Abstract
Recent molecular single-cell studies have shown that in approximately 95% of cases, Reed-Sternberg cells of classic Hodgkin disease (HD) are derived from B cells of germinal center origin. Attempts to determine the cellular nature of the remaining cases have so far failed. To clarify whether they are derived from T cells, this study examined 791 single CD30+ Hodgkin and Reed-Sternberg (HRS) cells from 13 T-cell marker-positive cases and from 6 cases with null-cell phenotype for rearranged T-cell receptor-gamma (TCR-γ) genes by single copy polymerase chain reaction. Monoclonally rearranged TCR-γ genes were detectable in 2 of the 13 classic HD cases with T-cell marker-positive HRS cells, with none detectable in the null-cell cases. Eight of the T-cell marker-positive cases and all 6 null-cell cases were also studied for rearrangements of immunoglobulin genes. Six of the 8 T-cell marker-positive cases harbored clonal immunoglobulin gene rearrangements. The 2 cases without rearranged immunoglobulin genes were those that contained clonal TCR-γ rearrangements and lacked expression of the B-cell-specific activator protein. From these findings we conclude that cases of classic HD with T-cell-derived HRS cells definitely exist, although their overall incidence at 1% to 2% is very low. Even within the T-cell marker-positive cases only a minority (15%) were derived from T cells. The majority (85%) originated from B cells, indicating that the T-cell antigens expressed by HRS cells are, in contrast to those expressed in non-Hodgkin lymphoma, not lineage specific.
Classic Hodgkin disease (HD) represents one of the most common types of malignant lymphomas in the Western world. It is characterized by the presence of a low number of tumor cells (usually < 1%), designated as Hodgkin and Reed-Sternberg (HRS) cells, residing in an abundant admixture of nonmalignant cells of different types.1 The identification of the normal cellular counterpart of HRS cells has been the focus of numerous investigations in the last few decades. Immunohistologic studies have disclosed an expression of either B-cell or T-cell antigens in 20% to 40% of the HD cases, mostly restricted to a small proportion of Reed-Sternberg cells.2-7 Attempts to detect clonal antigen receptor rearrangements in whole tissue DNA extracts by means of Southern blot analysis or polymerase chain reaction (PCR) have failed in most instances.8-12 Only when methods for the isolation of single cells became available,13-15 could it be demonstrated that HRS cells harbor clonal immunoglobulin gene rearrangements with somatic hypermutations in more than 90% of HD cases. This suggests that most cases of classic HD originate from B cells at the germinal or postgerminal center stage of differentiation.
An exclusive B-cell derivation of the HRS cells in all HD cases appears, however, to be unlikely because T-cell markers or cytotoxic molecules, or both, have been convincingly demonstrated on these cells in about 15% to 20% of the cases.2-6,16-18 Furthermore, it was reported that classic HD cases and T-cell lymphomas may arise from a common T-cell clone.19 A T-cell derivation of HRS cells in some instances is further supported by the finding that nearly half of the established HD-derived cell lines have a T-cell phenotype and a T-cell genotype.20 However, previous attempts to detect T-cell receptor rearrangements in DNA derived from whole tissue extracts8,10,12,21 or from single cells22provided no conclusive or negative results.
We have, therefore, initiated a study for the detection of rearranged T-cell receptor- γ (TCR-γ) chain genes in classic HD cases with a T-cell immunophenotype. Our results show that T-cell-derived HD cases, although at low frequency, really exist and that cases with a T-cell immunophenotype much more frequently harbor clonally rearranged immunoglobulin genes than TCR-γ rearrangements.
Methods
Tissue samples, cell lines, and immunostaining
From a series of 120 cases of classic HD stored at the files of the Institute of Pathology, Free University Berlin, we selected 13 cases in which all HRS cells, or a proportion of them, expressed T-cell antigens. The diagnosis of classic HD was made on the basis of the criteria of the Revised European-American lymphoma (REAL) classification.1 In 6 cases with T-cell phenotype HRS cells, the diagnosis of classic HD was confirmed by 7 experts on lymphoma pathology (H.S.; I.A.; Dr H.-D. Foss, Berlin, Germany; Prof P. G. Isaacson, London, GB; Prof S. Poppema, Groningen, NL; Prof Dr E. Jaffe, Bethesda, MD; and Prof N. L. Harris, Boston, MA). All experts had no prior knowledge of the phenotype or genotype of the HRS cells of these cases. In addition, 10 HD-derived cell lines (L428, L591, KM-H2, L540, Co, Ho, HDLM2, SUP-HD1, HD-MyZ, L1236) were included in this study and for control purposes, several specimens with reactively diseased lymphoid tissues, T-cell lymphomas as well as B-cell (Daudi, Raji) and T-cell (Peer, Jurkat) lines were investigated. Immunohistochemistry was performed as previously described using the APAAP technique.23
Single cell analysis
The CD30+ HRS cells were isolated from immunostained frozen tissue sections of the 19 selected cases using a hydraulic micromanipulation device.24 Initially, 2 to 6 single HRS cells were pooled (primary screening) to obtain higher PCR amplification rates. At least 12 cells were analyzed by this approach to enable conclusive results to be achieved. Those cases giving rise to TCR-γ amplification products were studied in further detail by the isolation and analysis of individual HRS cells. Single HRS cells of 8 cases with T-cell marker-positive tumor cells and of 6 cases with null-cell type tumor cells were also investigated for the presence of immunoglobulin heavy-chain and kappa light-chain rearrangements. All cases were analyzed at least twice in completely independent cell isolation and PCR assays.
PCR and DNA sequence analysis
Polymerase chain reaction; TCR-γ rearrangement.
Isolated cells were digested with proteinase K (1 hour, 50°, 0.1 mg/mL) and, after heat denaturation of the enzyme, subjected to PCR. For the detection of TCR-γ rearrangements a seminested PCR was performed. The first round of amplification was carried out with a consensus primer specific for the Vγ1-8 gene segments (VG-1)25 and 2 consensus primers (JGT1/2 and JGT3)26 specific for the Jγ gene segments. For reamplification the VG-1 primer was replaced by a nested Vγ1-8 consensus primer (VG-2)25 and the PCR was divided into 2 reactions performed in conjunction with the same JGT1/2 and JGT3 primers, respectively.
PCR; immunoglobulin heavy- and kappa light-chain rearrangements.
Immunoglobulin gene rearrangements in single cells were analyzed as previously described.24 For the amplification of immunoglobulin heavy-chain rearrangements, a PCR using a set of 6 frame work-1 family-specific primers in conjunction with a primer specific for the joining region was performed in the first round of amplification. For reamplification, a set of 6 frame work-2 family-specific primers was used together with a nested joining region-specific primer. Rearrangements of the immunoglobulin kappa light chain in single cells were detected by application of a fully nested PCR, using 2 different sets of frame work-1 family-specific primers in the first and second amplification, respectively, in conjunction with 2 different sets of nested primers specific for the immunoglobulin kappa light-chain joining segments.15
The resulting PCR products were isolated, sequenced by fluorescence chain termination technique (Applied Biosystems, Weiterstadt, Germany) and compared to published data bank sequences (GenBank, VBASE) and to our own antigen receptor sequences collected in our institute over the last 7 years.
Results
Immunophenotype
A series of 120 cases of classic HD were screened for cases with T-cell antigen-positive HRS cells. Thirteen cases contained HRS cells expressing 1 or more T-cell antigens (Table1; Figures 1and 2). In 3 cases the HRS cells carried in addition, the cytotoxic molecules granzyme B or perforin or both. In 1 of the 13 cases only granzyme B was detectable. For comparison, 6 additional cases were selected whose HRS cells were devoid of B- and T-cell antigens. None of the 19 cases expressed the anaplastic lymphoma kinase (ALK) protein indicative of a translocation (2;5) and 3 cases displayed Epstein-Barr virus (EBV)-infected HRS cells as revealed by their expression of the EBV-encoded latent membrane protein (LMP-1). The B-cell-specific activator protein (BSAP) was found in all but 3 cases (Table 1).
Antigen profile of the 13 classic HD cases of T-cell phenotype
| Case . | CD30 . | CD15 . | CD20 . | CD3 . | CD4 . | CD8 . | TCR-β . | Perforin . | Granzyme B . | ALK . | CD56 . | BSAP . | EBV . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | + | − | − | − | − | + | − | + | + | − | − | − | − |
| 2 | + | + | − | + | − | + | + | + | − | − | − | − | − |
| 3 | + | + | − | + | − | + | − | − | − | − | − | + | − |
| 4 | + | + | − | − | − | + | + | − | − | − | − | − | + |
| 5 | + | + | − | − | − | − | + | − | − | − | − | + | − |
| 6 | + | + | − | + | + | − | + | − | − | − | − | + | − |
| 7 | + | + | − | + | + | + | − | − | − | − | − | nd | − |
| 8 | + | + | − | + | + | − | + | − | − | − | − | + | − |
| 9 | + | + | − | + | − | + | + | − | − | − | − | + | − |
| 10 | + | + | − | − | + | − | + | − | − | − | − | + | − |
| 11 | + | + | − | + | + | + | − | − | − | − | − | + | + |
| 12 | + | + | − | + | + | − | + | nd | nd | nd | − | + | − |
| 13 | + | + | − | − | − | − | − | nd | + | − | − | + | + |
| Case . | CD30 . | CD15 . | CD20 . | CD3 . | CD4 . | CD8 . | TCR-β . | Perforin . | Granzyme B . | ALK . | CD56 . | BSAP . | EBV . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | + | − | − | − | − | + | − | + | + | − | − | − | − |
| 2 | + | + | − | + | − | + | + | + | − | − | − | − | − |
| 3 | + | + | − | + | − | + | − | − | − | − | − | + | − |
| 4 | + | + | − | − | − | + | + | − | − | − | − | − | + |
| 5 | + | + | − | − | − | − | + | − | − | − | − | + | − |
| 6 | + | + | − | + | + | − | + | − | − | − | − | + | − |
| 7 | + | + | − | + | + | + | − | − | − | − | − | nd | − |
| 8 | + | + | − | + | + | − | + | − | − | − | − | + | − |
| 9 | + | + | − | + | − | + | + | − | − | − | − | + | − |
| 10 | + | + | − | − | + | − | + | − | − | − | − | + | − |
| 11 | + | + | − | + | + | + | − | − | − | − | − | + | + |
| 12 | + | + | − | + | + | − | + | nd | nd | nd | − | + | − |
| 13 | + | + | − | − | − | − | − | nd | + | − | − | + | + |
Note: There were no differences in the T-cell antigen expression or the CD30 staining among the cases.
TCR indicates T-cell receptor; ALK, anaplastic lymphoma kinase; BSAP, B-cell specific activator protein; EBV, Epstein-Barr virus; +, positive; −, negative; nd, not done.
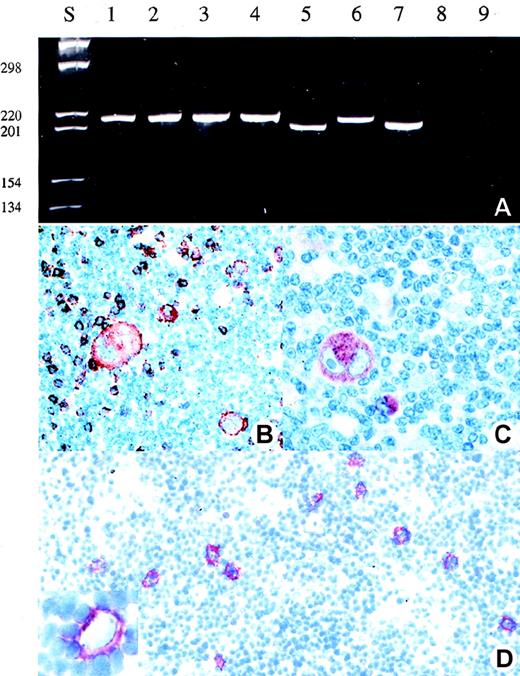
Detection of clonal TCR-γ rearrangement and expression of T-cell marker in the HRS cells of patient 1.
(A) Analysis of the amplificates obtained after single copy TCR-γ PCR in single HRS cells (lane 1-4), reactive T-cells (lane 5-7), buffer control (lane 8), and negative control (lane 9) (B-cell line Raji) (polyacrylamide gel stained with ethidium bromide). (B) and (C) Immunohistologic staining for perforin (B) and CD8 (C) (APAAP staining). (D) CD30 staining of a frozen section from which the single HRS cells or their nuclei were isolated. In the insert, a Hodgkin cell is shown following isolation of its nucleus by micromanipulation.
Detection of clonal TCR-γ rearrangement and expression of T-cell marker in the HRS cells of patient 1.
(A) Analysis of the amplificates obtained after single copy TCR-γ PCR in single HRS cells (lane 1-4), reactive T-cells (lane 5-7), buffer control (lane 8), and negative control (lane 9) (B-cell line Raji) (polyacrylamide gel stained with ethidium bromide). (B) and (C) Immunohistologic staining for perforin (B) and CD8 (C) (APAAP staining). (D) CD30 staining of a frozen section from which the single HRS cells or their nuclei were isolated. In the insert, a Hodgkin cell is shown following isolation of its nucleus by micromanipulation.
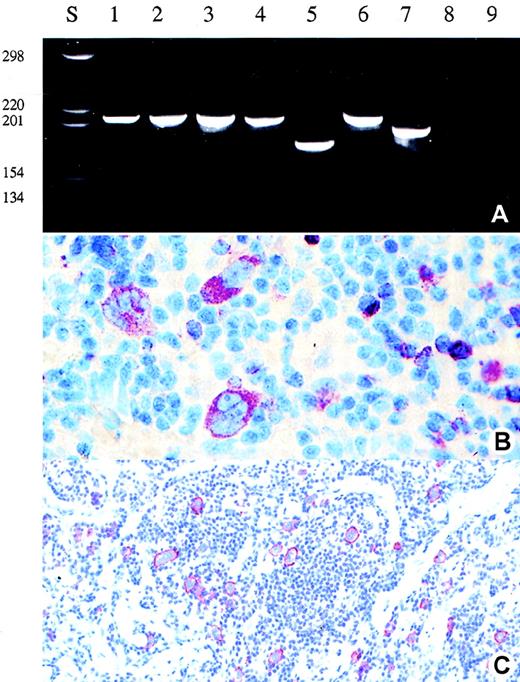
Detection of clonal TCR-γ rearrangement and expression of perforin in the Hodgkin and Reed-Sternberg cells of patient 2.
(A) Analysis of the amplificates obtained after single copy TCR-γ PCR in single HRS cells (lane 1-4), reactive T-cells (lane 5-7), buffer control (lane 8), and negative control (lane 9) (B-cell line Raji) (polyacrylamide gel stained with ethidium bromide). (B) and (C) Immunohistologic staining for perforin (B) and CD30 (C) (APAAP staining). Note the perifollicular and noncohesive growth pattern of the CD30+ tumor cells.
Detection of clonal TCR-γ rearrangement and expression of perforin in the Hodgkin and Reed-Sternberg cells of patient 2.
(A) Analysis of the amplificates obtained after single copy TCR-γ PCR in single HRS cells (lane 1-4), reactive T-cells (lane 5-7), buffer control (lane 8), and negative control (lane 9) (B-cell line Raji) (polyacrylamide gel stained with ethidium bromide). (B) and (C) Immunohistologic staining for perforin (B) and CD30 (C) (APAAP staining). Note the perifollicular and noncohesive growth pattern of the CD30+ tumor cells.
Rearrangements of T-cell and B-cell receptors
Rearranged TCR-γ genes were detectable in the isolated single HRS cells in 2 of the 13 T-cell marker-positive cases, but in none of the 6 null-cell cases (Table 2; Figures 1 and 2). The sequence analysis showed that the HRS-cell derived TCR-γ rearrangements were clonal in both cases (Table3). Mutations were not encountered. The analysis of the immunoglobulin gene loci revealed heavy- or light-chain rearrangements in all null-cell cases and in all T-cell phenotype cases investigated except the 2 cases in which rearranged TCR-γ genes were detectable (Table 2). All but one immunoglobulin gene rearrangement derived from T-cell marker-positive cases were functional, although they harbored highly mutated heavy-chain rearrangements. Interestingly, 3 of the 4 immunoglobulin heavy-chain rearrangements isolated from T-cell marker-positive cases involved the VH3 segment DP47; however, they differed in their clone specific complementarity determining region 3.
Detection of TCR-γ and immunoglobulin gene rearrangements in single Hodgkin and Reed-Sternberg cells of 19 cases of classic HD
| Case . | Age/Sex . | Phenotype . | TCR-γ . | Immunoglobulin Heavy-Chain Gene . | Immunoglobulin Light-Chain Gene . | |||
|---|---|---|---|---|---|---|---|---|
| Isolated Cells . | Rearrangements (I/N) . | Isolated Cells . | Rearrangements (I/N) . | Isolated Cells . | Rearrangements (I/N) . | |||
| 1 | 56/M | T | 35 | 12/0 | 19 | 0/0 | 15 | 0/0 |
| 2 | 72/M | T | 30 | 9/0 | 16 | 0/0 | 12 | 0/0 |
| 3 | 27/M | T | 25 | 0/0 | 12 | 0/0 | 20 | 8/0 |
| 4 | 34/M | T | 20 | 0/0 | 27 | 10/0 | 12 | 0/0 |
| 5 | Na | T | 24 | 0/0 | 21 | 6/0 | nd | |
| 6 | Na | T | 14 | 0/0 | 12 | 3/0 | nd | |
| 7 | 31/F | T | 12 | 0/0 | 34 | 3/0 | nd | |
| 8 | 12/F | T | 12 | 0/0 | 10 | 0/0 | 23 | 4/0 |
| 9 | 41/F | T | 13 | 0/0 | nd | nd | ||
| 10 | 24/M | T | 29 | 0/0 | nd | nd | ||
| 11 | 27/F | T | 12 | 0/0 | nd | nd | ||
| 12 | 23/M | T | 14 | 0/0 | nd | nd | ||
| 13 | 23/M | T | 12 | 0/0 | nd | nd | ||
| 14 | 32/M | 0 | 12 | 0/0 | 23 | 9/0 | 14 | 5/0 |
| 15 | 80/F | 0 | 12 | 0/0 | 16 | 6/0 | 13 | 4/0 |
| 16 | 35/M | 0 | 12 | 0/0 | 28 | 10/0 | 12 | 6/0 |
| 17 | 38/F | 0 | 12 | 0/0 | 18 | 6/0 | 18 | 0/0 |
| 18 | 30/M | 0 | 12 | 0/0 | 16 | 6/0 | 20 | 0/0 |
| 19 | 38/M | 0 | 29 | 0/0 | 25 | 0/0 | 14 | 5/0 |
| Case . | Age/Sex . | Phenotype . | TCR-γ . | Immunoglobulin Heavy-Chain Gene . | Immunoglobulin Light-Chain Gene . | |||
|---|---|---|---|---|---|---|---|---|
| Isolated Cells . | Rearrangements (I/N) . | Isolated Cells . | Rearrangements (I/N) . | Isolated Cells . | Rearrangements (I/N) . | |||
| 1 | 56/M | T | 35 | 12/0 | 19 | 0/0 | 15 | 0/0 |
| 2 | 72/M | T | 30 | 9/0 | 16 | 0/0 | 12 | 0/0 |
| 3 | 27/M | T | 25 | 0/0 | 12 | 0/0 | 20 | 8/0 |
| 4 | 34/M | T | 20 | 0/0 | 27 | 10/0 | 12 | 0/0 |
| 5 | Na | T | 24 | 0/0 | 21 | 6/0 | nd | |
| 6 | Na | T | 14 | 0/0 | 12 | 3/0 | nd | |
| 7 | 31/F | T | 12 | 0/0 | 34 | 3/0 | nd | |
| 8 | 12/F | T | 12 | 0/0 | 10 | 0/0 | 23 | 4/0 |
| 9 | 41/F | T | 13 | 0/0 | nd | nd | ||
| 10 | 24/M | T | 29 | 0/0 | nd | nd | ||
| 11 | 27/F | T | 12 | 0/0 | nd | nd | ||
| 12 | 23/M | T | 14 | 0/0 | nd | nd | ||
| 13 | 23/M | T | 12 | 0/0 | nd | nd | ||
| 14 | 32/M | 0 | 12 | 0/0 | 23 | 9/0 | 14 | 5/0 |
| 15 | 80/F | 0 | 12 | 0/0 | 16 | 6/0 | 13 | 4/0 |
| 16 | 35/M | 0 | 12 | 0/0 | 28 | 10/0 | 12 | 6/0 |
| 17 | 38/F | 0 | 12 | 0/0 | 18 | 6/0 | 18 | 0/0 |
| 18 | 30/M | 0 | 12 | 0/0 | 16 | 6/0 | 20 | 0/0 |
| 19 | 38/M | 0 | 29 | 0/0 | 25 | 0/0 | 14 | 5/0 |
Cases 1-13: Expression of T-cell antigens. Cases 14-19: Absence of B-cell and T-cell antigens. I, indicates identical rearrangements; N, nonidentical rearrangements; F, female; M, male; na, not available; nd, not done.
Clone-specific sequences of the two classic HD cases with TCR-γ gene rearrangements
| Case . | Vγ . | N Region . | Jγ . |
|---|---|---|---|
| 1 | ...TGTGCCACCTGGGACAGGCA | TTCCTTC | ATTATAAGAAACTCTTTGGC... |
| 2 | ...TGTGCCACCTGGGACGGGC | GAG | TTATAAGAAACTCTTTGGCA.. |
| Case . | Vγ . | N Region . | Jγ . |
|---|---|---|---|
| 1 | ...TGTGCCACCTGGGACAGGCA | TTCCTTC | ATTATAAGAAACTCTTTGGC... |
| 2 | ...TGTGCCACCTGGGACGGGC | GAG | TTATAAGAAACTCTTTGGCA.. |
Single cell controls
For control purposes, single nonneoplastic T cells and B cells as well as aliquots drawn from the buffer covering the tissue sections during the isolation process were analyzed for the presence of TCR-γ and immunoglobulin gene rearrangements, respectively. None of the 450 buffer controls gave rise to an amplification product. The PCR of 124 isolated reactive T cells led to the detection of nonidentical rearrangements of the TCR-γ gene in 37 cells (30%), whereas the 9 PCR products obtained from 35 neoplastic cells (26%) of a peripheral T-cell lymphoma case contained identical (clonal) rearrangements. In none of the 46 single nonneoplastic B cells was a TCR-γ rearrangement detectable, and vice versa, in none of the 46 single nonneoplastic T cells was an immunoglobulin gene rearrangement seen.
HD-derived cell lines
All 10 HD-derived cell lines were analyzed by PCR for the presence of antigen receptor gene rearrangements. Four cell lines (L1236, L428, KM-H2, and L591) displayed IgH rearrangements, a further 4 cell lines (L540, Co, Ho, and HDLM-2) showed TCR-γ rearrangements, and the remaining 2 cell lines (Sup-HD and HD-MyZ) were devoid of any antigen receptor gene rearrangement.
Discussion
The origin of HRS cells of classic HD is a long-standing enigma. Recent molecular biologic single-cell studies have provided convincing evidence that more than 90% of HRS cells are B-cell derived.14,15,27 In view of the observation that HRS cells of some cases express one or more T-cell antigen(s), it was speculated that the remaining cases originate from T cells.2,16-18,28To test this hypothesis we selected 13 cases with T-cell marker-positive HRS cells from a large series of 120 classic HD cases. Six cases with null-cell type HRS cells were also included in the study to disclose possible differences between Hodgkin lymphomas with T-cell marker-positive and -negative HRS cells. From these 19 cases we isolated CD30+ HRS cells and analyzed the configuration of their TCR-γ genes. The HRS cells from 2 of the 13 T-cell marker-positive cases, but none of the null-cell cases, proved to contain clonally rearranged TCR-γ genes. To determine whether HRS cells with rearranged TCR-γ genes additionally contain immunoglobulin gene rearrangements, and to clarify whether cases lacking detectable TCR-γ rearrangements carry rearranged immunoglobulin genes, we extended the study to the immunoglobulin gene loci. This revealed rearrangements of the immunoglobulin heavy- or light-chain genes in the HRS cells in 6 of the 8 T-cell marker-positive cases studied, and in all 6 cases of the null-cell type. The 2 negative cases were those whose HRS cells contained rearranged TCR-γ genes. This indicates that only 1 type of antigen receptor gene rearrangement is present in HRS cells. To further substantiate this conclusion we extended our studies to the BSAP, which has proven its B-cell specificity by recent immunohistochemical studies.29 All but one of the cases with immunoglobulin gene rearrangements analyzed displayed an expression of BSAP in the HRS cells, whereas both cases with rearranged TCR-γ genes were negative. This nearly excluded a second antigen receptor gene rearrangement that might have escaped by PCR. Taken together, these findings indicate that cases of classic HD with T cell-derived HRS cells really exist and the frequency of T cell-derived classic HD cases is between 1% or 2% and thus 10 times lower than that of T-cell derived neoplasms in non-Hodgkin lymphomas, and only one type of antigen receptor gene rearrangement is present in HRS cells.
The possibility that the 2 Hodgkin lymphomas with rearranged TCR-γ genes were confused with anaplastic large cell lymphomas, which may mimic HD was excluded by their negative labeling for ALK whose expression is restricted to anaplastic large-cell lymphoma.30 31 Moreover, a confusion of the 2 T-cell genotype Hodgkin lymphomas with ALK-negative anaplastic large-cell lymphomas or other lymphoma types was ruled out by the confirmation of the diagnosis of classic HD by 4 external internationally renowned lymphoma experts who had no prior knowledge about the phenotype and the genotype of these cases. Both cases showed no sinusoidal spread and the tumor cells grew separately from each other in all areas of the biopsies (Figures 1D and 2C).
Another interesting finding of this study is that most Hodgkin lymphomas with T-cell phenotype HRS cells were genotypically B-cell derived. This suggests that the transforming event leading to classic HD may be associated with a partial activation of genes encoding the T-cell antigens. The discordance, however, between the genotype and phenotype in HD is not restricted to genes coding for T-cell antigens, but also involves other lineage characteristic genes, for example, dendritic cell genes encoding fascin and thymus and activation-regulated chemokine (TARC).32 33
To test whether the 2 cases with TCR-γ rearrangements were distinct from the remaining cases we compared their HD subtype, their EBV-infection status, and the sex and the age of the patients. Interestingly, whereas the cases without TCR-γ rearrangements affected predominantly younger patients (average, 26.9 years; range, 12-41 years) the patients with TCR were 56 and 72 years old. Moreover, perforin was expressed in the HRS cells of both cases, whereas perforin was absent in the remaining cases. Finally, both cases were EBV negative although they belonged to the mixed cellularity subtype. However, a larger number of Hodgkin lymphomas with T-cell genotype needs to be studied to substantiate whether differences between the 2 groups do really exist.
The occasional derivation of HRS cells from T cells is in keeping with previous studies that reported that some Hodgkin lymphomas, mycosis fungoides, and lymphomatoid papulosis may have a common T-cell precursor.19,34,35 Further support for the existence of T-cell-derived Hodgkin lymphomas comes from the HD-derived cell lines 40% of which display a T-cell phenotype and genotype36; however, this high incidence is in contrast to the situation in vivo. This finding implies that either T-cell-type HRS cells are easier to cultivate than B-cell-type HRS cells, or that most, or all, T-cell-derived Hodgkin cell lines originate from cells other than HRS cells. Irrespective of which possibility is true, the T-cell-derived Hodgkin cell lines do not reflect the situation characteristic of HD. Therefore, conclusions for the pathogenesis of classic HD based on these cell lines must be regarded with caution. The demonstrated existence of T-cell-derived Hodgkin lymphomas points toward a possible relationship between the former and ALK-negative anaplastic large-cell lymphoma of T-cell type. It remains for future studies to show whether there is a well-defined border between the mentioned lymphomas categories or not.
To conclude, HRS cells with a genotype and phenotype of T cells exist. Most cases of classic HD, however, represent B-cell-derived lymphomas irrespective of their immunophenotype.
Note added in proof: During the review process, a report submitted in parallel to ours appeared, and it describes the presence of HRS cells with clonally rearranged T-cell receptor-beta chains in one case of classic HD.37
Acknowledgments
We thank H. Lammert, H.-H. Müller, and D. Jahnke for their excellent technical assistance and to L. Udvarhelyi for his editorial assistance. The work constitutes parts of the doctoral thesis of V.S. (FB Chemie).
Supported by grants from the Deutsche Forschungsgemeinschaft (DFG), Deutsche Krebshilfe, and by the Berliner Krebsgesellschaft.
Reprints:Harald Stein, Institute of Pathology, University Hospital Benjamin Franklin, Free University Berlin, Hindenburgdamm 30, 12200 Berlin, Germany; e-mail: stein@medizin.fu-berlin.de.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal