Abstract
The Wilms tumor (WT1) gene has been reported to be preferentially expressed in acute leukemia cells, regardless of leukemia subtype and chronic myelogenous leukemia cells in blast crisis, but not in normal cells. This finding suggests strongly that WT1 protein is a potential target of immunotherapy for human leukemia. In this study, we established a CD8+ cytotoxic T-lymphocyte (CTL) clone directed against a WT1-derived peptide and examined its immunologic actions on leukemia cells. A CD8+ CTL clone, designated TAK-1, which lysed autologous cells loaded with a WT1-derived 9-mer peptide consisting of the HLA-A24 (HLA-A*2402)-binding motifs was established by stimulating CD8+ T lymphocytes from a healthy individual repeatedly with WT1 peptide-pulsed autologous dendritic cells. TAK-1 was cytotoxic to HLA-A24–positive leukemia cells expressing WT1, but not to HLA-A24–positive lymphoma cells that did not express WT1, HLA-A24–negative leukemia cells, or HLA-A24–positive normal cells. Treating leukemia cells with an antisense oligonucleotide complementary to the WT1 gene resulted in reduced TAK-1-mediated cytotoxicity, suggesting that target antigen of TAK-1 on leukemia cells is the naturally processed WT1 peptide in the context of HLA-A24. TAK-1 did not inhibit colony formation by normal bone marrow cells of HLA-A24–positive individuals. Because WT1 is overexpressed ubiquitously in various types of leukemia cells, but not in normal cells, immunotherapy using WT1 peptide-specific CTL clones should be an efficacious treatment for human leukemia. (Blood. 2000;95:286-293)
Cytotoxic T lymphocytes (CTLs) undoubtedly play an important role in resistance to cancers, including various types of leukemia, and adoptive transfer of CTLs, which discriminate malignant and normal cells, should be an efficacious treatment for human leukemia.1 To develop effective immunotherapy, the specific tumor antigens that are recognized by the immune system must be identified. So far, various molecules have been proposed as candidates for the targets of leukemia-specific CTLs. Among them, fusion proteins resulting from chromosomal translocations are potential targets of immune responses, as fusion proteins are produced only by leukemia cells, not by normal cells. On the basis of this concept, generation of fusion protein–specific T lymphocytes has been attempted, and several investigators have succeeded in establishing T-lymphocyte clones and bulk cell lines that specifically recognize the synthetic peptide spanning the fusion point between 2 proteins, such as BCR-ABL in chronic myelogenous leukemia (CML),2-10 PML-RARα in acute promyelocytic leukemia,11 ETV6-AML1 in pre-B acute lymphoblastic leukemia (ALL),12 and DEK-CAN in acute myelogenous leukemia (AML).13
Another strategy for inducing tumor-specific T-lymphocyte responses is the use of synthetic peptides derived from normal proteins that are preferentially expressed or overexpressed in tumor cells. Although various proteins, such as tyrosinase, Pmel/gp100, Melan-A/Mart-1, HER-2/neu, p53, and PRAME, have been identified as the targets of melanoma- and solid tumor–specific CTLs, only proteinase 3 has been reported to be a potential target antigen of CTLs directed against human leukemia cells.14 In this study, we selected Wilms tumor protein (WT1) as a target of CTLs for the development of effective immunotherapy for leukemia, because WT1 recently was found to be overexpressed in a variety of leukemia cells but not in normal cells.15-17
The WT1 gene encodes a zinc finger transcription factor,18 and WT1 binds the early growth response-1 DNA consensus sequence present in growth factor gene promoters, such as platelet-derived growth factor A chain, colony-stimulating factor-1, transforming growth factor-β1, insulin-like growth factor II, and its own gene promoters.19 Although WT1 was initially shown to act as a transcriptional repressor, its specific functions in normal and neoplastic tissues remain to be elucidated. During normal ontogenesis, the WT1 gene is expressed in a time- and tissue-dependent manner mainly in the fetal kidney, testis, ovary, and supportive structures of mesodermal origin.20,21 Analysis of mice with homozygous deletion of the WT1 gene demonstrated the crucial role of this gene in early urogenital development.22 In adults, WT1 gene expression is limited to very few tissues, including the splenic capsule and stroma, the Sertoli cells of the testis, and the granulosa cells of the ovary23,24; the existence of WT1 expression in bone marrow hematopoietic CD34+ precursor cells is controversial.17,25 26
Recently, it was reported that most human leukemia cells aberrantly overexpress WT1 regardless of the leukemia subtype, and WT1 expression was proposed to be a leukemia cell marker useful for the diagnosis of minimal residual disease.16,27 28 The evidence that WT1 is preferentially expressed in a variety of human leukemia cells, but not in normal cells, suggests strongly that it may be possible to develop efficacious immunotherapy for patients with leukemia by targeting this protein. Therefore, our aim was to generate WT1-specific CTLs and examine their immunologic actions on leukemia cells. In this study, we succeeded in establishing a CD8+ CTL clone that recognized a 9-mer peptide derived from WT1 in the context of HLA-A24, the most common HLA class I type in all people of Japanese descent (over 60% are positive). The WT1 peptide-specific CTL clone efficiently lysed HLA-matched leukemia cells but not HLA-mismatched leukemia cells, HLA-matched lymphoma cells that did not express WT1, or HLA-matched normal cells. Furthermore, this CTL clone did not inhibit colony formation by HLA-matched normal bone marrow cells. In light of these findings, the possibility of developing immunotherapy for leukemia using WT1-specific CTLs is discussed.
Materials and methods
Cell lines
B-lymphoblastoid cell lines (B-LCLs) were established by transformation of peripheral blood B lymphocytes using Epstein-Barr virus. LCLs and C1R cell lines were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS). HLA-A*2402gene–transfected C1R cell line (C1R-A*2402)29 was cultured in RPMI 1640 medium supplemented with 10% FCS and 500 μg/mL hygromycin B (Sigma, St. Louis, MO). Primary human foreskin fibroblasts were prepared from punch skin biopsy specimens and cultured in Dulbecco's modified Eagle medium supplemented with 10% FCS.
Synthetic peptide
Because HLA-A24 (HLA-A*2402) is the most common HLA class I type in the Japanese population, 9-mer peptide sequences of WT1 consisting of binding motifs for HLA-A*2402 were designed. The peptide sequences of the 4 synthetic peptides, designated WT1-T1, T2, T3, and T4, used are as follows (the underlined amino acids indicate the binding motifs for HLA-A*2402): WT1-T1, QMTSQLECM (residues 228-236); WT1-T2, CMTWNQMNL (residues 235-243); WT1-T3, DFKDCERRF (residues 356-364); and WT1-T4, RWPSCQKKF (residues 417-425). Peptides derived from MAGE-3, IMPKAGLLI (residues 195-203)30 and human immunodeficiency virus-1 (HIV-1) gp41, RYLKDQQLL (residues 583-591)31 were also synthesized and used as positive controls, because they bind to HLA-A24 molecules and elicit the generation of peptide-specific and HLA-A24–restricted CTLs. The peptides were synthesized to a minimum purity of 90% using an automated peptide synthesizer (Model 432A Synergy; Applied Biosystems, Foster City, CA) with the 9-fluorenylmethoxycarbonyl (Fmoc) procedure.
HLA typing
HLA serotyping was performed using a microlymphocyte cytotoxicity test with local qualified antisera. According to the serologic typing results, HLA class I alleles were amplified by the polymerase chain reaction (PCR) using group-specific primers and were then typed at the nucleotide sequence level. In brief, for example, HLA-A9 group alleles, including HLA-A*2402, were amplified using HLA-A9–specific primers and then analyzed using 9 probes that can be used to distinguish 6 alleles (A*2301, A*2402-A*2406), as described previously.32 HLA-A24 expression on some leukemia and lymphoma cells was examined by flow cytometry using a fluorescein isothiocyanate (FITC)-conjugated anti-HLA-A24 monoclonal antibody (MoAb) (One Lambda, Canoga Park, CA) and FITC-conjugated mouse immunoglobulin (Ig) G as the control. HLA-A24 nucleotide sequencing was performed using the dideoxy-chain termination method, as described previously.33
Generation of WT1 peptide-specific CTL clones
Peripheral blood dendritic cells (DCs) were generated as follows. Monocyte-enriched peripheral blood mononuclear cell (PBMC) fractions were isolated, using a plastic adherence technique, from total PBMCs of HLA-A24–positive healthy individuals. The plastic-adherent cells were cultured further in RPMI 1640 medium supplemented with 10% FCS, 500 U/mL recombinant human interleukin (IL)-4 (Genzyme, Boston, MA), and 800 U/mL recombinant human granulocyte-macrophage colony-stimulating factor (GM-CSF) (Kirin Brewery, Tokyo, Japan). On day 3 of incubation, half of the medium was exchanged for fresh culture medium supplemented with IL-4 and GM-CSF, and culture was continued. On day 5, half of the medium was exchanged for culture medium supplemented with IL-4, GM-CSF, and 100 U/mL recombinant human tumor necrosis factor (TNF)-α (Dainippon Pharmaceutical, Osaka, Japan). On day 8 or 9, the cells were harvested and used as monocyte-derived DCs for antigen stimulation. The generated cells appeared to express DC-associated antigens, such as CD1a, CD80, CD83, CD86, and HLA class I and class II, on their cell surfaces (data not shown). CD8+ T lymphocytes were isolated, using magnetizable polystyrene beads coated with an anti-CD8 MoAb (DYNAL, Oslo, Norway), from the same donors. A total of 1 million CD8+ T lymphocytes were cultured with 1 × 105 autologous DCs treated with mitomycin C (MMC) (Kyowa Hakko, Tokyo, Japan) in RPMI 1640 medium supplemented with 10% heat-inactivated human AB type serum and 5 ng/mL recombinant human IL-7 (Genzyme) together with a WT1 synthetic peptide at a concentration of 10 μg/mL in a 16-mm well. After culture for 7 days, half of the medium was exchanged for fresh culture medium supplemented with IL-7, and the cells were stimulated again by adding 1 × 105 autologous DCs treated with MMC and WT1 peptide at a concentration of 10 μg/mL. After culture for a further 7 days, the cells were stimulated a third time, as they were for the second except for the addition of IL-7. After culture for a further 4 days (day 18 of culture), 10 U/mL recombinant human IL-2 (Boehringer Mannheim, Mannheim, Germany) was added to each well. The cytotoxicity of the growing cells was then examined, and the bulk of the cells that were cytotoxic to WT1 peptide-loaded autologous LCL was cloned by a limiting dilution method, as described previously.34 T-lymphocyte clones were cultured continuously in IL-2–containing culture medium, and MMC-treated autologous PBMCs and the required WT1 peptide were added to the wells every 2 weeks.
Cytotoxicity assays
Chromium-51 release assays were performed as described previously.35 Briefly, 1 × 10451Cr (Na251CrO4) (New England Nuclear, Boston, MA)-labeled target cells suspended in 100 μL RPMI 1640 medium supplemented with 10% FCS (assay medium) were seeded into round-bottomed microtiter wells and incubated with or without synthetic peptide for 2 hours. In some experiments, the target cells were preincubated with an anti-HLA class I MoAb, w6/32 (ATCC, Rockville, MD), at an optimal concentration (10 μg/mL) for 30 minutes to determine whether cytotoxicity was restricted by HLA class I. Various numbers of effector cells suspended in 100 μL assay medium were added to the well, incubated for 4 hours, and 100 μL supernatant was collected from each well. The percentage of specific lysis was calculated as follows: (cpm experimental release − cpm spontaneous release)/(cpm maximal release − cpm spontaneous release) × 100.
Northern blot analysis
Total RNAs were extracted from leukemia and lymphoma cell lines and freshly isolated leukemia cells; 20 μg of total RNAs were separated by electrophoresis on a 1% agarose gel containing 2.2 mol/L formaldehyde and transferred to nylon filters, which were hybridized with 32P ([32P]dCTP;) ICN Radiochemicals, Irvine, CA)-labeled WT1 or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probes. The WT1 probe used was the 1.4-kb DNA fragment derived from K562 cell line by the reverse transcription-PCR using 5′-CCGAATTCAGGATGTGCGACGTGTGCCT-3′ and 5′-CCGAATTCACCTGTATGAGTCCTGGTGTG-3′ primers. The extent of hybridization was quantified by scanning the autoradiographs using a Bio-Imaging Analyzer system (BAS1000; Fujifilm, Tokyo, Japan).
Treatment of leukemia cells with a WT1 antisense oligonucleotide
The 18-base antisense oligonucleotide phosphorothioate complementary to the sequence of the translation initiation site of the WT1gene was synthesized and purified by high-performance liquid chromatography, as described previously.36 Sense and random sequences of the same 18-base oligonucleotide were also synthesized and used as controls. The oligonucleotide sequences were as follows: sense, 5′-CAGCAAATGGGCTCCGAC-3′; antisense, 5′-GTCGGAGCCCATTTGCTG-3′. Random oligonucleotides were prepared by mixing 4 different 18-base oligonucleotides, heating the mixture at 80°C for 5 minutes, and allowing it to cool slowly to 37°C over 5 hours. The cells were suspended in FCS-free RPMI 1640 medium and placed in a 24-well culture plate. The oligonucleotides were added to the medium at a concentration of 20 μM and incubated at 37°C. After 2 hours, FCS was added to the culture medium at a final concentration of 10%. The same oligonucleotides were added to each well every 24 hours at half the dose of the initial concentration. Three days later, the cells were collected, and their expression levels of WT1 protein were determined by Western blot analysis. These oligonucleotide-treated cells were used as targets in the cytotoxicity assays.
Western blot analysis
Western blot analysis was performed according to the ECL protocol (Amersham Pharmacia Biotech, Little Chalfont, England, UK). The cells were washed twice with phosphate-buffered saline and then lysed with buffer containing 1% Triton X-100, 100 mg/mL phenylmethylsulfonyl fluoride, 2 mg/mL leupeptin, and 2 mg/mL aprotinin as protease inhibitors. Equal amounts (30 μg) of protein lysate were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and blotted onto Hybond ECL membranes (Amersham Pharmacia Biotech) using the Trans-Biot electrophoretic transfer system (Bio-Rad, Hercules, CA). The protein blots were incubated with a rabbit anti-human WT1 polyclonal antibody (Ab) (WT 180; Santa Cruz Biotechnology, Santa Cruz, CA) or a rabbit anti-human GAPDH polyclonal Ab (Trevigen, Gaithersburg, MD) for 1 hour at room temperature. After washing, each filter was incubated with horseradish peroxidase-labeled goat anti-rabbit IgG and subjected to the enhanced chemiluminescence assay using the ECL detection system (Amersham Pharmacia Biotech).
Colony-forming assay
The effect of the T-lymphocyte clone on the growth of normal bone marrow cells was examined by performing the colony formation assay described previously8 with a slight modification. Bone marrow cells were isolated from patients with malignant lymphoma without bone marrow invasion and those with nonhematologic disorders undergoing bone marrow examination, after obtaining their informed consent, and cryopreserved until required for use. Cloned T lymphocytes and bone marrow cells, which had been incubated in the presence or absence of WT1 peptide at a concentration of 10 μg/mL for 2 hours were suspended in assay medium at an effector cell to target cell ratio of 3:1, centrifuged at 1000 revolutions per minute for 3 minutes to ensure close cell contact, and then coincubated in assay medium at 37°C for 4 hours. Control bone marrow cells were centrifuged and incubated without the T-lymphocyte clone in the same manner. After incubation, 5 mL of Iscove's modified Dulbecco's medium containing 1% methylcellulose, 5% GCT-conditioned medium, 1% bovine serum albumin, 30% FCS, 100 μM 2-mercaptoethanol, and 3 U/mL erythropoietin (Stem Cell CFU Kit; Baxter, Deerfield, IL) was added to the cell pellet at the final bone marrow cell concentration of 1 × 105 cells/mL. Each cell suspension was then placed in triplicate 24-mm wells and cultured at 37°C for 12 to 14 days, after which, the numbers of colony-forming unit granulocyte-macrophage (CFU-GM) and burst-forming unit erythroid (BFU-E) were counted using an inverted microscope. The significance of differences between values for 2 groups was determined using the paired 2-tailed Student t test, and those atP < .05 were considered significant.
Results
Generation of a WT1 peptide-specific CTL clone
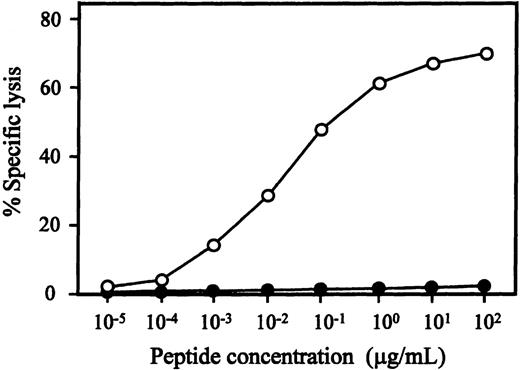
We attempted to generate WT1-specific CTLs from 3 healthy HLA-A24–positive individuals using WT1 peptide-pulsed DCs, as described above, and established 1 CTL clone, designated TAK-1, which lysed a WT1 peptide-loaded, but not unloaded, autologous LCL. Flow cytometric analysis demonstrated that more than 99% of TAK-1 cells were CD3+, CD4-, CD8+, and CD56-. The antigen specificity and HLA restriction of TAK-1–mediated cytotoxicity are summarized in Table1. TAK-1 lysed autologous LCL that was loaded with the WT1-T2 peptide, but was not cytotoxic to unloaded, WT1-T1–, WT1-T3–, or WT1-T4–loaded autologous LCL. TAK-1 also did not lyse autologous LCL loaded with an MAGE-3–derived or an HIV-1 gp41–derived peptide consisting of HLA-A24–high binding affinity. The restriction element of TAK-1 seemed to be HLA-A24, because only HLA-A24–positive allogeneic LCLs were lysed by TAK-1 and TAK-1-mediated cytotoxicity was inhibited by adding the anti-HLA class I MoAb to target cells (data not shown). To confirm HLA-A24 restriction of TAK-1, we examined the cytotoxic activity of TAK-1 to HLA-A*2402 transfectant cell line C1R-A*2402. In the presence of WT1-T2 peptide, TAK-1 was cytotoxic to C1R-A*2402 but not to its parent cell line, C1R. These data show that TAK-1–mediated cytotoxicity was WT1-T2 peptide-specific and restricted by HLA-A24. The peptide titration assay demonstrated that WT1-T2 peptide-specific cytotoxicity was detectable with final peptide concentrations of 1 ng/mL to 100 μg/mL, and the optimal peptide concentration range for TAK-1–mediated cytotoxicity was 1 to 10 μg/mL (Figure1).
HLA-A24-restricted and WT1-T2 peptide-specific cytotoxicity of TAK-1
| Target Cells . | HLA Class I Alleles . | Peptide . | % Specific Lysis* . | ||
|---|---|---|---|---|---|
| 10:1 . | 5:1 . | 2.5:1 . | |||
| Autologous LCL | A24/26, B62/−, Cw4/w9 | None | 1.0 | 0.9 | 1.0 |
| WT1-T2 | 72.6 | 68.4 | 58.8 | ||
| WT1-T1 | 1.2 | 1.4 | 1.1 | ||
| WT1-T3 | 0.9 | 0.6 | 0.5 | ||
| WT1-T4 | 2.1 | 0.3 | 0.8 | ||
| MAGE-3 | 1.1 | 0.3 | 0.7 | ||
| HIV-1 gp41 | 1.2 | 2.4 | 2.6 | ||
| Allogeneic LCL #1 | A24/2, B52/55, Cw1/− | None | 1.7 | 0.5 | 0.2 |
| WT1-T2 | 69.4 | 60.8 | 54.7 | ||
| Allogeneic LCL #2 | A24/−, B7/52, Cw7/− | None | 2.1 | 0.8 | 1.9 |
| WT1-T2 | 70.4 | 62.2 | 55.7 | ||
| Allogeneic LCL #3 | A24/−, B48/−, Cw8/− | None | 0.9 | 0.4 | 0.3 |
| WT1-T2 | 66.6 | 61.2 | 53.9 | ||
| Allogeneic LCL #4 | A24/33, B54/62, Cw1/w9 | None | 0.2 | 0 | −0.2 |
| WT1-T2 | 59.6 | 54.4 | 49.0 | ||
| Allogeneic LCL #5 | A24/−, B52/54, Cw1/− | None | 2.3 | 0.4 | 0.2 |
| WT1-T2 | 62.7 | 61.4 | 52.7 | ||
| Allogeneic LCL #6 | A26/31, B61/62, Cw3/− | None | 0.4 | 0.7 | 0.4 |
| WT1-T2 | 0.7 | 1.5 | 0.9 | ||
| Allogeneic LCL #7 | A2/33, B44/51, Cw−/− | None | 0.3 | 0.9 | 0.9 |
| WT1-T2 | 1.0 | 0.4 | 0.1 | ||
| Allogeneic LCL #8 | A2/26, B35/62, Cw3/− | None | 0.7 | 1.0 | 0.4 |
| WT1-T2 | 2.2 | 0.8 | 0.7 | ||
| Allogeneic LCL #9 | A2/11, B7/35, Cw3/w7 | None | 0.9 | 0.1 | 0.6 |
| WT1-T2 | 0.9 | 0.9 | 0.4 | ||
| Allogeneic LCL #10 | A1/26, B27/60, Cw−/− | None | 1.2 | 1.1 | 1.1 |
| WT1-T2 | 2.1 | 1.0 | 1.6 | ||
| C1R | Negative | None | 0.8 | 0 | 1.4 |
| WT1-T2 | 1.6 | 1.0 | 1.4 | ||
| C1R-A*2402 | A24 | None | 1.3 | 1.5 | 1.0 |
| WT1-T2 | 82.3 | 71.3 | 66.3 | ||
| Target Cells . | HLA Class I Alleles . | Peptide . | % Specific Lysis* . | ||
|---|---|---|---|---|---|
| 10:1 . | 5:1 . | 2.5:1 . | |||
| Autologous LCL | A24/26, B62/−, Cw4/w9 | None | 1.0 | 0.9 | 1.0 |
| WT1-T2 | 72.6 | 68.4 | 58.8 | ||
| WT1-T1 | 1.2 | 1.4 | 1.1 | ||
| WT1-T3 | 0.9 | 0.6 | 0.5 | ||
| WT1-T4 | 2.1 | 0.3 | 0.8 | ||
| MAGE-3 | 1.1 | 0.3 | 0.7 | ||
| HIV-1 gp41 | 1.2 | 2.4 | 2.6 | ||
| Allogeneic LCL #1 | A24/2, B52/55, Cw1/− | None | 1.7 | 0.5 | 0.2 |
| WT1-T2 | 69.4 | 60.8 | 54.7 | ||
| Allogeneic LCL #2 | A24/−, B7/52, Cw7/− | None | 2.1 | 0.8 | 1.9 |
| WT1-T2 | 70.4 | 62.2 | 55.7 | ||
| Allogeneic LCL #3 | A24/−, B48/−, Cw8/− | None | 0.9 | 0.4 | 0.3 |
| WT1-T2 | 66.6 | 61.2 | 53.9 | ||
| Allogeneic LCL #4 | A24/33, B54/62, Cw1/w9 | None | 0.2 | 0 | −0.2 |
| WT1-T2 | 59.6 | 54.4 | 49.0 | ||
| Allogeneic LCL #5 | A24/−, B52/54, Cw1/− | None | 2.3 | 0.4 | 0.2 |
| WT1-T2 | 62.7 | 61.4 | 52.7 | ||
| Allogeneic LCL #6 | A26/31, B61/62, Cw3/− | None | 0.4 | 0.7 | 0.4 |
| WT1-T2 | 0.7 | 1.5 | 0.9 | ||
| Allogeneic LCL #7 | A2/33, B44/51, Cw−/− | None | 0.3 | 0.9 | 0.9 |
| WT1-T2 | 1.0 | 0.4 | 0.1 | ||
| Allogeneic LCL #8 | A2/26, B35/62, Cw3/− | None | 0.7 | 1.0 | 0.4 |
| WT1-T2 | 2.2 | 0.8 | 0.7 | ||
| Allogeneic LCL #9 | A2/11, B7/35, Cw3/w7 | None | 0.9 | 0.1 | 0.6 |
| WT1-T2 | 0.9 | 0.9 | 0.4 | ||
| Allogeneic LCL #10 | A1/26, B27/60, Cw−/− | None | 1.2 | 1.1 | 1.1 |
| WT1-T2 | 2.1 | 1.0 | 1.6 | ||
| C1R | Negative | None | 0.8 | 0 | 1.4 |
| WT1-T2 | 1.6 | 1.0 | 1.4 | ||
| C1R-A*2402 | A24 | None | 1.3 | 1.5 | 1.0 |
| WT1-T2 | 82.3 | 71.3 | 66.3 | ||
The cytotoxicity of TAK-1 to various B-LCL cells loaded or not loaded with the WT1 or control peptide was determined by 4-hour51Cr release assays at E:T ratios of 10:1, 5:1, and 2.5:1.
WT1 peptide concentration-dependent cytotoxicity of TAK-1.
The cytotoxicity of TAK-1 to autologous (open circles) and HLA-A24–negative allogeneic (closed circles) LCLs preincubated with various concentrations of WT1-T2 peptide for 1 hour was determined by 4-hour 51Cr release assays at an E:T ratio of 5:1.
WT1 peptide concentration-dependent cytotoxicity of TAK-1.
The cytotoxicity of TAK-1 to autologous (open circles) and HLA-A24–negative allogeneic (closed circles) LCLs preincubated with various concentrations of WT1-T2 peptide for 1 hour was determined by 4-hour 51Cr release assays at an E:T ratio of 5:1.
Expression of WT1 mRNA in leukemia and lymphoma cell lines
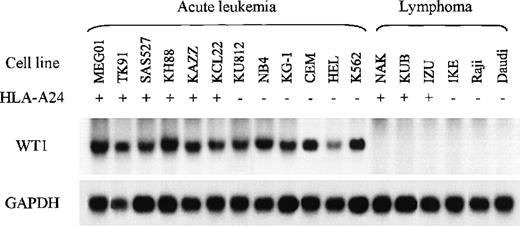
Previous studies demonstrated that WT1 is expressed in most acute leukemia cells and CML cells at blast crisis regardless of the leukemia subtype. First, to determine whether TAK-1 lysed leukemia cell lines in a WT1-specific manner, we carried out Northern blot analysis to evaluate the WT1 messenger RNA (mRNA) expression levels of various leukemia and lymphoma cell lines. Representative data are shown in Figure 2. All the cell lines established from AML, ALL, and CML at blast crisis examined appeared to express abundant WT1 mRNA. In contrast, all the lymphoma cell lines examined expressed no WT1 mRNA. These data affirm those reported previously.
Northern blot analysis of WT1 mRNAs in various leukemia and lymphoma cell lines.
Samples of total cellular RNA were hybridized with a32P-labeled WT1 cDNA probe (top), and each sample blot was also hybridized with a GAPDH cDNA probe (bottom).
Northern blot analysis of WT1 mRNAs in various leukemia and lymphoma cell lines.
Samples of total cellular RNA were hybridized with a32P-labeled WT1 cDNA probe (top), and each sample blot was also hybridized with a GAPDH cDNA probe (bottom).
Lysis of leukemia cell lines by TAK-1
We next examined whether TAK-1 was cytotoxic to leukemia cell lines that expressed abundant WT1. As shown in Table2, TAK-1 showed cytotoxicity to HLA-A24–positive leukemia cell lines despite the absence of added WT1-T2 peptide, whereas no cytotoxicity to HLA-A24–negative leukemia cell lines was detected. Furthermore, TAK-1 appeared to have no cytotoxic activity to lymphoma cell lines, which did not express WT1, regardless of their HLA-A24 expression status. These data suggest strongly that WT1 peptide is processed naturally in leukemia cells, expressed in the context of HLA-A24, and recognized by CD8+ CTLs.
Cytotoxicity of TAK-1 to leukemia and lymphoma cell lines
| Target Cells . | HLA-A24 . | WT1/ GAPDH* . | % Specific Lysis† . | ||
|---|---|---|---|---|---|
| 10:1 . | 5:1 . | 2.5:1 . | |||
| Leukemia cell lines | |||||
| TK91 | + | 0.31 | 46.2 | 32.8 | 24.6 |
| MEG01 | + | 0.45 | 52.6 | 40.1 | 28.7 |
| SAS527 | + | 0.18 | 39.2 | 22.4 | 14.1 |
| KH88 | + | 0.57 | 40.5 | 30.8 | 18.3 |
| KAZZ | + | 0.31 | 41.0 | 29.4 | 15.8 |
| KCL-22 | + | 0.33 | 33.0 | 22.0 | 12.4 |
| OUN-1 | + | 0.40 | 47.4 | 34.1 | 20.1 |
| CMK11-5 | + | 0.23 | 36.4 | 27.8 | 18.7 |
| KU812 | − | 0.29 | 0.7 | 0.8 | 0.2 |
| J-111 | − | 0.26 | 0.1 | 0.5 | 0.9 |
| NB4 | − | 0.44 | 2.4 | 2.2 | 2.7 |
| HEL | − | 0.13 | 0.2 | 0.1 | 0 |
| KG-1 | − | 0.40 | 3.1 | 1.6 | 1.9 |
| GANMO-1 | − | 0.21 | 0.2 | 0.1 | 0.7 |
| KT-1 | − | 0.57 | 0.3 | 0.4 | 0.2 |
| MZ93 | − | 0.15 | 1.7 | 1.4 | 1.7 |
| YS-1 | − | 0.34 | 2.4 | 0.7 | 0.4 |
| K562 | − | 0.42 | 0.1 | 0.9 | 0.3 |
| CEM | − | 0.49 | 0.4 | 0.6 | 0.1 |
| Jurkat | − | 0.04 | 1.3 | 1.3 | 1.3 |
| Lymphoma cell lines | |||||
| NAK | + | <0.01 | 1.0 | 1.4 | 1.1 |
| NIS | + | <0.01 | 0.3 | 0.1 | 0.4 |
| KUB | + | <0.01 | 0.2 | 1.0 | 0.7 |
| IZU | + | <0.01 | 2.0 | 1.8 | 1.4 |
| KAW | + | <0.01 | 1.6 | 0.8 | 0.4 |
| IKE | − | <0.01 | 1.0 | 0.4 | 1.1 |
| TKC | − | <0.01 | 0.3 | 0.5 | 1.0 |
| Raji | − | <0.01 | 0.4 | 0.3 | 0.3 |
| Daudi | − | <0.01 | 0 | 0.3 | 1.0 |
| Target Cells . | HLA-A24 . | WT1/ GAPDH* . | % Specific Lysis† . | ||
|---|---|---|---|---|---|
| 10:1 . | 5:1 . | 2.5:1 . | |||
| Leukemia cell lines | |||||
| TK91 | + | 0.31 | 46.2 | 32.8 | 24.6 |
| MEG01 | + | 0.45 | 52.6 | 40.1 | 28.7 |
| SAS527 | + | 0.18 | 39.2 | 22.4 | 14.1 |
| KH88 | + | 0.57 | 40.5 | 30.8 | 18.3 |
| KAZZ | + | 0.31 | 41.0 | 29.4 | 15.8 |
| KCL-22 | + | 0.33 | 33.0 | 22.0 | 12.4 |
| OUN-1 | + | 0.40 | 47.4 | 34.1 | 20.1 |
| CMK11-5 | + | 0.23 | 36.4 | 27.8 | 18.7 |
| KU812 | − | 0.29 | 0.7 | 0.8 | 0.2 |
| J-111 | − | 0.26 | 0.1 | 0.5 | 0.9 |
| NB4 | − | 0.44 | 2.4 | 2.2 | 2.7 |
| HEL | − | 0.13 | 0.2 | 0.1 | 0 |
| KG-1 | − | 0.40 | 3.1 | 1.6 | 1.9 |
| GANMO-1 | − | 0.21 | 0.2 | 0.1 | 0.7 |
| KT-1 | − | 0.57 | 0.3 | 0.4 | 0.2 |
| MZ93 | − | 0.15 | 1.7 | 1.4 | 1.7 |
| YS-1 | − | 0.34 | 2.4 | 0.7 | 0.4 |
| K562 | − | 0.42 | 0.1 | 0.9 | 0.3 |
| CEM | − | 0.49 | 0.4 | 0.6 | 0.1 |
| Jurkat | − | 0.04 | 1.3 | 1.3 | 1.3 |
| Lymphoma cell lines | |||||
| NAK | + | <0.01 | 1.0 | 1.4 | 1.1 |
| NIS | + | <0.01 | 0.3 | 0.1 | 0.4 |
| KUB | + | <0.01 | 0.2 | 1.0 | 0.7 |
| IZU | + | <0.01 | 2.0 | 1.8 | 1.4 |
| KAW | + | <0.01 | 1.6 | 0.8 | 0.4 |
| IKE | − | <0.01 | 1.0 | 0.4 | 1.1 |
| TKC | − | <0.01 | 0.3 | 0.5 | 1.0 |
| Raji | − | <0.01 | 0.4 | 0.3 | 0.3 |
| Daudi | − | <0.01 | 0 | 0.3 | 1.0 |
The WT1 mRNA expression levels are expressed as WT1 mRNA/GAPDH mRNA intensity ratios quantified by densitometric analysis.
The cytotoxicity of TAK-1 to various leukemia and lymphoma cell lines in the absence of the WT1-peptide was determined by 4-hour51Cr release assays at E:T ratios of 10:1, 5:1, and 2.5:1.
Lysis of freshly isolated leukemia cells by TAK-1
The cytotoxicity of TAK-1 to various kinds of leukemia cells, which were freshly isolated from patients, is shown in Table3. As observed with the cell lines, TAK-1 was cytotoxic to leukemia cells isolated from HLA-A24–positive patients with AML and ALL, except for some cases in which WT1 may not have been processed efficiently enough to be presented to the T lymphocytes and/or certain molecules important to CTL-mediated cytotoxicity may not have been expressed. Leukemia cells isolated from HLA-A24–negative patients were not lysed by TAK-1, which was not cytotoxic to normal foreskin fibroblasts or PBMCs isolated from HLA-A24–positive or HLA-A24–negative donors. Therefore, TAK-1 appeared to discriminate freshly isolated leukemia cells, as well as leukemia cell lines, from normal cells.
Cytotoxicity of TAK-1 to freshly isolated leukemia cells
| Target Cells (donor) . | HLA-A24 . | WT1/GAPDH3-150 . | % Specific Lysis3-151 . | ||
|---|---|---|---|---|---|
| 10:1 . | 5:1 . | 2.5:1 . | |||
| Leukemia cells | |||||
| AML, M2 (S. M.) | + | 0.29 | 42.1 | 29.9 | 20.0 |
| AML, M2 (M. M.) | + | 0.23 | 29.3 | 19.9 | 11.1 |
| AML, M2 (I. K.) | + | 0.47 | 51.7 | 43.3 | 36.2 |
| AML, M4 (K. O.) | + | 0.22 | 39.7 | 24.7 | 18.8 |
| AML, M4 (T. F.) | + | 0.19 | 40.3 | 32.9 | 27.5 |
| AML, M5 (T. O.) | + | ND | 15.4 | 10.5 | 7.0 |
| AML, M5 (F. O.) | + | 0.15 | 0.9 | 0.9 | 0.1 |
| ALL, L2 (K. I.) | + | ND | 32.1 | 27.3 | 13.5 |
| ALL, L2 (M. H.) | + | 0.34 | 3.9 | 1.1 | 0.8 |
| AML, M2 (E. O.) | − | <0.01 | 0.1 | 0.8 | 0 |
| AML, M3 (S. N.) | − | ND | 2.9 | 0.5 | 1.2 |
| AML, M4 (K. N.) | − | 0.30 | 0.7 | 0.4 | 0.9 |
| AML, M5 (M. O.) | − | 0.29 | 1.8 | 1.0 | 0.5 |
| ALL, L1 (N. Y.) | − | 0.43 | 1.5 | 0.5 | 1.0 |
| ALL, L2 (T. M.) | − | 0.32 | 1.3 | 0.5 | 0.2 |
| ALL, L2 (Y. I.) | − | ND | 0.4 | 0.5 | 1.3 |
| Normal foreskin fibroblasts | |||||
| #1 (A. S.) | + | <0.01 | 0.8 | 0.6 | 0.2 |
| #2 (T. A.) | + | <0.01 | 1.6 | 0.9 | 1.1 |
| #3 (Y. N.) | + | <0.01 | 0.7 | 1.0 | 0.4 |
| #4 (N. G.) | − | <0.01 | 0.2 | 0.7 | 1.2 |
| #5 (M. Y.) | − | <0.01 | 0.8 | 0 | 1.9 |
| Normal PBMCs | |||||
| #1 (H. O.) | + | <0.01 | 0.6 | 0.5 | 0.1 |
| #2 (J. A.) | + | <0.01 | 1.4 | 0.2 | 0.8 |
| #3 (T. N.) | − | <0.01 | 0.1 | 0.5 | 0.7 |
| Target Cells (donor) . | HLA-A24 . | WT1/GAPDH3-150 . | % Specific Lysis3-151 . | ||
|---|---|---|---|---|---|
| 10:1 . | 5:1 . | 2.5:1 . | |||
| Leukemia cells | |||||
| AML, M2 (S. M.) | + | 0.29 | 42.1 | 29.9 | 20.0 |
| AML, M2 (M. M.) | + | 0.23 | 29.3 | 19.9 | 11.1 |
| AML, M2 (I. K.) | + | 0.47 | 51.7 | 43.3 | 36.2 |
| AML, M4 (K. O.) | + | 0.22 | 39.7 | 24.7 | 18.8 |
| AML, M4 (T. F.) | + | 0.19 | 40.3 | 32.9 | 27.5 |
| AML, M5 (T. O.) | + | ND | 15.4 | 10.5 | 7.0 |
| AML, M5 (F. O.) | + | 0.15 | 0.9 | 0.9 | 0.1 |
| ALL, L2 (K. I.) | + | ND | 32.1 | 27.3 | 13.5 |
| ALL, L2 (M. H.) | + | 0.34 | 3.9 | 1.1 | 0.8 |
| AML, M2 (E. O.) | − | <0.01 | 0.1 | 0.8 | 0 |
| AML, M3 (S. N.) | − | ND | 2.9 | 0.5 | 1.2 |
| AML, M4 (K. N.) | − | 0.30 | 0.7 | 0.4 | 0.9 |
| AML, M5 (M. O.) | − | 0.29 | 1.8 | 1.0 | 0.5 |
| ALL, L1 (N. Y.) | − | 0.43 | 1.5 | 0.5 | 1.0 |
| ALL, L2 (T. M.) | − | 0.32 | 1.3 | 0.5 | 0.2 |
| ALL, L2 (Y. I.) | − | ND | 0.4 | 0.5 | 1.3 |
| Normal foreskin fibroblasts | |||||
| #1 (A. S.) | + | <0.01 | 0.8 | 0.6 | 0.2 |
| #2 (T. A.) | + | <0.01 | 1.6 | 0.9 | 1.1 |
| #3 (Y. N.) | + | <0.01 | 0.7 | 1.0 | 0.4 |
| #4 (N. G.) | − | <0.01 | 0.2 | 0.7 | 1.2 |
| #5 (M. Y.) | − | <0.01 | 0.8 | 0 | 1.9 |
| Normal PBMCs | |||||
| #1 (H. O.) | + | <0.01 | 0.6 | 0.5 | 0.1 |
| #2 (J. A.) | + | <0.01 | 1.4 | 0.2 | 0.8 |
| #3 (T. N.) | − | <0.01 | 0.1 | 0.5 | 0.7 |
ND indicates not determined.
The WT1 mRNA expression levels are expressed as WT1 mRNA/GAPDH mRNA intensity ratios quantified by densitometric analysis.
The cytotoxicity of TAK-1 to various freshly isolated leukemia cells, normal foreskin fibroblasts, and normal PBMCs in the absence of the WT1-peptide was determined by 4-hour 51Cr release assays at E:T ratios of 10:1, 5:1, and 2.5:1.
Lack of cytotoxicity to an HLA-A24 mutant cell line
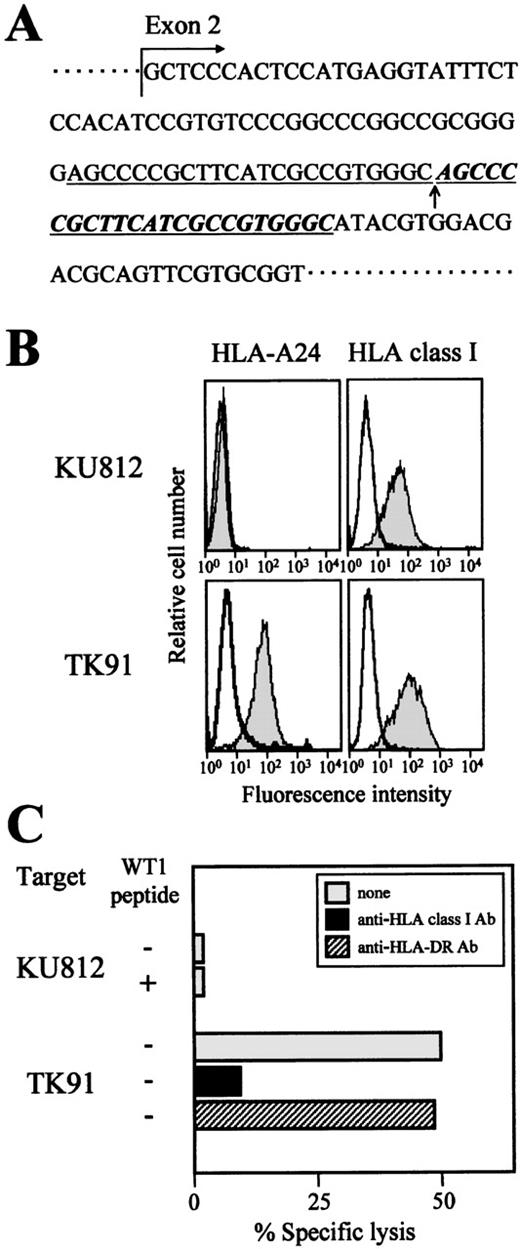
We addressed the question of whether the cytotoxicity of TAK-1 to leukemia cells was restricted by HLA-A24, as it was in the experiment on WT1 peptide-loaded target cells, by performing a conventional inhibition assay using an anti-HLA class I MoAb. As expected, the addition of an anti-HLA class I framework MoAb, w6/32, to the assay medium inhibited the cytotoxicity of TAK-1 to leukemia cell lines (Figure 3C). Furthermore, confirmation that the cytotoxicity of TAK-1 to leukemia cells was restricted by HLA-A24 was provided by the experiment using the HLA-A24 mutant cell line KU812.37 Sequence analysis of the HLA-A gene demonstrated that the 23-base repeated insertion occurred inHLA-A24 gene exon 2 (Figure 3A). This insertion resulted in a frameshift of the HLA-A24 gene sequence and the failure of HLA-A24 molecule expression (Figure 3B). As shown in Figure 3C, TAK-1 did not lyse KU812, even though KU812 expressed WT1, whereas TK91, an HLA-A24–positive myeloid leukemia cell line, was lysed by TAK-1. These data suggest strongly that the cytotoxocity of TAK-1 to leukemia cells is restricted by HLA-A24.
Lack of cytotoxicity of TAK-1 to the HLA-A24 mutant cell line KU812.
(A) The sequence of the HLA-A24 gene of the myeloid leukemia cell line KU812 was determined. The inserted 23-base pair repeated nucleotides are shown by underlined italics, and the insertion point is marked with an arrow. (B) The cell surface expression levels of HLA-A24 and HLA class I on the myeloid leukemia cell lines KU812 and TK91 were measured by flow cytometry. The cells were stained with an FITC-conjugated mouse anti-HLA-A24 MoAb, FITC-conjugated mouse anti-HLA class I MoAb (shaded histograms), or FITC-conjugated mouse IgG (open histograms). (C) The cytotoxicity of TAK-1 to KU812 and TK91 cells was determined by 51Cr release assays for 4 hours at an E:T ratio of 10:1 in the presence or absence of the WT1 peptide. The cytotoxicity of TAK-1 to TK91 cells preincubated with an anti-HLA class I MoAb or an anti-HLA-DR MoAb was also examined.
Lack of cytotoxicity of TAK-1 to the HLA-A24 mutant cell line KU812.
(A) The sequence of the HLA-A24 gene of the myeloid leukemia cell line KU812 was determined. The inserted 23-base pair repeated nucleotides are shown by underlined italics, and the insertion point is marked with an arrow. (B) The cell surface expression levels of HLA-A24 and HLA class I on the myeloid leukemia cell lines KU812 and TK91 were measured by flow cytometry. The cells were stained with an FITC-conjugated mouse anti-HLA-A24 MoAb, FITC-conjugated mouse anti-HLA class I MoAb (shaded histograms), or FITC-conjugated mouse IgG (open histograms). (C) The cytotoxicity of TAK-1 to KU812 and TK91 cells was determined by 51Cr release assays for 4 hours at an E:T ratio of 10:1 in the presence or absence of the WT1 peptide. The cytotoxicity of TAK-1 to TK91 cells preincubated with an anti-HLA class I MoAb or an anti-HLA-DR MoAb was also examined.
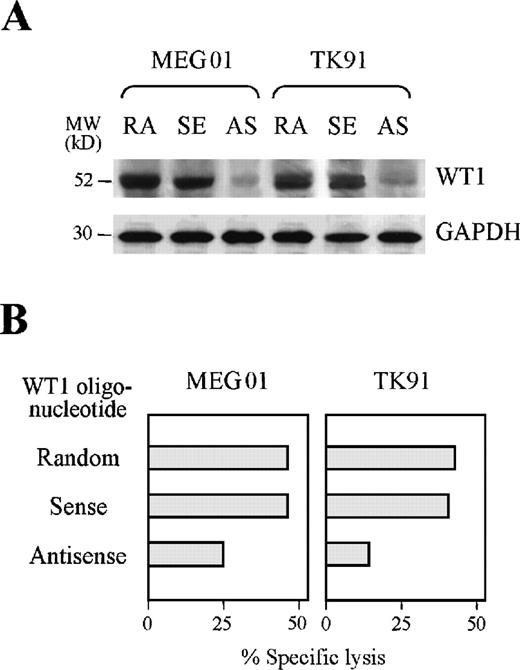
Reduction of TAK-1–mediated cytotoxicity to leukemia cells by treatment with a WT1 antisense oligonucleotide
We carried out further investigations to establish whether TAK-1 recognizes endogenously processed WT1 peptide using an antisense oligonucleotide complementary to WT1 gene. As shown in Figure4A, the WT1 protein expression levels of the leukemia cell lines TK91 and MEG01 were reduced considerably by treatment with an antisense, but not a random or sense oligonucleotide, complementary to the WT1 gene. Inhibition of WT1 synthesis by leukemia cells reduced the degrees of TAK-1–mediated cytotoxicity (Figure 4B). Treatment of leukemia cell lines with an antisense oligonucleotide complementary to the WT1 gene affected neither cell viability nor the expression of surface molecules important for CTL recognition, including HLA class I, ICAM-1, B7, and CD40 (data not shown). Therefore, these data indicate that TAK-1 lyses leukemia cells by recognizing the WT1 peptide in the context of the HLA-A24 molecule.
Reduction of TAK-1–mediated cytotoxicity by treating target cells with a WT1 antisense oligonucleotide.
(A) WT1 protein expression in the leukemia cell lines MEG01 and TK91 treated with random (RA), WT1 sense (SE), or WT1 antisense (AS) oligonucleotides was examined by Western blot analysis (top). GAPDH protein expression was also examined by analyzing the same blot (bottom). (B) The cytotoxicity of TAK-1 to leukemia cell lines pretreated with random, WT1 sense, or WT1 antisense oligonucleotides for 72 hours was determined by 4-hour 51Cr release assays at an E:T ratio of 10:1.
Reduction of TAK-1–mediated cytotoxicity by treating target cells with a WT1 antisense oligonucleotide.
(A) WT1 protein expression in the leukemia cell lines MEG01 and TK91 treated with random (RA), WT1 sense (SE), or WT1 antisense (AS) oligonucleotides was examined by Western blot analysis (top). GAPDH protein expression was also examined by analyzing the same blot (bottom). (B) The cytotoxicity of TAK-1 to leukemia cell lines pretreated with random, WT1 sense, or WT1 antisense oligonucleotides for 72 hours was determined by 4-hour 51Cr release assays at an E:T ratio of 10:1.
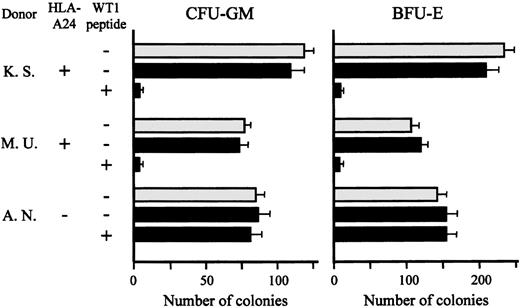
Lack of effect of TAK-1 on colony formation by normal bone marrow cells
Recently, WT1 was reported to be expressed in normal bone marrow CD34+ hematopoietic progenitors, but this is still controversial.17,25 26 Therefore, we addressed the issue of whether TAK-1 recognizes WT1 peptide expressed on normal bone marrow progenitor cells and suppresses their growth. As shown in Figure5, after coculture with TAK-1 in the absence of WT1 peptide, the numbers of CFU-GM and BFU-E generated from bone marrow cells of 2 HLA-A24–positive individuals were almost the same as those generated from bone marrow cells cultured alone. However, the numbers of CFU-GM and BFU-E decreased significantly when HLA-A24–positive bone marrow cells were pretreated with WT1 peptide and then cocultured with TAK-1. As expected, TAK-1 had no effect on colony formation by HLA-A24–negative bone marrow cells, which had been pretreated with WT1 peptide or left untreated. These data suggest strongly that WT1 is not expressed in normal bone marrow progenitors or their expression levels of WT1 are not high enough to be recognized by T lymphocytes.
Lack of effect of TAK-1 on colony formation by normal bone marrow cells.
The numbers of CFU-GM and BFU-E generated from normal bone marrow cells, which had been treated or not treated with WT1 peptide and then cocultured with (black bars) or without (gray bars) TAK-1, are shown. Cells in triplicate wells were cultured, and the data are expressed as mean colony counts ± standard deviation.
Lack of effect of TAK-1 on colony formation by normal bone marrow cells.
The numbers of CFU-GM and BFU-E generated from normal bone marrow cells, which had been treated or not treated with WT1 peptide and then cocultured with (black bars) or without (gray bars) TAK-1, are shown. Cells in triplicate wells were cultured, and the data are expressed as mean colony counts ± standard deviation.
Discussion
Evidence that human T lymphocytes discriminate leukemia and normal cells has been accumulating. The target molecules of T-lymphocyte–mediated immune responses against human leukemia cells reported most frequently in the previous papers are fusion proteins, such as BCR-ABL,2-10 PML/RARα,11ETV6-AML1,12 and DEK-CAN.13 Only 1 report of human CTLs specific for an antigen that is overexpressed in leukemia cells (proteinase 3) has been published.14 Proteinase 3–specific CTLs have been reported to inhibit colony formation by CML cells but not that by normal bone marrow cells.38 In light of the recent finding that WT1 is aberrantly overexpressed in cells from patients with various types of leukemia, we undertook this study to establish WT1-specific CTLs, and we are, we believe, the first to succeed in establishing a WT1 peptide-specific human CD8+CTL clone, designated TAK-1. TAK-1 lysed leukemia cell lines and freshly isolated leukemia cells, as well as WT1 peptide-loaded target cells, in the context of HLA-A24. Because leukemia and lymphoma cells that did not express WT1 and HLA-A24–negative leukemia cells were not lysed by TAK-1, the cytotoxicity of TAK-1 to leukemia cells seemed to be mediated in a WT1-specific and HLA-A24–restricted manner. The evidence that TAK-1 lysed leukemia cells via recognition of naturally processed WT1 peptide in the context of HLA-A24 was demonstrated by the following findings. First, treating leukemia cells with a WT1gene antisense oligonucleotide reduced TAK-1–mediated cytotoxicity. Second, the HLA-A24 mutant myeloid cell line that expressed abundant WT1 was not lysed by TAK-1.
Although the WT1 gene was originally identified as a tumor suppressor gene, its significance in tumorigenesis is still obscure. Unlike other tumor suppressor genes, such as Rb andp53, WT1 is expressed in a tissue-specific manner. Although it is well known that WT1 is expressed strongly in fetal kidneys and plays animportant role in genitourinary development, its expression in adults is very limited; it is expressed weakly only in the splenic parenchyma, testis, and ovary.23,24 Recently, aberrant expression of WT1 in various types of acute leukemia cells and CML cells at blast crisis has been reported,15-17 and our results confirm this. Furthermore, a correlation between the WT1 expression levels of leukemia cells and a poor prognosis for patients with acute leukemia has been reported.28 39 These findings suggest strongly that WT1 is the optimal target of immunotherapy for human leukemia.
Recently, the WT1 expression levels of normal bone marrow CD34+ hematopoietic progenitors and acute leukemia cells were reported to be the same.26,40 However, this finding is still controversial. Sugiyama et al reported that the WT1 expression level of normal CD34+ cells in bone marrow was < 0.01 that of acute leukemia cells.25 If as much WT1 is expressed in normal hematopoietic progenitors as in leukemia cells, the adoptive transfer of WT1-specific CTLs may cause severe hematopoietic failure. To explore this possibility, we examined the effects of TAK-1 on colony formation by HLA-matched normal bone marrow cells and found that TAK-1 had no effect on normal bone marrow cell growth. Because the growth of HLA-A24–positive bone marrow cells was inhibited markedly by TAK-1 in the presence of exogenous WT1 peptide, either WT1 is not expressed in normal hematopoietic progenitors or the WT1 expression level in normal bone marrow cells is not high enough to be recognized by WT1-specific CTLs. These findings also suggest that immunotherapy for leukemia using WT1-specific CD8+ CTLs should be safe.
In this study, we used synthetic WT1 peptides consisting of HLA-A24–binding motifs, because more than 60% of Japanese persons are HLA-A24–positive.41 Of the 4 synthetic peptides used, only 1, WT1-T2, elicited generation of WT1-specific CTLs. The reason that WT1-specific CTLs could not be generated using the other 3 peptides is unkown. High-affinity anchor residues for HLA-A24 have been reported to be Y and F at position 2 and F, W, L, and I at the C terminus.42 Although we did not examine the binding affinities of these peptides to HLA-A24 molecules, the WT1-T2 peptide does not seem to possess high affinity, because its anchor residue at position 2 is methionine. Recent studies demonstrated that intermediate- or low-affinity peptides may not be expressed at high enough levels to induce T-lymphocyte tolerance,43suggesting that WT1-T2 may be more effective for eliciting generation of WT1-specific CTLs from peripheral blood than other WT1 peptides. Because WT1-T2 lies in exon 4 of the WT1 gene, its expression is not deleted by alternative splicing of the 17 amino acids of exon 5 or 3 amino acids from the end of exon 9 of WT1. Therefore, we expect the sequence of WT1-T2 to be conserved in all 4 isoforms of WT1.
We attempted to generate WT1-specific CTLs from 3 HLA-A24–positive individuals, and a WT1-specific CTL clone was generated from only 1 of them. A relatively low frequency of WT1 peptide-specific CTL precursors in healthy individuals may be one reason for the failure to establish WT1-specific CTLs from the other 2. We have not tried to generate WT1-specific CTLs from patients with acute leukemia because a large amount of blood is needed. However, it is essential to generate WT1-specific CTLs from patients with acute leukemia to evaluate clinically adoptive immunotherapy using WT1-specific CTL clones. Therefore, the frequency of WT1-specific CTL precursors in the peripheral blood of patients with acute leukemia must be examined. We investigated whether WT1 protein contains binding motifs for HLA class I alleles, which are common worldwide, such as HLA-A*0201, A*0301, A*1101, B*0701, B*3501, and B*5101, and identified the binding motifs for each HLA class I allele in the amino acid sequence of WT1 (data not shown). This finding suggests that WT1-specific CD8+ CTLs can be generated from most of the general population regardless of race, and we are now attempting to establish WT1-specific CTL clones from individuals with various HLA class I alleles other than HLA-A24.
We used monocyte-derived DCs as antigen-presenting cells to elicit the generation of peptide-specific T-lymphocyte clones. DCs are known as the professional antigen-presenting cells that induce cytotoxic responses from naive T lymphocytes, and adoptive transfer of DCs into patients with various kinds of malignancy has been performed to induce antitumor immunity.44,45 Recently, it has been reported that DCs were generated from CML and AML cells by adding cytokines to the cultures, and they elicited generation of leukemia-specific CTLs in vitro.46-48 It would be of considerable interest to examine whether WT1 is expressed in leukemia cell-derived DCs and whether these DCs can present naturally processed WT1 peptide to T lymphocytes.
In summary, we have established the WT1 peptide-specific human CTL clone for the first time and have demonstrated its cytotoxic activity to leukemia cells. Our findings may contribute to the development of novel immunotherapy for leukemia. Recent studies have demonstrated that WT1 is also aberrantly expressed by and may be important in the pathogenesis of certain malignant tumors, such as renal cell carcinoma,49 malignant mesothelioma,50 and ovarian tumors.51 These findings suggest that adoptive immunotherapy using WT1-specific CTLs should also be effective against these tumors as well as against leukemia.
Acknowledgments
We thank Drs Masafumi Takiguchi (Kumamoto University, Kumamoto, Japan), Toshihiko Kaneshige (Shionogi Biomedical Laboratory, Osaka, Japan), Kenji Kishi (Tokai University, Kanagawa, Japan), Masuhiro Takahashi (Niigata University, Niigata, Japan), and Hayato Yamauchi (Ehime University, Ehime, Japan) for providing the cell lines and for their helpful suggestions. We also thank Kirin Brewery Co Ltd, Dainippon Pharmaceutical Co Ltd, and Kyowa Hakko Kogyo Co Ltd, for providing GM-CSF, TNF-α, and MMC, respectively.
Supported by grants from the Ministry of Education, Science, Sports, and Culture of Japan; the Ministry of Health and Welfare of Japan; the Mochida Foundation for Medical and Pharmaceutical Research, Japan; the Inamori Foundation, Japan; the Suzuken Memorial Foundation, Japan; and the Naito Foundation, Japan.
Reprints:Masaki Yasukawa, the First Department of Internal Medicine, Ehime University School of Medicine, Shigenobu, Ehime 791-0295, Japan; e-mail: yasukawa@m.ehime-u.ac.jp.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal