Abstract
Anaplastic lymphoma kinase (ALK)-positive lymphomas are characterized by expression of a hybrid protein, comprising the cytoplasmic portion of the ALK tyrosine kinase fused to a partner protein. This hybrid kinase is often encoded by the nucleophosmin (NPM)NPM-ALK fusion gene resulting from the (2;5)(p23;q35) chromosomal translocation. However, the ALK gene at 2p23 may also be involved in 2 variant translocations, namely t(1;2)(q25;p23) and t(2;3)(p23;q21), which create the TPM3-ALK andTFG-ALK fusion genes, respectively. We report here 2 lymphomas with an unusual finely granular cytoplasmic ALK staining pattern, clearly different from the pattern observed in ALK-positive lymphomas carrying NPM-ALK or its variants. A cloned complementary DNA sequence from 1 of these 2 lymphomas contained the ALK gene fused to the second clathrin heavy chain gene (also referred to as clathrin heavy polypeptide-like gene) (CLTCL). The distinctive granular cytoplasmic staining pattern for ALK was likely to be due to binding of the fusion protein to clathrin-coated vesicles. TheCLTCL gene is constitutively expressed in lymphoid cells and therefore presumably contributes an active promoter for theCLTCL-ALK gene. The fusion protein had a molecular weight (250 kd) that differs from all known ALK products, and it was autophosphorylated in an in vitro kinase assay, confirming that it is constitutively active and hence capable of contributing to malignant transformation. These 2 cases, therefore, represent a hitherto undescribed mechanism of ALK activation in lymphoma and further illustrate the diversity of fusion partners for the ALKgene.
In 1998, Benharroch et al1 and Falini et al2 defined a lymphoma category (anaplastic lymphoma kinase [ALK]-positive lymphoma or ALK-oma) characterized by the presence of a novel chimeric protein in which the cytoplasmic portion of the ALK tyrosine kinase is linked to a partner protein. ALK-positive lymphomas show a range of morphologic appearances and only some of them have the classical morphologic features of anaplastic large cell lymphoma. However, they are usually of cytotoxic T-cell (or occasionally null) phenotype, tend to occur in young patients, and are associated with a good prognosis.
Expression of the ALK protein in these tumors is usually due to the (2;5)(p23;q35) chromosome translocation, which fuses the ALKgene at 2p23 to the NPM (nucleophosmin) gene at 5q35.3-7 The ALK gene, which encodes a member of the insulin growth factor receptor superfamily, is silent in normal lymphoid cells.7 In contrast, the NPM gene encodes a nucleolar phosphoprotein (involved in shuttling ribonucleoproteins from the cytoplasm to the nucleus), which is widely expressed.8,9 Its ubiquitous presence in normal lymphoid cells means that the NPM-ALK gene will be transcribed if cells acquire the (2;5) translocation. Furthermore, the NPM moiety contains motifs that mediate homodimerization and can thus activate the kinase portion of the fusion NPM-ALK protein.10 11
The t(2;5) translocation, causing fusion of the ALK andNPM genes, is found in about two thirds of ALK-positive lymphomas. However, among the remaining cases at least 3 different cytogenetic abnormalities can activate the ALKgene,12-17 as summarized in Table1. NPM-ALK protein typically accumulates in the cytoplasm and also the nuclei and nucleoli of lymphoma cells. In contrast, other ALK fusion proteins are not found in the nucleus (Table1), and ALK immunostaining can be used for screening ALK-positive variants of the classical (2:5) translocation.
Genetic abnormalities in ALK-positive lymphoma that create fusion genes
| Chromosomal Anomaly . | ALK Partner . | M wt of ALK-Hybrid . | Immunostaining Pattern for ALK . |
|---|---|---|---|
| t(2;5)(p23;q35) | NPM7 | 80 | Nuclear, diffuse cytoplasmic |
| t(1;2)(q25;p23) | TPM314 | 104 | Diffuse cytoplasmic with peripheral intensification |
| t(2;3)(p23;q21) | TFG17 | 97 | Diffuse cytoplasmic |
| Inv (2)(p23q35) | ATIC* | 96 | Diffuse cytoplasmic |
| Chromosomal Anomaly . | ALK Partner . | M wt of ALK-Hybrid . | Immunostaining Pattern for ALK . |
|---|---|---|---|
| t(2;5)(p23;q35) | NPM7 | 80 | Nuclear, diffuse cytoplasmic |
| t(1;2)(q25;p23) | TPM314 | 104 | Diffuse cytoplasmic with peripheral intensification |
| t(2;3)(p23;q21) | TFG17 | 97 | Diffuse cytoplasmic |
| Inv (2)(p23q35) | ATIC* | 96 | Diffuse cytoplasmic |
Own unpublished results (ATIC: 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase).
M wt indicates molecular weight.
In the present study, 2 large cell lymphomas showed a unique granular ALK cytoplasmic staining pattern. Fresh tissue was available from 1 case, which showed that the ALK gene was fused to theCLTCL gene, which encodes the second clathrin heavy chain (also known as the clathrin heavy polypeptide-like protein). We conclude that the unique ALK staining pattern arises when the CLTCL-ALK fusion protein associates with clathrin-coated vesicles. These 2 cases provide further evidence for the diversity of partners that can fuse to the intracytoplasmic domain of ALK and promote its activation.
Patients, materials, and methods
Patients
Case 1.
A girl, aged 3 years, presented with a skin nodule and axillary lymphadenopathy. Physical examination, blood count, and bone marrow examination were normal and no disease spread was found on staging assessment. Lymph node and skin biopsies showed the histologic features of anaplastic large cell lymphoma (ALCL). Complete remission was achieved after 2 courses of chemotherapy (VED-B1) and the patient is still in complete remission 1 year after finishing treatment.
Case 2.
A 52-year-old man presented with prominent left inguinal lymphadenopathy associated with fever and poor performance status. A diagnosis of ALCL was made following lymph node biopsy. The patient showed a good response to chemotherapy (CHOP) and is still on treatment (8 months follow-up).
Materials
Tissue samples and cell lines.
In addition to the diagnostic formalin-fixed paraffin-embedded biopsy specimens, frozen samples of lymph node and of the skin tumor were available from case 1. Normal tonsil was obtained from the ENT department of the Radcliffe Infirmary, Oxford. The SU-DHL-1 and Rh30 cell lines were kindly provided by Dr M.L. Cleary (Stanford University Medical Center, Stanford, CA) and Dr S. W. Morris (St Jude Children's Research Hospital, Memphis, TN). CEM, Jurkat, and HSB-2 cell lines (all acute T leukemia, ATCC CCL 119, ATCC TIB 153, and ATCC CCL 120.1, respectively) were used to investigate the constitutive expression of the CLTCL gene.
Methods
Immunostaining.
RNA extraction for reverse transcriptase-polymerase chain reaction (RT-PCR).
5′RACE (rapid amplification of cDNA ends).
PolyA+ RNA from about 100 μg of tissue from the lymph node biopsy was prepared with the Oligotex TM Direct messenger RNA (mRNA) purification kit (Qiagen) following the manufacturer's recommendations. The RACE procedure was performed using the Marathon TM cDNA Amplification Kit (Clontech, Saint Quentin Yvelines, France). One microgram of polyA+ RNA was reverse transcribed using AMV reverse transcriptase and ALKoligonucleotide specific primer (ALK1) (5′-GCCAGCAAAGCAGTAGTTGGGGTTG-3′). Second strand synthesis was carried out using Rnase H, DNA polymerase I, DNA ligase, T4 DNA polymerase mix, and extraction was performed with 100 μL of phenol-chloroform-isoamyl alcohol (25:24:1) followed by adding 100 μL chloroform-isoamyl alcohol (24:1) to the aqueous phase and by precipitating with 2.5 volumes of 95% ethanol. After centrifugation, the pellets were rinsed with 80% ethanol and dissolved in 10 μL water. This double-strand cDNA was used for the ligation of the Marathon TM cDNA Adaptor (included in the kit).
The first round of PCR was performed for 35 cycles with the AmpliTaq Gold (Perkin Elmer, Courtaboef, France), using a DNA Thermal Cycler 480 (Perkin Elmer) and with adaptor primer AP1: 5′-CCATCCTAATACGACTCACTATAGGGC-3′ and ALK1 specific primer. A touch-down protocol was used to meet the requirements of different primers, which enabled the specific amplification of hybrid ALKcDNA. The second round consisted of a nested PCR performed using 5 μL of the primary PCR, with the nested adaptor primer AP2: 5′ACTCACTATAGGGCTCGAGCGGC-3′ and an inner nestedALK gene-specific primer (ALK2: 5′-GTCGAGGTGCGGAGCTTGCTCAGC-3′). PCR products were analyzed on a 1% agarose gel, followed by ethidium bromide staining.
Sequencing of PCR products.
Products of the nested PCR were extracted and purified on a 1.5% agarose gel (Tris borate/EDTA) using the QIAquick Gel Extraction Kit (Qiagen) following the manufacturer's recommendations. The purified PCR products were directly sequenced using the Dye Terminator Cycle Sequencing method (Perkin Elmer) with AP2 and ALK2 primers. After purification, samples were electrophoresed and analyzed with a Perkin Elmer ABI 373A DNA sequencer.
Detection of hybrid CLTCL-ALK transcripts.
Two nested CLTCL primers, CLA1-L (5′-GAGTGCTTTGGAGCTTGTCTGTT TACCTG-3′) (position 4861-4890) and CLA3 (5′-CGATCTTTTAAGGCCAGAT GTCGTCC-3′) (position 4894-4919) were chosen according to the cDNA sequence of CLTCL (clathrin heavy polypeptide-like mRNA) deposited in the Genbank database (accession number D21260). Synthesis of the first cDNA strand and PCR amplification were performed in a single tube using the RT-PCR Access Kit (Promega France, Charbonnières, France), following the manufacturer's recommendations. The first step consisted of an ALK gene-specific reverse transcription at 48°C for 45 minutes, followed by 30 cycles comprising a denaturation step at 94°C for 45 seconds, an annealing step at 64°C for 45 seconds, and an elongation step at 72°C for 45 seconds. The first round of PCR was performed using CLA1-L (position 4861-4890) and ALK-1 (position 4211-4187 on the cDNA ALK sequence) primers. The second round was performed on a 5-μL aliquot from the first amplification, using nested primers CLA3 (position 4894-4919) and ALK-2 (position 4171-4148), yielding a RT-PCR product of 270 bp.
Detection of CLTCL transcripts.
The first round of PCR was performed as described above, using CLA1-L and CLA-R2 (5′-GGTGACTACAGGATCAGCGCTTCA-3′) primers (position 5201-5226). The second round (nested PCR) was performed using 1 μL from the first amplification, using nested primers CLA3 (position 4894-4919) and CLA-R1 (5′-GTACCCAAAGCCAGGCTGTGGCTG-3′) (position 5168-5191) yielding a PCR product of 298 bp.
Biochemical assays.
Cryostat sections (6 μm) were cut from a lymph node biopsy sample (case 1) and from tonsil. Western blotting and in vitro kinase assays using anti-ALK monoclonal antibodies were performed on proteins extracted from these tissues and from cell samples, as previously described.15
Results
Immunomorphologic features
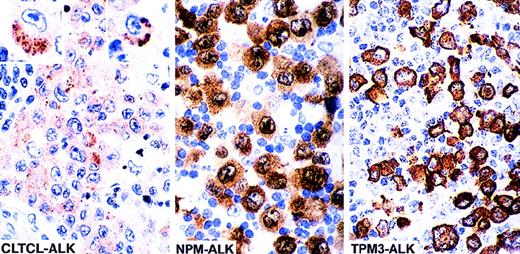
The lymph node and skin lesions from case 1 showed infiltration by large cells, some of which had horseshoe- or kidney-shaped eccentric nuclei similar to those of the “hallmark cells” previously described as typical of ALK-positive lymphoma.1 Some cells contained large cytoplasmic vacuoles. The malignant cells coexpressed CD30 and epithelial membrane antigen (EMA) and were also weakly positive for CD3. All tumor cells were also positive for ALK protein but, in contrast to classical t(2;5) positive tumors (Table 1), staining was limited to the cytoplasm and consisted of clusters of fine granules (Figure 1).
ALK-staining patterns in anaplastic large cell lymphoma.
The cellular localization of the fusion protein is influenced by the ALK partner. The majority of anaplastic large cell lymphomas (70%-80%) are associated with the (2;5)(p23;q35) translocation and express NPM-ALK fusion protein. In such cases ALK staining is cytoplasmic, nuclear, and nucleolar associated. Approximately 10% to 20% (unpublished data) of cases carry the (1;2)(q25;p23) translocation and express the TPM3-ALK fusion protein, which is restricted to the cytoplasm (diffuse staining). In clear contrast, the 2 cases in the present study show intracytoplasmic finely granular clusters of staining, a pattern that differs clearly from those previously described (the 2 insets show single cells positive for CLTCL-ALK, at high power).
ALK-staining patterns in anaplastic large cell lymphoma.
The cellular localization of the fusion protein is influenced by the ALK partner. The majority of anaplastic large cell lymphomas (70%-80%) are associated with the (2;5)(p23;q35) translocation and express NPM-ALK fusion protein. In such cases ALK staining is cytoplasmic, nuclear, and nucleolar associated. Approximately 10% to 20% (unpublished data) of cases carry the (1;2)(q25;p23) translocation and express the TPM3-ALK fusion protein, which is restricted to the cytoplasm (diffuse staining). In clear contrast, the 2 cases in the present study show intracytoplasmic finely granular clusters of staining, a pattern that differs clearly from those previously described (the 2 insets show single cells positive for CLTCL-ALK, at high power).
The lymph node biopsy from case 2 was massively involved by large cells that resembled, both morphologically and phenotypically, those seen in case 1. They were positive for CD43, perforin, and TiA1 but negative for CD3. ALK staining in this case also took the form of finely granular intracytoplasmic clusters, some of which showed small central vacuoles.
Molecular cloning of the ALK breakpoint
A 300 bp PCR product was amplified by the 5′-RACE technique from a lymph node sample (case 1) using ALK1-specific oligonucleotide. Sequencing showed that this comprised a portion of the second human clathrin heavy chain gene (also referred to as clathrin heavy polypeptide-like gene or CLTCL) fused in frame to ALKat nucleotide 4083 (Figure 2B). TheALK breakpoint in this chimeric sequence was in the same intron as that involved in both the classical (2;5) translocation and in the recently characterized (1;2)14 and in (2;3)17translocations. The breakpoint in the CLTCL transcript was close to its 3′ extremity, being situated at nucleotide 5075.
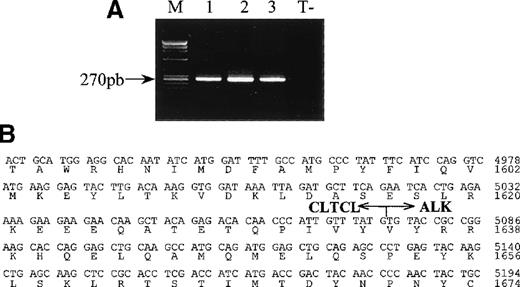
RT-PCR analysis of CLTCL-ALK and the sequence of the cloned CLTCL-ALK cDNA fusion junction.
(A) All samples show the expected 270 bp CLTCL-ALK RT-PCR product (M, size markers).Total RNA prepared from the 3 biopsy specimens (lane 1 and 2: axillary lymph nodes; lane 3: skin tumor) of case 1. RNA obtained from the NPM-ALK-positive SU-DHL-1 cell line was used as negative control (T−). RT-PCR was performed using 2 rounds of PCR. (B) Nucleotide and deduced amino acid sequences ofCLTCL-ALK cDNA. The arrow shows the translocation breakpoint. Nucleotides are numbered from the first nucleotide of the CLTCL transcript.
RT-PCR analysis of CLTCL-ALK and the sequence of the cloned CLTCL-ALK cDNA fusion junction.
(A) All samples show the expected 270 bp CLTCL-ALK RT-PCR product (M, size markers).Total RNA prepared from the 3 biopsy specimens (lane 1 and 2: axillary lymph nodes; lane 3: skin tumor) of case 1. RNA obtained from the NPM-ALK-positive SU-DHL-1 cell line was used as negative control (T−). RT-PCR was performed using 2 rounds of PCR. (B) Nucleotide and deduced amino acid sequences ofCLTCL-ALK cDNA. The arrow shows the translocation breakpoint. Nucleotides are numbered from the first nucleotide of the CLTCL transcript.
Detection of hybrid CLTCL-ALK transcripts
The RT-PCR technique showed that a 270 bp CLTCL-ALK band could be amplified from each of the 3 tissue samples available from case 1 (2 lymph node samples and a skin biopsy) (Figure 2A). Sequencing of the nested RT-PCR products confirmed the presence of hybridCLTCL-ALK transcripts.
CLTCL transcripts in normal lymphoid cells
Analysis by RT-PCR showed that the CLTCL transcript was present in the T-cell lines Jurkat, CEM, HSB-2, and SU-DHL-1, and it was also detected in the patient's biopsy specimens, presumably from the second CLTCL allele.
Characteristics of the CLTCL-ALK fusion protein
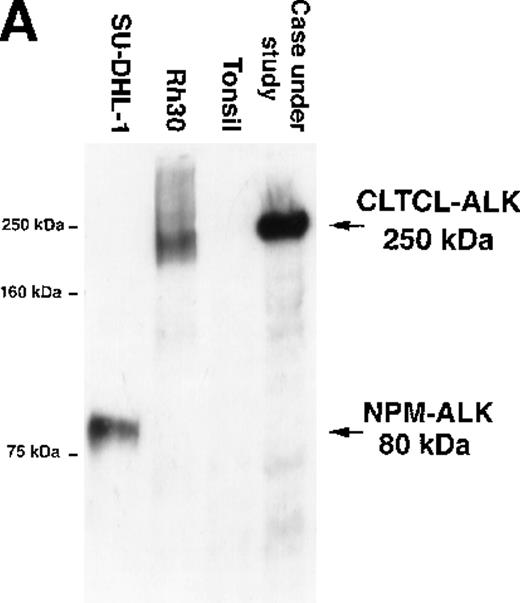
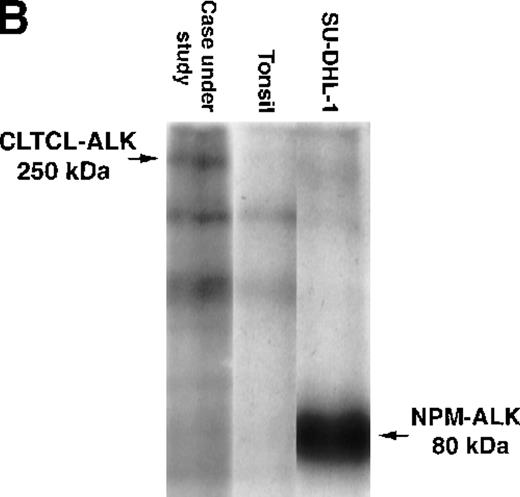
The predicted CLTCL-ALK protein comprised 2197 amino acids, of which 1634 were encoded by the CLTCL gene and 562 by the ALKgene. The valine at position 1635 is encoded by a new codon created by the rearrangement. The predicted protein product is of relatively high molecular weight (248 kd) and Western blot analysis showed a band of 250 kd in very close agreement (Figure 3A). An in vitro kinase assay performed on proteins immunoprecipitated with anti-ALK from the lymph node sample revealed a prominent phosphorylated protein that was also of 250 kd molecular weight, presumably representing autophosphorylated CLTCL-ALK fusion protein (Figure 3B).
Biochemical assays of proteins extracted from cryostat tissue sections.
(A) Western blotting using a 5.5% acrylamide gel. Anti-ALK (ALKc) detects a 250 kd band in lysates from the ALCL case expressing the CLCTL-ALK transcripts. This ALK protein is of a higher molecular weight than the normal 200 kd ALK protein present in the Rh30 cell line and the oncogenic 80 kd NPM-ALK expressed by the SU-DHL-1 cells. No ALK proteins were detected in tonsil lysates used as negative controls. (B) In vitro kinase assay using a 7.5% acrylamide gel. A phosphorylated 250 kd protein is present in the CLTCL-ALK-positive ALCL case. The 80 kd NPM-ALK protein expressed by SU-DHL-1 cells is used as a positive control. No corresponding bands were present in tonsil lysates.
Biochemical assays of proteins extracted from cryostat tissue sections.
(A) Western blotting using a 5.5% acrylamide gel. Anti-ALK (ALKc) detects a 250 kd band in lysates from the ALCL case expressing the CLCTL-ALK transcripts. This ALK protein is of a higher molecular weight than the normal 200 kd ALK protein present in the Rh30 cell line and the oncogenic 80 kd NPM-ALK expressed by the SU-DHL-1 cells. No ALK proteins were detected in tonsil lysates used as negative controls. (B) In vitro kinase assay using a 7.5% acrylamide gel. A phosphorylated 250 kd protein is present in the CLTCL-ALK-positive ALCL case. The 80 kd NPM-ALK protein expressed by SU-DHL-1 cells is used as a positive control. No corresponding bands were present in tonsil lysates.
Discussion
The unusual granular intracytoplasmic ALK staining pattern in the 2 cases in this study raised the suspicion that a novel ALKfusion gene might be present.
This was confirmed in 1 case by using the RACE technique to amplify cDNA in which the ALK gene was fused to the CLTCL gene. This gene, located on chromosome 22q11.2,19,20 encodes a clathrin heavy polypeptide-like chain and should be distinguished from the clathrin heavy chain gene (CLTC), which maps to chromosome 17 q11-qter.21 Although we could not analyze the second case (for want of fresh material), we assume that the same fusion gene was present, because the staining pattern was so clearly different from that seen in any of the other ALK-positive lymphoma (approximately 200 cases) that we have studied.
No cytogenetic data were available but it is likely that these tumors carry a (2;22)(p23;q11.2) translocation. A CD30−diffuse large B-cell lymphoma carrying this translocation22has been reported but, all other ALK fusion genes are found in T-cell or “null” lymphomas (rather than in B-cell neoplasms), and the breakpoint on chromosome 22 was the site of the immunoglobulin lambda light chain gene. The clathrin heavy polypeptide-like gene has also been implicated in a translocation in DiGeorge syndrome/velocardiofacial syndrome. This balanced t(21;22)(p12;q11) translocation disrupts the clathrin heavy polypeptide-like gene, leading to a truncated CLTCLtranscript.23
Clathrin, which is highly conserved in mammalians,24comprises a heavy and a light chain and is the main structural protein of coated vesicles, structures that are involved in the selective intracellular transport of molecules such as hydrolytic enzymes.25 These vesicles originate either by invagination of coated regions of plasma membrane (“coated pits”) or from the Golgi apparatus26-28 and their unique polyhedral basket-like structure (seen by electron microscopy) is accounted for by trimolecular clathrin “triskelions” that constitute the basic unit of the polyhedral coat.29 30
In the present cases, the presence of almost all of the clathrin heavy chain in CLTCL-ALK hybrid, including the motifs responsible for triskelion assembly, presumably accounts for the unique granular ALK cytoplasmic immunostaining pattern reflecting an ALK-positive polyhedral coat on the surface of vesicles. Furthermore, clathrin coat formation is likely to mimic ligand binding and induce activation of the kinase function of CLTCL-ALK.
The overall 10-year survival rates of ALK-positive lymphomas is better than 70% versus less than 40% for ALK-negative anaplastic large cell lymphomas.31-33 The limited data available have shown no significant survival difference between lymphomas carrying theNPM-ALK gene and those with variant ALK fusion genes,31 34 these results suggesting that proteins that fuse to ALK are important in terms of activating the kinase but have little other influence on neoplastic cell behavior.
The present study further demonstrates that the expression of ALK protein by lymphoid cells (where ALK gene is normally silent) is always due to translocations that place the ALK gene under the control of genes encoding constitutively expressed proteins. The resultant fusion proteins contain oligomerization domains, which mimic ligand binding and thereby activate the ALK catalytic domain. Moreover, the present case of ALCL, associated with a novel CLTCL-ALKhybrid gene, indicates that different partners can promote oncogenic expression of the ALK intracytoplasmic domain and that the cellular localization of the fusion protein is strongly dependent on ALK partners. This study further emphasizes the usefulness of anti-ALK antibodies for detecting cases containing variant translocations involving the ALK gene.
Supported by the Projet Hospitalier de Recherche Clinique (PHRC 98), Ligue Nationale Contre le Cancer: Comités de la Haute Garonne, de l'Aveyron et du Gers, and the Leukemia Research UK Fund.
Reprints:Georges Delsol, Laboratoire d'Anatomie Pathologique, CHU Purpan, Place du Dr Baylac, 31059, Toulouse Cedex, France; e-mail:delsol.g@chu-toulouse.fr.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal