The Janus kinase-signal transducer and activator of transcription (Jak-Stat) pathway stands as a paradigm of how diverse extracellular signals can elicit rapid changes in gene expression in specific target cells. This pathway is widely used by members of the cytokine receptor superfamily, including those for the clinically important cytokines granulocyte colony-stimulating factor (G-CSF), erythropoietin, thrombopoietin, the interferons, and numerous interleukins, which makes it central to hematopoietic cell biology and hematologic therapy alike. Indeed, study of the Jak-Stat pathway has provided a wealth of information on hematopoiesis and hematopoietic disease, and conversely, studies of hematopoietic disorders have yielded new insights into the functions of Jaks and Stats. This review aims to detail the role of the Jak-Stat pathway in the normal development and function of hematopoietic cells and to describe how several hematopoietic disorders are caused, at least in part, by perturbations of this pathway.
Jaks
Jaks are cytoplasmic tyrosine kinases that participate in signaling from a range of cell-surface receptors, particularly members of the cytokine receptor superfamily, which lack intrinsic tyrosine kinase activity.1,2 There are 4 mammalian Jaks: Jak1, Jak2, Jak3, and Tyk2. These associate with the weakly conserved “box 1” and “box 2” recognition motifs in the membrane-proximal region of cytokine receptors3,4 and are responsible for a range of phosphorylation events on stimulation of such receptors with their specific ligand. In addition, some receptors that have tyrosine kinase activity, such as those for macrophage colony-stimulating factor and stem cell factor, also activate Jaks, though it is unclear what role they play in these instances.5 6
Cell lines deficient for either Jak1 or Jak2 are unable to mediate a response to interferon-γ, whereas those deficient in Tyk2 fail to respond to interferon-α/β.7-9 In addition, the expression of kinase-deficient Jaks or the introduction of mutations that prevent Jak binding and activation abolishes the proliferative and anti-apoptotic signaling from a number of other cytokine receptors.10-14 The essential role of Jaks in mediating the effects of these hematopoietic regulators was recently confirmed by targeted disruption of the corresponding murine genes (Table1). Jak1-deficient mice exhibited perinatal lethality, apparently because of defective neural function, and defective lymphoid development.15 Targeted disruption of the Jak2 gene resulted in an embryonic lethal phenotype caused by a block in definitive erythropoiesis but with intact lymphoid development.16,17 In both cases, a number of other specific cytokine-induced biologic responses were absent or impaired. Finally, Jak3 knockout mice exhibited severe combined immunodeficiency, with markedly reduced numbers of functional T and B lymphocytes,18,19 and dysregulated myelopoiesis.20 Thus, the Jaks collectively are vital for normal hematopoietic function, which can be explained by their nonredundant role in the signaling of specific cytokines (Table1).
Jaks in hematopoiesis: phenotype of mouse knockouts
| Jak Type . | Relevant Phenotypes . | Cytokines Affected . | Refs . |
|---|---|---|---|
| Jak1 | • Impaired lymphoid development | IL-2, IL-4, IL-6, IL-7, IL-9, IL-10, IL-15, LIF, all interferons | 15 |
| • Defective responses to class 2 cytokines and those using γc or gp130 receptor subunits | |||
| Jak2 | • No definitive erythropoiesis | EPO, TPO, IL-3, IL-5, GM-CSF, IFN-γ | 16, 17 |
| Jak3 | • Defective lymphoid development | IL-4, IL-7, IL-9, IL-15 | 18-20 |
| • Dysregulated myelopoiesis |
| Jak Type . | Relevant Phenotypes . | Cytokines Affected . | Refs . |
|---|---|---|---|
| Jak1 | • Impaired lymphoid development | IL-2, IL-4, IL-6, IL-7, IL-9, IL-10, IL-15, LIF, all interferons | 15 |
| • Defective responses to class 2 cytokines and those using γc or gp130 receptor subunits | |||
| Jak2 | • No definitive erythropoiesis | EPO, TPO, IL-3, IL-5, GM-CSF, IFN-γ | 16, 17 |
| Jak3 | • Defective lymphoid development | IL-4, IL-7, IL-9, IL-15 | 18-20 |
| • Dysregulated myelopoiesis |
EPO, erythropoietin; IFN, interferon; IL, interleukin; TPO, thrombopoietin.
Stats
Stats are latent cytoplasmic transcription factors that become activated after recruitment to an activated receptor complex. Subsequently, these active Stats translocate to the nucleus to affect gene expression. Seven Stat proteins have been identified in mammalian cells—Stats1 to 6, including Stat5a and Stat5b, which are encoded by distinct genes. In addition, different isoforms of several Stats have been identified.21
Targeted inactivation of Stat genes in the mouse also resulted in severe effects on the development and function of hematopoietic cells (Table 2). Stat1 knockout mice showed defective innate immune responses to viruses and bacteria because of the absence of interferon signaling,22,23whereas Stat2 knockout mice showed defective responses to interferon α/β (C. Schindler, personal communication, 1999). Targeted disruption of the Stat3 gene produced early embryonic lethality.24 Subsequently, T-cell–specific Stat3-deficient mice were generated that were severely impaired in IL-6–induced proliferation because of enhanced apoptosis.25Stat4 knockout mice were defective in the formation of Th1 cells, largely a result of disrupted IL-12 receptor function.26,27 Mice with both Stat5a andStat5b genes disrupted showed multiple defects, with responses to IL-2, IL-3, granulocyte-macrophage colony-stimulating factor (GM-CSF), and granulocyte colony-stimulating factor (G-CSF) affected,28,29 whereas the respective single Stat5knockouts also exhibited defective proliferative responses to specific cytokines.30,31 Finally, a block in Th2 cell development and IgE class switching was observed in Stat6 knockout mice.32 33
Stats in hematopoiesis: phenotypes of mouse knockouts
| Stat Type . | Relevant Phenotypes . | Cytokines Affected . | Refs . |
|---|---|---|---|
| Stat1 | • Interferon responses absent: — innate immune responses absent — highly sensitive to viral/microbial infection — IFN-responsive genes not activated | IFNs only | 22, 23 |
| Stat 2 | • Type 1 interferon responses impaired | IFN-α/β | C. Schindler, pers. comm. |
| Stat 3 | • Embryonic lethal | 24 | |
| Stat 4 | • IL-12 responses absent ↓ production of high IFN-γ, low IFN-γ Rα Th1 cells ↓ priming for high level IFN-γ production ↓ lymphocyte proliferation ↓ enhancement of NK cell-mediated cytotoxicity ↑ Th2 cells | IL-12 only | 26, 27 |
| Stat5ab | • Proliferation signaling affected: ↓ CFU-Mix, Eos, G, GM, Pre-B ↓ peripheral T cells Absence of NK cells | IL-2, IL-3, IL-7, GM-CSF, G-CSF | 28, 29 |
| Stat6 | • IL-4 responses absent: -Absence of IL-4 producing Th2 cells -Block in B-cell IgE class switching ↓ lymphocyte proliferation (partial) ↓ expression of IL-4-induced cell surface markers | IL-4 only | 32, 33, 179 |
| Stat Type . | Relevant Phenotypes . | Cytokines Affected . | Refs . |
|---|---|---|---|
| Stat1 | • Interferon responses absent: — innate immune responses absent — highly sensitive to viral/microbial infection — IFN-responsive genes not activated | IFNs only | 22, 23 |
| Stat 2 | • Type 1 interferon responses impaired | IFN-α/β | C. Schindler, pers. comm. |
| Stat 3 | • Embryonic lethal | 24 | |
| Stat 4 | • IL-12 responses absent ↓ production of high IFN-γ, low IFN-γ Rα Th1 cells ↓ priming for high level IFN-γ production ↓ lymphocyte proliferation ↓ enhancement of NK cell-mediated cytotoxicity ↑ Th2 cells | IL-12 only | 26, 27 |
| Stat5ab | • Proliferation signaling affected: ↓ CFU-Mix, Eos, G, GM, Pre-B ↓ peripheral T cells Absence of NK cells | IL-2, IL-3, IL-7, GM-CSF, G-CSF | 28, 29 |
| Stat6 | • IL-4 responses absent: -Absence of IL-4 producing Th2 cells -Block in B-cell IgE class switching ↓ lymphocyte proliferation (partial) ↓ expression of IL-4-induced cell surface markers | IL-4 only | 32, 33, 179 |
CFU, colony-forming unit: G-CSF, granulocyte colony-stimulating factor; GM-CSF, granulocyte-macrophage colony-stimulating factor; IFN, interferon; IL, interleukin; NK, natural killer.
Further evidence of the vital role of Stats in cytokine receptor signaling has been obtained from studies in cell lines. For example, a mutant cell line deficient in Stat1 expression showed a block in interferon signaling similar to that in Stat1 knockout mice.34 Other studies using dominant-negative Stats or specific receptor mutants have shown, for example, that Stat3 activation plays a key role in the differentiation responses to IL-6 and G-CSF,35-37 whereas Stat5 appears very important for proliferative responses to IL-3, IL-5, G-CSF, and GM-CSF38-40 and for neutrophilic differentiation in response to G-CSF.40 Interestingly, these latter studies differ from the relative mild effects on hematopoiesis seen inStat5ab double knockout mice,28 and they serve to highlight some of the potential problems in interpreting both model systems. For example, compensatory overlapping pathways can mask the physiological consequences of a gene disruption in a whole animal, whereas effects seen in cell lines may represent physiologically irrelevant endpoints or nonspecific consequences of a presumed dominant-negative protein acting on other pathways. However, both types of studies have together yielded great insight into the biologic function of Stats.
Jak-Stat pathway
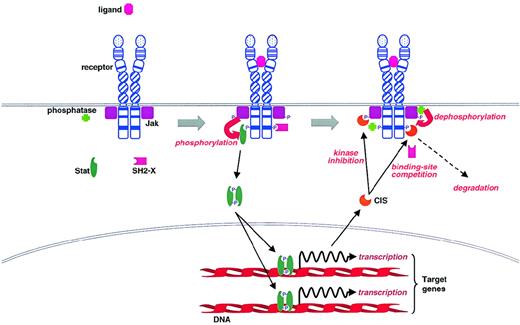
A general model of Jak-Stat activation from cytokine receptors has been proposed,2,21,41 though there are several exceptions and variations on the basic theme first elucidated for the interferon receptors.42 Binding of ligand to a cytokine receptor leads to the activation of Jaks, presumably through autophosphorylation on tyrosines (Figure 1). Activated Jaks then phosphorylate the receptor, creating docking sites for specific signaling proteins, including Stat proteins, which the Jaks can then phosphorylate on a conserved tyrosine residue at their C-terminus. Subsequently, the Stats form stable homodimers and heterodimers by interactions between the Src homology 2 (SH2) domain of one Stat protein and the phosphotyrosine of another before translocation to the nucleus, where they influence transcription of target genes by binding to specific regulatory sequences.21 41
Activation of the Jak-Stat pathway by cytokine receptors and its regulation by CIS family members and tyrosine phosphatases.
Activation of the Jak-Stat pathway by cytokine receptors and its regulation by CIS family members and tyrosine phosphatases.
Specificity in the Jak-Stat pathway
Specificity in cytokine signaling is largely determined by the combination of activated Jaks and Stats. A wide range of cytokines and growth factors activate Jak1, Jak2, and Tyk, whereas Jak3 is only activated by cytokines that have the common γ-chain as a component of their receptor complex.43 Receptors show more specificity in their ability to recruit and activate Stats. For example, stimulation with interferon (IFN)-α or IFN-β leads to phosphorylation of Stats 1 and 2, which form a complex with a third protein, p48.44 In contrast, G-CSF stimulation leads to the activation of Stat3 and Stat5 homodimers, some Stat1 homodimers, and Stat1/3 and Stat3/5 heterodimers.45,46 Such studies have led to the suggestion that diversity within the type of Stat complexes activated contributes to the nature of the cellular responses to a given cytokine or growth factor.47 The subsequent gene knockout and dominant-negative mutant studies described above have largely supported this hypothesis.
The specificity of Stat activation is partially mediated through their recruitment to specific cytoplasmic tyrosines of particular receptors through their SH2 domains. For example, tyrosine 440 in the cytoplasmic domain of the IFN-γ receptor α chain is responsible for recruitment and activation of Stat1,48 whereas tyrosines 578 and 606 of the IL-4-R are required for phosphorylation and activation of Stat6.49 Similarly, numerous studies have identified the YxxQ motif as a consensus Stat3 docking site, though it can bind to other motifs.45,50,51 In addition, several examples of Stat activation that do not require direct docking to receptor tyrosines have been reported. For instance, full activation of Stat1 by G-CSF or growth hormone and of Stat5 by G-CSF and GM-CSF occurs in the complete absence of receptor tyrosines.38,45,51,52 It has been proposed that Jak1 and Jak2 can specifically recruit and phosphorylate Stat1 and Stat5, respectively,53,54 which could explain their activation in these cases. However, there is growing evidence that other receptor components can also act as docking sites for Stats.45,55 For example, Stat1 is recruited to the IFN-α/β receptor complex by binding to a Stat2 molecule already docked to the activated receptor.21 Thus, Stat specificity is determined by recruitment to the receptor complex in toto rather than simply to the linear sequence of each receptor. However, the particular Jaks and Stats activated may also be dependent on the cell-type or its state of differentiation,21,56-58 and receptor “cross-talk” may further modify the response elicited. For example, IL-4 inhibits IL-2–mediated Stat5 activation,59 IL-10 suppresses interferon-mediated Stat activation,60 and cyclic adenosine monophosphate impairs IL-2–dependent signaling by downregulating levels of the Jak3 protein itself.61
Individual Stats bind to similar DNA response elements, mostly related to a γ interferon-activated site, a regulatory element in the promoter of IFN-γ–inducible genes.42 However, the recognition sites are not identical,62-64 and so different genes are targeted for induction by different Stats (Table3). In addition, Stats can mediate transcriptional repression at specific promoters.65,66 Some Stats are able to form both homodimers and heterodimers, which can further broaden the range of Stat/DNA-binding specificities.21 The duration of Stat activation is also able to influence the transcriptional program induced.21,46In addition, various Stat isoforms are differentially expressed in specific cell types, which can also have an impact on the expression of Stat-responsive genes.21,57,58,65 It has recently been shown that Stats can interact with a range of other nuclear factors and coactivators, including CBP, Nmi, the glucocorticoid receptor c-Jun, and MCM5,21,67-69 which increases the range of transcriptional responses in which Stats can participate. Finally, though phosphorylation of the C-terminal tyrosine is critical for Stat activation, serine phosphorylation probably also modulates the transcriptional response.70-73 Indeed, it was shown that Ser727 of Stat1α is directly involved in the recruitment of MCM5 as part of IFN-γ–induced transcriptional activation.68Together, this complex control of specificity enables an individual hematopoietic cell to elicit the appropriate transcriptional response to incoming signals from cytokines, growth factors, and other stimuli.
Genes induced by Stat proteins
| Stat . | Genes Encoding . | Refs . |
|---|---|---|
| Stat1 | ISG54, IRF-1, CIITA, mig, 2,3 dioxygenase, GBP, p21 | 22, 23, 64 |
| Stat3 | JunB, SAA3, JAB, C-reactive protein, BclxL, p21 | 63, 64, 98, 134, 136 |
| Stat4 | IFN-γ, IRF-1, Fc-γRI, CD23, MHC class II | 33, 180 |
| Stat5 | β-casein, IL-2R-α, CIS, osm, pim1, p21, cyclin D1 | 92, 181-183 |
| Stat6 | IL-4R-α, Fc-εRIIa, C-ε, C-γ1, C-γ4 | 21, 63, 64 |
| Stat1/Stat1/p48 | GBP | 64 |
| Stat1/Stat2/p48 (ISGF3) | ISG15, ISG54, 6-16, 2′,5′ oligoadenylate synthetase, 2,3-dioxygenase | 64 |
| Stat . | Genes Encoding . | Refs . |
|---|---|---|
| Stat1 | ISG54, IRF-1, CIITA, mig, 2,3 dioxygenase, GBP, p21 | 22, 23, 64 |
| Stat3 | JunB, SAA3, JAB, C-reactive protein, BclxL, p21 | 63, 64, 98, 134, 136 |
| Stat4 | IFN-γ, IRF-1, Fc-γRI, CD23, MHC class II | 33, 180 |
| Stat5 | β-casein, IL-2R-α, CIS, osm, pim1, p21, cyclin D1 | 92, 181-183 |
| Stat6 | IL-4R-α, Fc-εRIIa, C-ε, C-γ1, C-γ4 | 21, 63, 64 |
| Stat1/Stat1/p48 | GBP | 64 |
| Stat1/Stat2/p48 (ISGF3) | ISG15, ISG54, 6-16, 2′,5′ oligoadenylate synthetase, 2,3-dioxygenase | 64 |
IFN, interferon; IL, interleukin; MHC, major histocompatibility complex.
Independent functions of Jaks and Stats
The major function of Jaks is generally considered to be Stat activation. However, this is clearly not the only role that Jaks play in signaling. For example, Jaks are directly implicated in the activation of the kinase Pyk2,74 stimulation of the Ras-MAPK pathway,13,75,76 and the induction of the c-fos and c-myc genes.77 Conversely, there is considerable evidence that some activation of Stats occurs independently of Jaks. For example, cell lines deficient in Jak2 or Tyk2 showed no effect on G-CSF–dependent Stat activation, and a cell line deficient in Jak1 showed only partial reduction in Stat3 activation.55 Similar results were obtained withJak knockout mice for a range of factors.16Furthermore, Stat6 activation after CD40 engagement occurs independently of detectable Jak phosphorylation.78 Such data suggest that other kinases are probably also involved in mediating Stat activation. In support of this, Src has been shown to bind and activate Stat3 directly,79 whereas Bcr-Abl can also recruit and activate Stat5 by the interaction of Stat5 with the adaptor CrkL, which itself docks to Bcr-Abl.80 Other work implicates a number of non-Jak kinases as responsible for the serine phosphorylation of Stats.73 81
Negative regulation of the Jak-Stat pathway
As further evidence of the importance of the Jak-Stat pathway, negative feedback mechanisms have been identified that control its activation (Table 4). These include endosomal degradation of Jak/receptor complexes through receptor-mediated endocytosis82,83 and the dominant-negative effects of several naturally occurring Stat variants.84,85 In addition, the PIAS proteins have been identified. They seem to bind directly to Stats and to inhibit DNA binding, though their exact biologic role remains unclear.72 86 Two other means of negative regulation, by CIS/SOCS/SSI family members and by tyrosine phosphatases, have been studied in more detail.
Negative regulatory mechanisms of the Jak-Stat pathway
| Mechanism . | Refs . |
|---|---|
| Receptor-mediated endocytosis | 82, 83 |
| Dominant-negative Stats | 84, 85 |
| PIAS family of proteins | 72, 86 |
| CIS/SOCS/SSI family of proteins | 88, 89 |
| Tyrosine phosphatases | 114, 115 |
| Mechanism . | Refs . |
|---|---|
| Receptor-mediated endocytosis | 82, 83 |
| Dominant-negative Stats | 84, 85 |
| PIAS family of proteins | 72, 86 |
| CIS/SOCS/SSI family of proteins | 88, 89 |
| Tyrosine phosphatases | 114, 115 |
CIS/SOCS/SSI Family
This family, variously called CIS, SOCS, or SSI, is a group of small proteins containing SH2 and CIS homology (CH) domains (also called SOCS boxes). At least 8 family members have been identified (CIS1 to CIS7 and JAB),87,88 which are involved in the relatively specific regulation of cytokine signaling.89
The first member of this family, originally denoted CIS (now called CIS1) for cytokine-induced SH2-containing protein, was cloned as an immediate early gene that was induced by IL-2, IL-3, and erythropoietin.90,91 The induced CIS protein can subsequently associate with specific tyrosines of the activated receptors,92 including one of the major Stat5 binding sites of the erythropoietin receptor.93 Forced expression of CIS1 partially suppresses IL-3 or erythropoietin-induced proliferation and Stat5 activation.92 Conversely, Stat5 activates the CIS1 promoter through interaction with tandem TTCNNNGAA motifs,92 with CIS1 expression absent in the ovaries ofStat5ab double knockout mice.28 Thus, CIS1 appears to act as a negative feedback regulator of the Jak-Stat5 pathway. This is supported by recent observations in CIS1-transgenic mice that exhibited a phenotype similar to that observed in Stat5abdouble knockout mice.94
The second CIS/SOCS/SSI family member, JAB (also called SOCS-1, SSI-1, and TIP-3), was identified independently because of its ability to interact with the kinase domains of Jak2 and Tec,95,96 to inhibit the IL-6–induced differentiation and growth arrest of a leukemic cell line,97 and to be recognized by an antibody to Stat SH2 domains.98 Overexpression of JAB can inhibit virtually any signal using Jaks, such as Stat5 activation by erythropoietin, Stat3 activation by leukemia inhibitory factor or IL-6, and c-fos induction by IL-2. However, JAB does not inhibit fibroblast growth factor (FGF)-induced c-fos activation or c-Kit phosphorylation, though it binds to the FGF receptor and c-Kit.87 Although JAB was found to suppress Tec kinase activity, this effect was marginal compared with its effect on Jaks. Therefore, JAB-mediated kinase inhibition seems to be specific for Jak tyrosine kinases; binding does not always imply inhibition. However, recently JAB has been shown to bind inducibly to the c-Kit receptor tyrosine kinase through its SH2 domain.95,99 Although JAB did not inhibit the catalytic activity of the c-Kit tyrosine kinase, it inhibited c-Kit–mediated proliferation signals, probably by interaction with the SH3 domains of the signaling proteins Grb2 and Vav, thereby suppressing their function.99 In certain circumstances JAB may also be able to suppress signals from non-Jak tyrosine kinases. JAB knockout mice displayed growth retardation, fatty degeneration of the liver, and monocytic infiltration of several organs. They died before weaning within 3 weeks of birth.100,101 Lymphocytes in the thymus and spleen of these mice exhibited accelerated apoptosis, and at 10 days of age their numbers were 20% to 25% of those in wild-type mice.100Among various pro-apoptotic and anti-apoptotic molecules examined, an upregulation of Bax was found in lymphocytes of the spleen and thymus of knockout mice.100 In addition, there was a progressive loss of maturing B lymphocytes in the bone marrow, spleen, and peripheral blood, whereas constitutive activation of Stat1 was found in the liver of JAB knockout mice.101 Part of this phenotype clearly resembles that found in IFN-γ–transgenic mice.102 In addition, hematopoietic progenitor cells from JAB knockout mice were hyperresponsive to IFN-γ, with the degree of inhibition varying markedly with the stimulating factor used.103 It is important to note that many of the pathologic conditions observed in JAB knockout mice can be eliminated by antibody injections or by crossing them with IFN-γ knockout mice.104 Therefore, JAB appears to act as a negative regulator of IFN-γ/Stat1 and probably functions by preventing apoptosis induced by Stat1. However, other studies suggest that JAB may also inhibit other pathways that induce Bax expression, thereby preventing Bax-induced apoptosis in vivo.100
So how do the CIS family members exert their negative effects on Jak-Stat signaling? Several possibilities, which are not mutually exclusive, are shown in Figure 1. Among CIS family members, JAB and CIS3 are able to bind the Jak2 catalytic (JH1) domain,87leading to direct inhibition of the Jak kinase.105 Binding requires the SH2 domain plus an additional N-terminal 12 amino acids (extended SH2 subdomain) containing 2 residues (Ile68 and Leu75) that are conserved in the CIS family.14,105 This subdomain interacts with the tyrosine residue Y1007 in the activation loop of Jak2, whose phosphorylation is critical for the induction of kinase activity.106,107 Other CIS family members bind directly to receptors, where they may function by preventing stimulatory signaling pathways coupled to specific phosphotyrosine motifs on receptors.92,93 Alternatively, given that CIS proteins have a relatively short half-life, they may act as scavengers of activated receptor complexes, targeting them for degradation.93 In support of this, it has been shown that CIS1 itself is ubiquitinylated,93 whereas the CH domain appears to interact with components of the proteasomal degradation pathway.108-110 However, other studies have shown that the CH domain of JAB may actually protect this molecule from degradation.108 111 Clearly more work is required to unravel the various intracellular functions of the CIS family members.
Tyrosine phosphatases
Several studies have shown that an important negative regulatory mechanism of the Jak-Stat pathway involves the recruitment of tyrosine phosphatases containing tandem SH2 domains (SHP-1 and SHP-2) to receptor complexes. Both phosphatases can bind either activated receptors or to Jak family members themselves, leading to dephosphorylation of the kinase (Figure 1).112-115 This, in turn, leads to reduced activation of Jak-Stat pathway components.114,116 The potential in vivo importance of this mechanism is strongly suggested by the phenotype of motheaten(me/me) mice lacking SHP-1, which die of a disease with components of autoimmunity and inflammation.117 However, it remains to be elucidated whether enhanced Jak kinase activity is entirely responsible for the motheaten phenotype because SHP-1 has also been shown to regulate negatively a number of receptor and nonreceptor tyrosine kinases.118,119 In addition, though it is clear that the major means by which Stat activity is attenuated is through dephosphorylation by protein tyrosine phosphatases,21 it is unknown whether SHP-1 or SHP-2 or some other phosphatase is responsible. However, it appears that SHP-1 can associate directly with Stat5, implicating it in the direct dephosphorylation and deactivation of this Stat.120
Jaks, Stats, and hematopoietic diseases
The wide use of the Jak-Stat pathway by hematologically important factors, the severity of artificially disrupting the Jak-Stat pathway on hematopoiesis, and the number of key genes with Stat-response elements already provides some appreciation of the importance of this pathway in hematopoiesis and the regulation of hematopoietic cell function. We will now summarize the studies showing that several diverse hematopoietic disorders exhibit perturbations in the Jak-Stat pathway. Indeed, in a number of these cases, experiments have directly implicated the altered Jak or Stat signaling, or both, in the pathogenesis of the disease. Such molecular investigations provide a foundation on which to build an understanding of these conditions and a framework for rational improvements in therapy.
Hematopoietic malignancies
Aberrant activation of Jaks and Stats
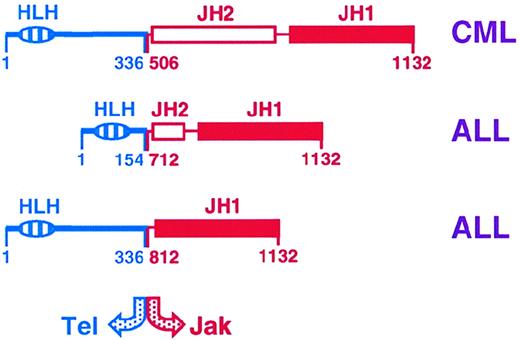
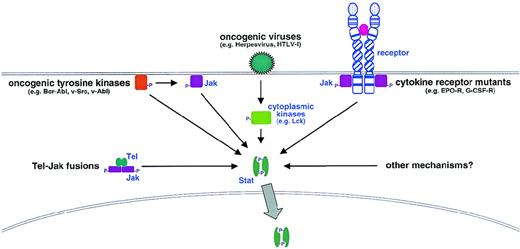
The most direct evidence implicating dysregulation of the Jak-Stat pathway in hematopoietic malignancies was the identification of Tel-Jak2 fusions in lymphoid and myeloid leukemias.121,122In early B-precursor acute lymphoblastic leukemia, t(9;12)(p24;p13) translocations were responsible, whereas in the case of atypical chronic myeloid leukemia there was a complex t(9;15;12)(p24;q15;p13) translocation. In each case, the helix-loop-helix oligomerization domain of the transcription factor Tel is fused to the catalytic JH1 domain of Jak2 (Figure 2), which leads to constitutive association and hence activation of the kinase and constitutive activation of Stat proteins.123,124 However, Jaks and Stats are also known to be constitutively activated in hematopoietic cells transformed by diverse oncogenic tyrosine kinases (Table 5), as well as in a variety of lymphomas and leukemias, including those transformed by oncogenic viruses (Table 6). For the oncogenic tyrosine kinases, the activation of Stats may be direct or occur through Jaks, whereas the oncogenic viruses activate a number of cytoplasmic kinases to mediate the constitutive Stat activation observed (Figure 3).125
Tel-Jak2 fusions observed in myeloid (CML; chronic myeloid leukemia) and lymphoid (ALL; acute lymphoblastic leukemia) leukemias.121 122
The relative position of the fusions, as well as the helix-loop-helix (HLH) and Jak homology (JH) domains are shown.
Stat activation by specific oncoproteins
| Cell type . | Oncoproteins . | Activated Stats . | Refs . |
|---|---|---|---|
| Primary bone marrow | Bcr-Abl Tel-Jak2 | Stat5 Stat5 | 184, 185 123, 124 |
| Myeloid cells | v-Src | Stats1,3,5 | 130 |
| Basophil/mast cells | Bcr-Abl | Stats1,5 | 129, 186 |
| Erythroleukemia/blast cells | Bcr-Abl | Stats1,5 | 129, 186, 187 |
| Pre-B lymphocytes | v-Abl | Stats1,5,6 | 188 |
| Cell type . | Oncoproteins . | Activated Stats . | Refs . |
|---|---|---|---|
| Primary bone marrow | Bcr-Abl Tel-Jak2 | Stat5 Stat5 | 184, 185 123, 124 |
| Myeloid cells | v-Src | Stats1,3,5 | 130 |
| Basophil/mast cells | Bcr-Abl | Stats1,5 | 129, 186 |
| Erythroleukemia/blast cells | Bcr-Abl | Stats1,5 | 129, 186, 187 |
| Pre-B lymphocytes | v-Abl | Stats1,5,6 | 188 |
Activation of Jaks and Stats in hematopoietic malignancies
| Type of Malignancy . | Activated Jaks and Stats . | Refs . |
|---|---|---|
| Lyphoma | ||
| LSTRA T cell line | Jaks1,2; Stats3,5 | 115, 189 |
| Cutaneous T-cell lymphoma | Jak3; Stats3,5 | 190 |
| Mycosis fungoides | Stat3 | 191 |
| Herpesvirus saimiri dependent | Stats1,3 | 131, 192 |
| EBV-related | Stats1,3 | 193 |
| Leukemia | ||
| Erythroleukemia | Stats1,5 | 186 |
| Acute myelocytic leukemia | Stats1,3,5 | 185, 193-196 |
| Chronic myelocytic leukemia | Stat5 | 184, 185 |
| Acute lymphocytic leukemia | Stats1,5 | 193, 194 |
| Megakaryocytic leukemia | Stat5 | 197 |
| HTLV-I dependent T-cell leukemia | Jaks1,3; Stats3,5 |
| Type of Malignancy . | Activated Jaks and Stats . | Refs . |
|---|---|---|
| Lyphoma | ||
| LSTRA T cell line | Jaks1,2; Stats3,5 | 115, 189 |
| Cutaneous T-cell lymphoma | Jak3; Stats3,5 | 190 |
| Mycosis fungoides | Stat3 | 191 |
| Herpesvirus saimiri dependent | Stats1,3 | 131, 192 |
| EBV-related | Stats1,3 | 193 |
| Leukemia | ||
| Erythroleukemia | Stats1,5 | 186 |
| Acute myelocytic leukemia | Stats1,3,5 | 185, 193-196 |
| Chronic myelocytic leukemia | Stat5 | 184, 185 |
| Acute lymphocytic leukemia | Stats1,5 | 193, 194 |
| Megakaryocytic leukemia | Stat5 | 197 |
| HTLV-I dependent T-cell leukemia | Jaks1,3; Stats3,5 |
EBV, Epstein-Barr virus; HTLV, human T-cell lymphoma virus.
Mechanisms of aberrant Stat activation in hematopoietic malignancy and disease.
Evidence for Jak-Stat involvement
Although the data above are suggestive of a positive role for constitutive activation of the Jak-Stat pathway in leukemia, the results are largely correlative. However, other studies have provided more direct evidence for this hypothesis. For example, overexpression of a Tel-Jak2 fusion is sufficient to render Ba/F3 cells factor-independent,121,123 and mice transplanted with retrovirus expressing this fusion develop a fatal mixed myeloproliferative and T-cell lymphoproliferative disorder with a latency of 2 to 10 weeks.123 In addition, 2 mutants of theDrosophila Jak kinase have been identified that lead to leukemia-like defects through hyperactivation of the kinase.126,127 Murine homologues of this Jak mutant have been further shown to induce leukemia in mice.127 Finally, inhibition of constitutive Jak2 phosphorylation in primary pre-B leukemic cells with the Jak2 inhibitor AG490 is able to inhibit cell proliferation.128 However, other studies suggest that the constitutive Jak activation seen in transformed cells is not actually required for transformation. Thus, dominant-negative Jaks are unable to inhibit either Stat5 activation or factor-independent cell proliferation induced by Bcr-Abl.129 In addition, v-Src can directly activate Stat3,130 whereas the Herpesvirus Tip protein co-associates Lck and Stat3, leading to constitutive Stat3 activation in T cells transformed by this virus,131suggesting that in these cases Jaks are also superfluous.
In contrast, numerous recent studies have provided strong evidence for a role of Stats in the transformation process. For example, a dominant-negative Stat5 was able to inhibit apoptosis-resistant, growth factor-independent proliferation and leukemic potential of Bcr-Abl transformed cells132,133 and of the growth factor-independent colony formation of primary mouse bone marrow progenitor cells transduced with Bcr-Abl retrovirus.132 In addition, a constitutively active Stat5 mutant could restore these functions to a mutant Bcr-Abl deficient in Stat activation.132 Similarly, the abrogation of IL-3 dependence of myeloid cells by v-Src requires the SH2 and SH3 domains, which specifies the activation of Stats,130 and dominant-negative Stat3 has been shown specifically to block v-Src transformation in other cell systems.134,135 Furthermore, studies in multiple myeloma cells that show constitutive Stat activation have revealed that dominant-negative Stat3 induces apoptosis,136 again implicating Stat3 in the transformation process. Finally, a constitutively active mutant of Stat5 is sufficient to induce factor independence of Ba/F3 cells.137 Thus, constitutive Stat activation appears necessary, and perhaps sufficient, for the transformation process.
The results of the above studies imply that a permanent alteration in the genetic program of transformed cells, achieved by the constitutive activation of Stat proteins, is a critical step in the transformation process. Recent studies have begun to shed light on those changes that may be important. Thus, constitutive expression of Jak2 in Ba/F3 cells has been shown to lead to the induction of Bcl-2, resulting in delayed cell death,138 but the constitutively activated Stat3 observed in bone marrow mononuclear cells from patients with multiple myeloma also confers resistance to apoptosis, this time through the induction of Bcl-xL.136 Identification of other genes involved in the transformation process remains an important goal for future research.
Other hematologic disorders
Alterations in the Jak-Stat pathway have been associated either directly or indirectly with other hematologic disease states.
Severe combined immunodeficiency
In the most common form of severe combined immunodeficiency (SCID), X-linked SCID, both cellular and humoral immunity are severely affected: T-cell development is arrested in the thymic cortex, there is an almost complete lack of circulating T lymphocytes, and, though B lymphocytes are present, they do not undergo class switching.139 In X-linked SCID, mutations have been identified in the gene encoding the common γ chain (γc), a constituent of a number of cytokine receptor complexes.140,141 Although these mutations occur at multiple positions, all mutant receptors are defective in the activation of Jak3.142,143 Indeed, Jak3 knockout mice display a SCID phenotype that is virtually indistinguishable from that of γc null mice.18,144 Moreover, in a less common autosomal-recessive form of SCID, patients have been reported with inactivating mutations in the Jak3 gene itself.4,145 146 Together these findings show that abrogation of the Jak-Stat pathway is sufficient to account for SCID in humans.
Severe congenital neutropenia/acute myeloid leukemia
Patients with severe congenital neutropenia (SCN) exhibit a severe reduction in circulating neutrophils and a maturation arrest of bone marrow progenitor cells at the promyelocyte/myeloid stage.147,148 Such patients have an increased risk for myelodysplasia, acute myeloid leukemia, or both, and a poor prognosis for survival.149,150 A subset of patients with SCN has been identified with acquired nonsense mutations in the gene encoding the G-CSF receptor, which truncate its carboxyl-terminus.151,152 This subset has a strong (around 50%) predisposition to acute myeloid leukemia. Mice carrying a similar G-CSF-R truncation also show reduced basal levels of circulating neutrophils, but on continuous G-CSF treatment, neutrophil counts become elevated to above those of wild-type controls because of the increased proliferation of myeloid progenitors.154 This suggests that the G-CSF-R truncation may contribute to SCN and to the subsequent development of acute myeloid leukemia in these patients. Bone marrow cells from these mutant mice show reduced Stat3 activation in response to G-CSF, even under saturating conditions. In addition, there is an altered dose-response of Stat3 compared to Stat5 activation, such that at lower G-CSF concentrations the Stat3 deficiency is even more pronounced, a result confirmed in myeloid 32D cells.45,155 Because Stat3 appears indispensable for differentiation responses to G-CSF, the reduced Stat3:Stat5 ratio in cells with truncated receptors at low G-CSF concentrations may contribute to the reduced maturation observed.45,154,155 In addition, molecular mechanisms have been identified recently that explain the hyperproliferative function of truncated G-CSF-R. Such receptors show defective internalization compared with wild-type receptors46,155,156 and have a concomitant extension in the activation of Stats, particularly Stat5,46 consistent with a previous report of enhanced Jak2 activation in patients with SCN (Figure 3).157 It has recently been shown in 32D cells expressing truncated G-CSF-R that dominant-negative Stat5 inhibits whereas dominant-negative Stat3 actually enhances G-CSF–mediated growth, implicating perturbed Stat5 activation as a key molecular determinant of the hyperproliferative responses elicited from truncated G-CSF-R (Ward et al, manuscript in preparation). In addition, a novel G-CSF-R mutation has been identified in a patient with SCN who was unresponsive to G-CSF therapy—in this case Stat5 activation was substantially reduced,158 again consistent with an important role of Stat5 in controlling proliferative responses to G-CSF.
Benign erythrocytosis
Benign erythrocytosis is a dominant autosomal condition characterized by a mild increase in red blood cell counts and normal serum levels of erythropoietin because of hypersensitivity to erythropoietin.159,160 In addition, there is an increased and a sustained activation of Jak2 and Stat5 after erythropoietin stimulation.112,160 A number of pedigrees have been identified, all of which lead to erythropoietin (EPO)-R truncations161,162 that invariably result in the loss of the binding site for SHP-1 at Tyr 449 of the EPO-R.112Because SHP-1 is a negative regulator of Jak2 activation by EPO, it appears that lack of SHP-1 activation is responsible for the altered Jak-Stat kinetics and enhanced EPO responses in these patients (Figure3).
Fanconi anemia
Fanconi anemia (FA) is an autosomal recessive chromosome instability syndrome characterized by progressive bone marrow failure and an increased susceptibility to malignancy.163,164 The FA group C gene (FAC) has been identified, with its disruption leading to profound hypersensitivity of hematopoietic precursor cells to IFN-γ in mice165 and in patients with FA group C.166This appears to be the result of sustained Stat1 activation leading to apoptosis of these cells.166 Other researchers have reported that the FAC protein is involved in the recruitment of Stat1 to the IFN-γ receptor complex,167 which further suggests that perturbed Stat1 activation contributes to the phenotype of this disease.
Interferon resistance
Interferons, particularly IFN-α, have important therapeutic applications in the treatment of hematologic malignancies, including CML, hairy cell leukemia, and cutaneous T-cell lymphoma (CTCL).168,169 However, the efficacy is limited by the development of clinical resistance to IFN therapy in these patients.169 Efforts to understand the molecular basis of IFN resistance have been made by generating somatic cell mutants resistant to IFN, which showed that defects in the IFN receptor, Jaks, or Stats could contribute to this phenomenon.21,42 Similar analysis of IFN-α–resistant derivatives of CTCL cells also revealed a defect in normal Jak-Stat responses caused by a total absence of Stat1 expression,170 as previously observed in patients with IFN-resistant melanoma.171 However, a recent study also suggests a possible role for JAB in IFN resistance, especially for patients with a dominant phenotype.172 Stable expression of JAB in either NIH-3T3 or M1 leukemic cells leads to resistance to IFN-γ– and IFN-β–induced growth arrest. In both cell systems, IFN-γ did not induce tyrosine phosphorylation and DNA-binding activity of Stat1. In addition, IFN-resistant clones derived from LoVo cells and Daudi cells were found to express high endogenous levels of JAB without stimulation, with a concomitant reduction in IFN-induced Stat1 and Jak phosphorylation.172
Other diseases with altered Jak-Stat activation
Other hematologic diseases also show defects in the normal activation and regulation of Jak-Stat pathway components. For example, bone marrow cells from patients with myelodysplastic syndrome show impaired erythropoietin-induced Stat5 activation,173whereas reduced Tyk2/SHP-1 interaction has been observed in a kindred of familial hemophagocytic lymphohistiocytosis.174 However, additional experiments will be required to identify mechanisms by which these perturbations in the Jak-Stat pathway may contribute to the pathogenesis of disease.
Future directions
It is clear from this review that the Jak-Stat pathway is perturbed in a variety of malignancies and hematopoietic disorders. There is also now solid evidence that constitutive activation of Jak-Stat pathway components plays an important role in transformation by Tel-Jak, Bcr-Abl, and v-Src and in multiple myeloma. Furthermore, the importance of defective Jak3 activation in SCID and of extended Stat5 activation in the hyperproliferative responses of truncated G-CSF-R is now established. However, the significance of altered Jak-Stat activation in the other disorders remains less clear. In each case, the judicious expression of dominant-negative or constitutively active Jak-Stat pathway components in either cell line or mouse models of these disorders should enable the relative contribution of altered Jak-Stat signaling to the disease phenotype to be established. In addition, the availability of numerous mouse strains either deficient or transgenic for specific Jak-Stat components provides additional opportunity for assessing their importance in vivo.
Conclusions
Much remains to be learned about structure/function relationships of Jaks and Stats and about the cross talk between the Jak-Stat pathway and other signaling pathways in hematopoietic cells. In addition, as outlined above, the definitive roles played by Stats in growth control and transformation must be determined. Furthermore, a complete understanding of the mechanisms by which the Jak-Stat pathway is negatively regulated remains an important goal. However, there is already much promise in applying the knowledge obtained on the Jak-Stat pathway and its perturbation for the development of innovative hematologic treatment strategies. As mentioned above, specific Jak inhibitors may have important clinical applications in acute lymphoblastic leukemia,128 and there is clear potential for using gene therapy to remedy Jak3 deficiency in SCID.175,176 Recent data providing the crystallographic structure of Stat1 and Stat3 complexed with DNA177 178opens the way for a finer understanding of Stat specificity at the molecular level, increasing the knowledge base required for designing suitable compounds for pharmacologic intervention. It is anticipated that future developments will further facilitate the translation of the basic science of the Jak-Stat pathway into the hematology clinic.
Acknowledgment
The authors apologize to colleagues whose works were not cited because of the size restrictions of the review.
Supported by an EMBO Long Term Fellowship and the NWO (A.C.W.) and by grants from the Ministry of Science, Education and Culture of Japan, the TORAY Research Foundation, and the Uehara Memorial Research Foundation (A.Y.).
Reprints:Alister C. Ward, Institute of Hematology, Erasmus University Rotterdam, P. O. Box 1738, 3000 DR Rotterdam, Netherlands; e-mail: ward@hema.fgg.eur.nl.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal