant pulse-chase. (B) The radioactivity of protein S bands on the dried gels was measured using an image analyzer. The amount of radioactive protein S in cell lysates at the beginning of the experiment was assigned a value of 100%. The graphs represent the radioactivity recovered from cell lysates (circles), medium (squares), or total (triangles) at each time point. Total radioactivity was calculated as the sum of radioactivity recovered from media and lysate. The values represent the mean ± SD of 3 or 4 independent experiments. The P-value of an unpaired t test comparing the radioactivity in the medium at 24 hours for each mutant compared with wild type protein S is given in the right axis.
Quantitative analysis of pulse-chase experiments using transient expression in COS 1 cells.
Radiolabeled media and cell lysates were immunoprecipitated and electrophoresed on SDS-PAGE. (A) A representative experiment of wild type protein S and each mutant pulse-chase. (B) The radioactivity of protein S bands on the dried gels was measured using an image analyzer. The amount of radioactive protein S in cell lysates at the beginning of the experiment was assigned a value of 100%. The graphs represent the radioactivity recovered from cell lysates (circles), medium (squares), or total (triangles) at each time point. Total radioactivity was calculated as the sum of radioactivity recovered from media and lysate. The values represent the mean ± SD of 3 or 4 independent experiments. The P-value of an unpaired t test comparing the radioactivity in the medium at 24 hours for each mutant compared with wild type protein S is given in the right axis.
Effect of alternative amino acid substitutions in selected mutants
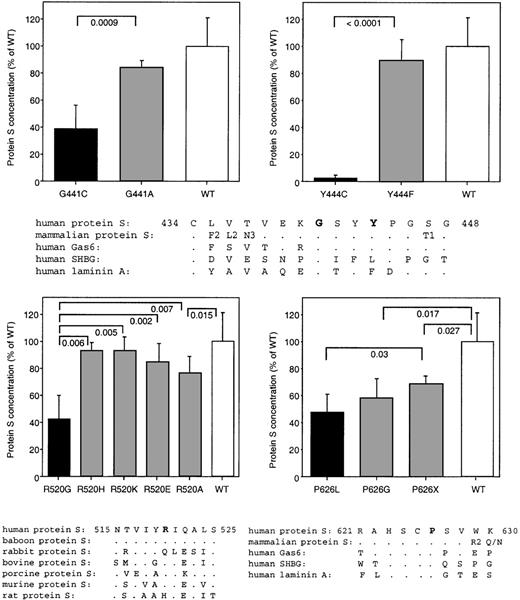
Next, we studied alternative amino acid changes at the mutated positions to test whether more conservative substitutions could recover the effect observed in the mutants in groups A and B. For these experiments, we compared the sequences of proteins with structure similar to protein S. All mutations in groups A and B are in residues belonging to the G domains, which form the C-terminal SHBG-like region of protein S. Therefore, in our sequence comparison, we used the 7 mammalian protein S sequences reported, as well as the human sequences of the structurally related proteins Gas6, SHBG, and laminin A.47 First, we changed the residues affected in group A mutations to amino acids with characteristics more similar to the amino acids found in the wild type sequence: G441A and Y444F. In both cases, the concentration of protein S in the media increased to almost normal levels, 84% and 89% respectively (Figure4). The remaining difference to wild type protein S observed in the case of G441A could be explained by the fact that Gly seems to be a structural feature of the G module at this position, as it is present in G modules distantly related to protein S, such as those in laminin A (Figure 4). At the position of Y444, mouse Gas6 and human laminin have a Phe, whereas human SHBG has a Leu, indicating that several side chains are compatible with the structure at this residue. Group B mutations were also changed to alternative amino acids. When R520 was changed to other charged amino acids, either positive or negative, the concentration in the media increased (R520H, 93%; R520K, 93%; and R520E, 84%) in comparison to that of the R520G mutant. An intermediate effect was produced by changing the residue to Ala (77%), which is more similar to Gly. Finally, at position 626, we located a Gly residue but the levels were only slightly increased compared with the original mutation (58% vs 48%). Interestingly, changing Pro for a stop codon seemed to be comparatively better for the secretion of the mutant protein (68%).
Transient expression of wild type and alternative recombinant mutant protein S in COS 1 cells.
The original natural occurring mutants are also shown. Bars represent the mean ± SD of 8 different protein S determinations corresponding to duplicates from 4 different transfections per mutant. A value of 100% was arbitrarily given to the mean value of the wild type expression. Comparison of protein S secretion in the alternative mutants and that in the wild type, as well as that in the original mutants was performed using an unpaired t test. OnlyP-values considered significant (≤ .05) are shown. A comparison of the sequences containing the mutated residues in human and other mammalian protein S and related proteins (human Gas6, human SHBG, and the G4 and G5 repeats in human laminin A) is also shown. Given the high conservation of the protein S sequence around G441 and Y444 in mammals, a consensus sequence from baboon, rabbit, bovine, porcine, murine, and rat PS is shown as mammalian protein S. When there is a difference between human protein S and other mammalian species, this is indicated in the consensus sequence by the different residue followed by the number of species, when more than 1, where this difference is observed. Dots indicate conserved amino acids. For a more complete sequence comparison, see reference 47. The mutated residues are in bold.
Transient expression of wild type and alternative recombinant mutant protein S in COS 1 cells.
The original natural occurring mutants are also shown. Bars represent the mean ± SD of 8 different protein S determinations corresponding to duplicates from 4 different transfections per mutant. A value of 100% was arbitrarily given to the mean value of the wild type expression. Comparison of protein S secretion in the alternative mutants and that in the wild type, as well as that in the original mutants was performed using an unpaired t test. OnlyP-values considered significant (≤ .05) are shown. A comparison of the sequences containing the mutated residues in human and other mammalian protein S and related proteins (human Gas6, human SHBG, and the G4 and G5 repeats in human laminin A) is also shown. Given the high conservation of the protein S sequence around G441 and Y444 in mammals, a consensus sequence from baboon, rabbit, bovine, porcine, murine, and rat PS is shown as mammalian protein S. When there is a difference between human protein S and other mammalian species, this is indicated in the consensus sequence by the different residue followed by the number of species, when more than 1, where this difference is observed. Dots indicate conserved amino acids. For a more complete sequence comparison, see reference 47. The mutated residues are in bold.
Discussion
The molecular basis of protein S deficiency is not known for most type I missense mutations. An exception is protein S Nagoya (R474C), where the mutation produces an 8-fold reduction in the secretion of protein S by transfected cells.44 This difference is due to impaired secretion and intracellular degradation of the mutant protein S. Phenotypically, the mutation is manifested as a type I deficiency. In this study we extend this characterization to PROS1mutations found in different types of protein S deficiencies to analyze the molecular fate of these mutants.
In kindreds where cosegregation of protein S deficiency and aPROS1 mutation existed, there was always an impairment in secretion of the mutant in the transiently transfected cell cultures (groups A and B). Decreased secretion efficiency probably reflects the role of the mutated residues in the folding of protein S. Accordingly, when the same residues where mutated to more conservative amino acids, secretion levels were recovered. This was the case of G441A, Y444F, and R520K or R520H. Residue R520 was predicted to be solvent exposed.47 The fact that secretion was recovered also in the R520E mutant indicates that R520 is not forming a structurally important salt bridge, because this would not be formed by a negatively charged amino acid. Interestingly, substitution by Ala gave an intermediate effect (77%). It is possible that R520 is stabilizing a secondary structure element in such a way that a Gly cannot do at this position. R520 is predicted to form part of a beta strand,47 and only polar residues are found in protein S from different species at this position. The 3 mutations studied at residue 626 (P626L, P626G, and P626X) affected the secretion of protein S to different extents. A Pro is present in all sequences of the protein family at this position (Figure 4), suggesting that the residue has structural importance. As P626 is located after a cysteine, it is possible that this residue affects the efficiency of formation of the disulfide bond. Interestingly, P626 could be substituted by a stop codon with only a 30% loss of secretion, indicating that the C-terminal part of protein S is not necessary for efficient secretion. This stands in contrast to protein C, where the last 10 amino acids are essential for secretion.48 To sum up, all mutations found to affect secretion are located in residues conserved in related sequences and likely to be involved in determination of the structure.
For mutations studied, there was an overall correlation between the experimentally determined effects in protein S secretion of each mutation and the laboratory measurements in carriers. The association was particularly good for the concentration of protein S in the media (Figure 2 and 3) and the free protein S concentration measured in plasma of the carriers (Table 1). Protein S secretion was more affected in group A than in group B mutants, mirroring the effect seen in carriers of those mutations. Furthermore, Y444C carriers had lower free protein S levels than G441C carriers (13% vs. 26%, Table 1). In turn, G441C secretion was similar to R520G, and both only slightly lower than P626L. The same tendency was observed in free protein S values for carriers of these mutations (26% for G441C and R520G, 38% for P626L), but not in total protein S values (41% for G441C, compared with 82% in R520G and 62% for P626L). A better correlation between secretion in cell culture and free (rather than total) protein S levels would suggest that in vivo, the level of protein S synthesis by the liver is better reflected in its free plasma concentration. Although a mechanism for this hypothesis is lacking, there are several studies that indicate that free protein S concentration is more tightly regulated than total protein S in situations such as acute phase, warfarin treatment, and sepsis.15,16 49-51 However, given the limited number of individuals in each family studied, the observed correlation between laboratory measurements and secretion should be considered as an overall trend.
With the exception of Y444C, the protein S missense mutants studied were detected at appreciable levels in the media of transfected cells. This suggests that protein S mutants would form part of the pool of plasma protein S in carriers of missense mutations. In fact, family PS 214 illustrates this phenomenon in vivo. The propositus is homozygous for the R520G mutation and has low but clearly detectable levels of total protein S (36%), indicating that the mutant R520G protein is present in plasma. All heterozygous carriers of the R520G mutation had total protein S levels well above borderline values (71%, 77%, and 99%), whereas their respective free protein S values were low (24%, 35%, and 20%). The difference between homozygous and heterozygous carriers likely reflects a gene dosage effect of this mutation on the plasma levels of protein S. The fact that the propositus did not suffer from perinatal thrombosis, as other homozygous cases reported in the literature, suggests that the mutant protein is functional to some extent.23 24 Comparison of this kindred with the other pedigrees studied here also illustrates the effects of other genetic or circumstantial factors on the observed phenotype. Although R520G, P626L, and G441C had similar levels of expression in the transfection experiments, only heterozygous carriers of the R520G mutation were diagnosed as type III protein S deficients.
In conclusion, we found decreased secretion as the underlying cause of protein S deficiency in 4 different PROS1 missense mutations manifesting either as type I or III in their carriers (groups A and B). In contrast, there was no significant effect on protein S secretion by 2 protein S mutations not cosegregating with protein S deficiency (group C), suggesting that they are uncommon and apparently neutral DNA variants. Nevertheless, from the current study we cannot exclude that these PROS1 variants could lead to minor effects in secretion, difficult to evaluate with our experimental setting, which could act as predisposing factors for protein S deficiency under certain conditions. The observed correlation between secretion of protein S in cell culture and protein S levels suggests that different causative mutations have quantitatively distinct effects on plasma protein S. Mutations in which a significant amount of mutant protein S is produced would be more likely to manifest as type III in some or all carriers, although the type of deficiency does not seem to be dictated only by PROS1.
Acknowledgment
We acknowledge Helena Kruyer for her helpful assistance with the manuscript.
Supported by grants from the Tore Nilson Trust (to P.G. de F.); the Albert Pählsson Trust, and the Fondation Louis-Jeantet de Mèdecine (to B.D.); the Swedish Medical Research Council (grants 07143 and 13000), research funds from the University Hospital, Malmö, the Johan and Greta Kock Trust, the AlfredÖsterlund Trust (to P.G. de F. and B.D.) and by the Spanish DGICYT (grant PB94-1233) and research funds from the Servei Català de la Salut (to N.S.). Y.E-P. was supported by a Marie Curie fellowship from the EU (grant ERB/4001/GT/97/1535).
Reprints:Björn Dahlbäck, Wallenberg Laboratory, University Hospital, Malmö, S-20502 Malmö, Sweden.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal