Abstract
Abnormality in the machinery of apoptosis is associated with a resistant phenotype of the tumor cell to chemotherapy. To determine the molecular basis of resistance to antitumor agent-induced apoptosis, we performed a complementary DNA (cDNA) subtractive hybridization with messenger RNA (mRNA) from human monocytic leukemia U937 and its variant UK711, which is resistant to apoptosis induced by antitumor agents. We found that glyoxalase I (GLO1), an enzyme that detoxifies methylglyoxal, is selectively overexpressed in the apoptosis-resistant UK711 cells. The GLO1 enzyme activity was significantly elevated in UK711 and UK110 cells, another drug-resistant mutant, as well as in K562/ADM, adriamycin-resistant leukemia cells, compared with their parental cells. When overexpressed in human Jurkat cells, GLO1 inhibited etoposide- and adriamycin-induced caspase activation and apoptosis, indicating the involvement of GLO1 in apoptosis suppression caused by these drugs. Moreover, cotreatment withS-p-bromobenzylglutathione cyclopentyl diester (BBGC), a cell-permeable inhibitor of GLO1, enhanced etoposide-induced apoptosis in resistant UK711 cells but not in parental U937 cells. Taken together, these results indicate that GLO1 is a resistant factor to antitumor agent-induced apoptosis in human leukemia cells and that the GLO1 inhibitor could be a drug resistance-reversing agent.
Emergence of resistance to antitumor agents is a serious problem in cancer chemotherapy. One mechanism of drug resistance is the modulation of gene expression. Overexpression of drug transporter proteins, such as P-glycoprotein and multidrug resistance-associated protein (MRP), has been observed in a number of tumor cells resistant to antitumor agents.1,2 Other reports show that the quantitative reduction of DNA topoisomerase I or II, molecular targets of antitumor agents such as camptothecin (CPT) and etoposide (VP-16), respectively, causes resistance to these agents to develop.3 4 In clinical situations, however, many tumors have been reported to acquire resistance to chemotherapy without undergoing these changes, suggesting that another mechanism could be involved.
Apoptosis is an active cell death mechanism that plays a role in several biologic processes. Various antitumor agents have been reported to elicit apoptosis in tumor cells.5,6 This implies that a blockade of the apoptosis signaling could be another mechanism for multidrug resistance. We previously isolated UK711,5 a mutant from human monocytic leukemia U937 cells, which showed resistance to apoptosis induced by such antitumor agents as VP-16, CPT, adriamycin (ADM), mitomycin C, and 1-(β-D-arabinofuranosyl) cytosine (Ara-C). In UK711 cells, the expressions of P-glycoprotein and DNA topoisomerase were comparable with those in parental U937 cells, and the initial DNA damage was induced equally in both cell lines after drug treatment. However, the drug-induced activation of caspases was significantly suppressed in UK711 cells.7-9 These observations suggest that the UK711 cells decreased susceptibility to apoptosis could be due to abnormality in cellular signaling, subsequent to initial DNA damage caused by the drugs.
To identify genes that are responsible for apoptosis resistance of the mutant cells upstream of caspase protease activation, we used a complementary (cDNA) subtraction approach using messenger (mRNA) from U937 and UK711 cells. As a result of the screening, we found that theglyoxalase I (GLO1) gene was overexpressed in UK711 cells. Here, we report that GLO1 was involved in apoptosis resistance to antitumor agents in human leukemia cells. Moreover, we demonstrated that the GLO1 inhibitor sensitized cells resistant to the chemotherapeutic agent.
Materials and methods
Materials
VP-16 was kindly provided by Bristol Myers-Squibb Co, Ltd (Tokyo, Japan). ADM was purchased from Sigma Chemical (St Louis, MO).S-p-bromobenzylglutathione cyclopentyl diester (BBGC) was synthesized, as described previously.10-12
Cell lines and cell culture
Human monocytic leukemia U937 cells, the human myelogenous leukemia K562 cells and the human T-cell leukemia Jurkat cells were grown in RPMI 1640 (Nissui Co, Ltd, Tokyo, Japan) supplemented with 10% heat-inactived fetal bovine serum and 100 μg/mL of kanamysin in a humidified atmosphere of 5% CO2 and 95% air. Antitumor drug-resistant variants, UK711, UK110, and K562/ADM were established and characterized as described.5,15 16 In brief, the mutagenized cells were exposed to VP-16 or ADM, and viable cells were cloned. The 3 resistant sublines displayed stable resistance.
Complementary DNA subtractive hybridization
mRNA was prepared from 108 of U937 or UK711 cells using a Fast Track mRNA isolation kit (Invitrogen, Carlsbad, CA). Subtracted cDNA fragments were obtained from mRNA of both U937 and UK711 cells using a PCR-Select cDNA subtraction kit (Clontech Laboratories, Tokyo, Japan). The cDNA fragments were further tested for differential expression by dot-blot analysis. Finally, the expression was confirmed by Northern blot analysis, as described previously.14Positive cDNA fragments were subcloned into pCRII vector (Invitrogen) and sequenced with an ABI PRISM Dye Primer Cycle Sequencing Kit (Applied Biosystems, Chiba, Japan) and an ABI PRISM 310 Genetic Analyzer (Applied Biosystems).
Glyoxalase I assay
Cytosolic fractions were prepared as previously described.15 Briefly, the cells were lysed in phosphate-buffered saline (PBS) containing 1 mmol/L phenylmethylsulfonyl fluoride (PMSF) by freezing and thawing and sonication, and then being centrifuged at 12 000g for 20 minutes. The supernatant was used as the cytosolic fraction. The GLO1 assay was performed in 7.9 mmol/L methylglyoxal, 1 mmol/L glutathione, 14.6 mmol/L magnesium sulfate, and 182 mmol/L imidazole-HCl (pH 7.0). An increase in absorbance at 240 nm due to the formation ofS-D-lactoylglutathione was measured with each cytosolic fraction.
Analysis of DNA fragmentation
After treatment with the drugs, cells were suspended in 20 μL of 50 mmol/L Tris-HCl (pH8.0), 10 mmol/L ethylenediamine tetraacetic acid (EDTA), and 0.5 mg/mL proteinase K (Sigma). They were incubated at 50°C for 1.5 hours, then 10 μL of 2 μg/mL RNase A solution was added, and the suspension was incubated for another 1.5 hours. The sample was mixed with 10 μL of the preheated (70°C) solution containing 10 mmol/L EDTA (pH 8.0) 1% low-melting point agarose, 0.25% bromphenol blue, and 40% sucrose. DNA was analyzed by electrophoresis in 2% agarose gels, stained with ethidium bromide, and then photographed on an ultraviolet transilluminator.
Assay of drug sensitivity
The sensitivity of tumor cell lines to drugs was evaluated by the inhibition of cell growth after 24-hour incubation with various concentrations of drugs. The number of viable cells was estimated by the [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] (MTS) method17 using CellTiter 96 AUueous 1 solution cell proliferation assay (Promega, Tokyo, Japan). The 50% inhibitory concentration (IC50) values were determined graphically from growth inhibition.
Transfection of glyoxalase I
cDNA of GLO1 open-reading frame was inserted into pFLAG (5a)-CMV expression vector. The pFLAG (5a)-CMV-GLO1 plasmid DNA was transiently transfected using DMRIE-C reagent (Gibco BRL), according to manufacturer's instructions, together with pHook-1 plasmid DNA (Invitrogen), which encodes a single-chain antibody fusion protein directed to the hapten phOx (4-ethoxymethlene-2-phenyl-2-oxazolin-5-1) and thereby allows the selective isolation of transfected cells with magnetic beads coated with phOx.18 GLO1-transfected cell populations were isolated on phOx-coated magnetic beads with the Capture-Teckit (Invitrogen). Cotransfection efficiency was more than 90%, as determined by β-galactosidase staining.
Flow cytometry and nuclear staining assays
After drug treatment, harvested cells were fixed in 70% ethanol. After treatment with 2 mg/mL RNase A solution, the cells were stained in 50 μg/mL of propidium iodide (PI) solution and then analyzed with a Becton Dickinson FACScan flow cytometer (Braintree, MA). To assay the nuclear morphology, harvested cells were washed with PBS and stained with 1 mmol/L Hechest 33342 for 15 minutes. The nuclear morphology of the cells was visualized using a fluorescene microscope (UFX-II A; Nikon, Tokyo, Japan).
Measurement of caspase activity
Drug-treated cell lysates were incubated with 20 mmol/L Acetyl-Asp-Glu-Val-Asp-α-(4-methyl-coumaryl-7-amide) (Ac-DEVD-MCA) at 37°C for 120 minutes, and the release of AMC was monitored by a spectrofluorometer (F-2500; Hitachi, Tokyo, Japan), using an excitation wavelength of 380 nm and an emission wavelength of 460 nm.
Western blot analysis
Western blot analysis was performed using an anti-FLAG (M2) monoclonal antibody (Sigma) according to the instructions of the manufacturer. Briefly, cell lysates were electrophoresed by SDS-PAGE and then transferred to a nitrocellulose membrane. After blocking, the membrane was incubated with primary antibody for 2 hours at 25°C. Detection was accomplished using an antimouse Ig-peroxidase conjugated and the enhanced chemiluminescene detection system (Amersham, Tokyo, Japan).
Statistics
Values are reported as means ± SD in triplicate. Statistical analysis of the data were performed using an unpaired Student ttest.
Results
Overexpression of glyoxalase I in drug-induced apoptosis-resistant leukemia cells
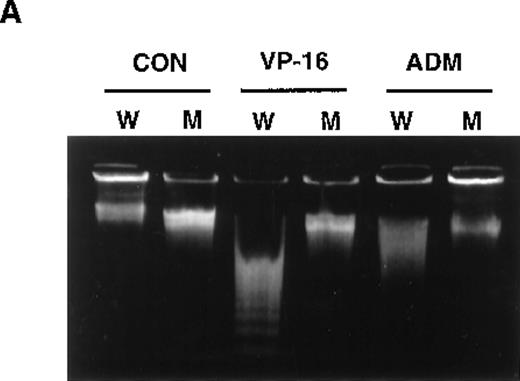
Previously, we isolated several mutants from U937 cells that showed resistance to multiple apoptosis inducers. Among them, UK711 showed significant resistance to apoptosis induced by several antitumor agents, including VP-16 and ADM (Figure1A), but not by death-receptor stimulation such as TNF-α or anti-Fas antibody (data not shown).5Because the initial DNA damage caused by the drugs was comparable in U937 and UK711 cells, cellular signaling leading to apoptosis could differ between the 2 cell lines.5
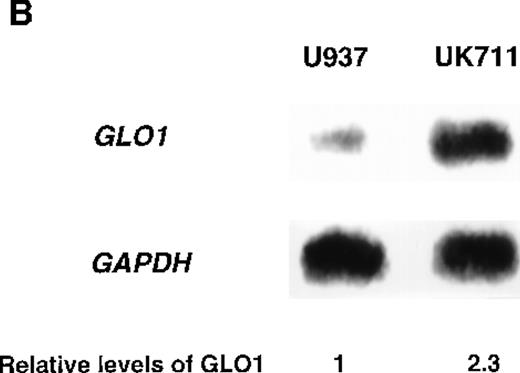
Overexpression of GLO1 in apoptosis-resistant UK711 cells.
(A) Wild-type U937 (W) and the mutant UK711 (M) cells were incubated with 5 μmol/L of VP-16 for 6 hours or 0.6 μmol/L of ADM for 12 hours. After the harvest of the cells, nuclear DNA fragmentation was analyzed by 2% agarose gel electrophoresis. (B) Northern blot analysis was carried out with mRNAs prepared from U937 and UK711 cells. The RNAs were transferred to nylon membranes and probed with the GLO1gene fragment (upper panels). Blots were also rehybridized with a GAPDH probe as a control (lower panels). Expression of the mRNA was quantified by densitometric analysis.
Overexpression of GLO1 in apoptosis-resistant UK711 cells.
(A) Wild-type U937 (W) and the mutant UK711 (M) cells were incubated with 5 μmol/L of VP-16 for 6 hours or 0.6 μmol/L of ADM for 12 hours. After the harvest of the cells, nuclear DNA fragmentation was analyzed by 2% agarose gel electrophoresis. (B) Northern blot analysis was carried out with mRNAs prepared from U937 and UK711 cells. The RNAs were transferred to nylon membranes and probed with the GLO1gene fragment (upper panels). Blots were also rehybridized with a GAPDH probe as a control (lower panels). Expression of the mRNA was quantified by densitometric analysis.
To identify genes that were responsible for apoptosis resistance of the mutant cells, we used a cDNA subtraction approach with mRNA from U937 and UK711 cells. Because treating the cells with 10 μg/mL of VP-16 for 1 hour was enough to commit U937 cells to apoptosis, mRNA was extracted from both U937 and UK711 cells after treatment with 10 μg/mL of VP-16 for 1.5 hours. After subtractive amplification of cDNAs overexpressed in UK711, the differential expression was further tested by dot-blot analysis, using the amplified gene fragments as probes. Of the 500 fragments, 8 were overexpressed in UK711. Among them, a fragment recognized about a 2.0-kilobase (kb) transcript on Northern blot analysis (Figure 1B), and it was identified with cDNA encoding human GLO1, an enzyme that detoxifies methylglyoxal, by comparison to standard sequencing databases in public domain (BLAST). Next, we examined the activity of GLO1 in the cells. As shown in Table 1, GLO1 enzyme activity was significantly elevated in UK711 cells, compared with parental U937 cells. The activity was also higher in UK110 cells, another mutant that showed resistance to apoptosis induced by VP-16, as well as by Fas receptor stimulation. Further studies with other leukemia cells revealed that K562/ADM (P-glycoprotein overexpressing cells),16 an ADM-resistant variant of human myelogenous leukemia K562, also expressed a higher GLO1 activity than wild-type K562 cells (Table 1). These results indicated that GLO1 is frequently overexpressed in drug-resistant leukemia cells.
GLO1 activity, drug resistance and P-glycoprotein expression in U937, K562 and their resistant cell lines
| Cell Lines . | GLO1 Activity (μmol/min/mg protein)* . | Degree of Resistance (fold)‡ . | P-Glycoprotein Expression1-153 . | |
|---|---|---|---|---|
| VP-16 . | ADM . | |||
| U937 | 8.79 ± 0.16 | 1.0 | 1.0 | − |
| UK711 | 13.63 ± 0.14† | 5.1 | 3.3 | − |
| UK110 | 18.65 ± 0.56† | 4.6 | 3.5 | − |
| K562 | 1.85 ± 0.20 | 1.0 | 1.0 | − |
| K562/ADM | 3.58 ± 0.27† | 9.4 | 27.3 | ++ |
| Cell Lines . | GLO1 Activity (μmol/min/mg protein)* . | Degree of Resistance (fold)‡ . | P-Glycoprotein Expression1-153 . | |
|---|---|---|---|---|
| VP-16 . | ADM . | |||
| U937 | 8.79 ± 0.16 | 1.0 | 1.0 | − |
| UK711 | 13.63 ± 0.14† | 5.1 | 3.3 | − |
| UK110 | 18.65 ± 0.56† | 4.6 | 3.5 | − |
| K562 | 1.85 ± 0.20 | 1.0 | 1.0 | − |
| K562/ADM | 3.58 ± 0.27† | 9.4 | 27.3 | ++ |
Cytosolic fractions were isolated, and enzyme assays were performed as described in “Materials and methods.” Values are means ± SD determined in triplicate.
P < 0.01 versus wild type (U937 or K562) cells, evaluated by an unpaired Student t test.
Cells were treated with VP-16 or ADM for 24 h. The number of viable cells was estimated by MTS tetrazolium method. Degree of resistance; IC50 resistant/IC50 parental cell line.
The expression of P-glycoprotein detected by Western blot analysis. The level of P-glycoprotein was then classified as follows: −, not observed; ++, overexpression.
Effect of overexpression of glyoxalase I on antitumor agent-induced apoptosis
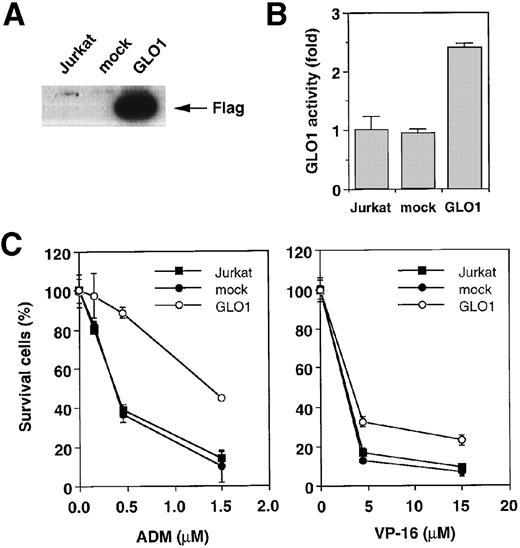
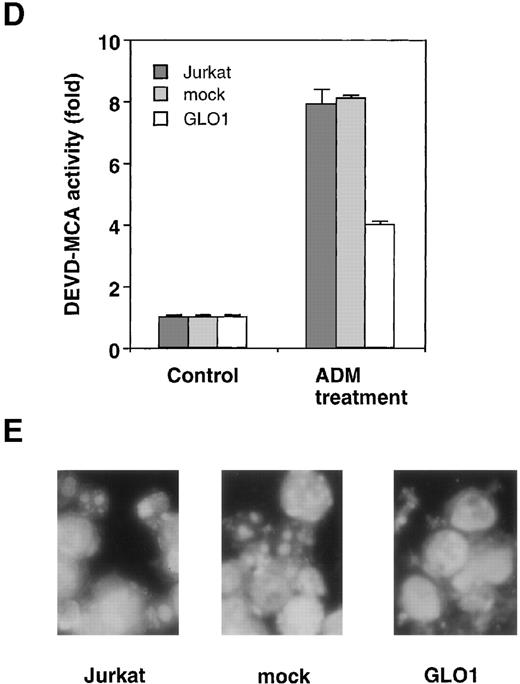
To explore the involvement of GLO1 in the resistance to antitumor agent-induced apoptosis, we introduced GLO1 cDNA with a FLAG epitope-tag at the C-terminus into Jurkat cells. GLO1 cDNA or an empty vector (mock) was transfected, together with pHook-1, and the transfected cells were selected using magnetic beads coated with phOx. Figure 2A shows the expression of FLAG-tagged GLO1 by Western blot analysis. In GLO1-transfected Jurkat cell population, there was an approximately 2.4-fold increase in GLO1 activity over that in parental cell line (Figure 2B). When the isolated transfectants were treated with ADM or VP-16, the cells expressing GLO1 showed significant resistance to these agents. As shown in Figure 2C, when the mock-transfected cells were treated with ADM at 0.45 μmol/L, 64% of the cells died. In contrast, the treatment killed only 11% of cells expressing GLO1. Similarly, the GLO1-expressing cells showed resistance to VP-16 treatment. To test whether the induction of apoptosis was suppressed, we further examined the activation of a caspase-3–like protease, as well as nuclear morphologic changes after drug treatment. As shown in Figure 2D, caspase activation was inhibited in GLO1-overexpressed cells after the treatment with ADM, and correspondingly, the apoptotic nuclear condensation was less eminent in GLO1-expressing cells than in mock-transfected cells (Figure 2E). These results indicated that GLO1 suppresses drug-induced apoptosis at a step upstream of caspase-3 activation.
Effect of overexpression of GLO1 on antitumor agent-induced apoptosis.
pFLAG (mock) or pFLAG-GLO1 (GLO1) expression plasmids were transiently cotransfected into Jurkat cells with pHook-1 as described in “Materials and methods,” and the transfected cells were selected using magnetic beads coated with phOx. (A) The expression of FLAG-tagged GLO1 in Jurkat cells was examined by anti-FLAG Western blot analysis. (B) Cytosolic fractions were isolated, and GLO1 activity was performed as described in “Materials and methods.” The means ± SD of triplicate cultures are shown. (C) Cells were treated with various concentrations of ADM or VP-16 for 24 hours, and cytotoxicity was determined by MTS method. The means ± SD of triplicate cultures are shown. (D) Cells were treated with 1.2 μmol/L of ADM for 14 hours, and caspase-3–like protease activity was measured with the specific fluorgenic substrate Ac-DEVD-MCA. Results are the means ± SD in triplicate. (E) Cells were treated with 0.5 μmol/L of ADM for 24 hours and were stained with Hechest 33342.
Effect of overexpression of GLO1 on antitumor agent-induced apoptosis.
pFLAG (mock) or pFLAG-GLO1 (GLO1) expression plasmids were transiently cotransfected into Jurkat cells with pHook-1 as described in “Materials and methods,” and the transfected cells were selected using magnetic beads coated with phOx. (A) The expression of FLAG-tagged GLO1 in Jurkat cells was examined by anti-FLAG Western blot analysis. (B) Cytosolic fractions were isolated, and GLO1 activity was performed as described in “Materials and methods.” The means ± SD of triplicate cultures are shown. (C) Cells were treated with various concentrations of ADM or VP-16 for 24 hours, and cytotoxicity was determined by MTS method. The means ± SD of triplicate cultures are shown. (D) Cells were treated with 1.2 μmol/L of ADM for 14 hours, and caspase-3–like protease activity was measured with the specific fluorgenic substrate Ac-DEVD-MCA. Results are the means ± SD in triplicate. (E) Cells were treated with 0.5 μmol/L of ADM for 24 hours and were stained with Hechest 33342.
Reversal of apoptosis resistance to antitumor agents by glyoxalase I inhibitor
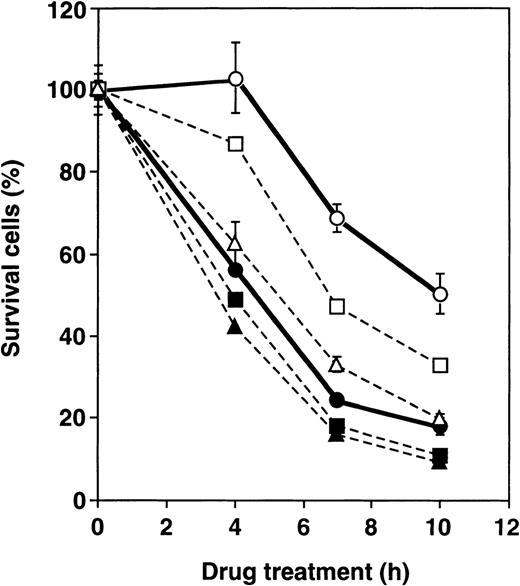
To verify the role of GLO1 further in apoptosis inhibition, we examined the effect of BBGC, a cell-permeable inhibitor of GLO1, on antitumor agent-induced apoptosis. Table 2shows the percentage of apoptotic cells after treatment with VP-16 alone, VP-16 in combination with BBGC and BBGC alone in U937, UK711, and UK110 cells. Both UK711 and UK110 cells that expressed higher levels of GLO1 than U937 cells showed resistance to VP-16–induced apoptosis, which was partially, but significantly, reversed in the presence of BBGC. Treatment of cells with BBGC alone for 4 hours at this concentration did not cause apoptosis induction. Consistently, caspase activation was enhanced in UK711 cells by cotreatment with BBGC and VP-16 (data not shown). Time course studies further revealed that, in UK711 cells, cotreatment with BBGC markedly accelerated VP-16–induced apoptosis in a concentration-dependent manner, whereas BBGC treatment was less effective in parental U937 cells (Figure3). These data indicate that inhibiting GLO1 with BBGC could serve as an effective strategy to kill drug-resistant cells that are overexpressing GLO1.
Reversal of apoptosis resistance by treatment with VP-16 in combination with BBGC
| Cell Lines . | Treatment . | Apoptotic Cell (%) . |
|---|---|---|
| U937 | none | 3.0 ± 0.1 |
| VP-16 | 39.1 ± 0.4 | |
| VP-16 + BBGC | 40.0 ± 0.3 | |
| BBGC | 5.3 ± 0.3 | |
| UK711 | none | 1.9 ± 0.1 |
| VP-16 | 5.6 ± 0.3 | |
| VP-16 + BBGC | 13.1 ± 1.4 | |
| BBGC | 3.8 ± 0.0 | |
| UK110 | none | 2.3 ± 0.1 |
| VP-16 | 26.4 ± 0.5 | |
| VP-16 + BBGC | 37.5 ± 0.4 | |
| BBGC | 4.3 ± 0.0 |
| Cell Lines . | Treatment . | Apoptotic Cell (%) . |
|---|---|---|
| U937 | none | 3.0 ± 0.1 |
| VP-16 | 39.1 ± 0.4 | |
| VP-16 + BBGC | 40.0 ± 0.3 | |
| BBGC | 5.3 ± 0.3 | |
| UK711 | none | 1.9 ± 0.1 |
| VP-16 | 5.6 ± 0.3 | |
| VP-16 + BBGC | 13.1 ± 1.4 | |
| BBGC | 3.8 ± 0.0 | |
| UK110 | none | 2.3 ± 0.1 |
| VP-16 | 26.4 ± 0.5 | |
| VP-16 + BBGC | 37.5 ± 0.4 | |
| BBGC | 4.3 ± 0.0 |
Cells were treated with 15 μmol/L of VP-16, 15 μmol/L of VP-16 in combination with 12.5 μmol/L BBGC, and 12.5 μmol/L BBGC alone for 4 h. The percentages of apoptotic cells were determined as subdiploid cells by flow cytometric analysis after staining with PI. The means ± S.D. of triplicate cultures are shown.
Effect of BBGC on sensitivity of U937 and UK711 cells to VP-16.
U937 (closed) and UK711 (open) cells were treated with 15 μmol/L of VP-16 combined with 0 (circle), 5 (squares), and 15 (triangles) μmol/L BBGC for the indicated periods. Cellular viability was determined by the MTS method. The means ± SD of triplicate cultures are shown.
Effect of BBGC on sensitivity of U937 and UK711 cells to VP-16.
U937 (closed) and UK711 (open) cells were treated with 15 μmol/L of VP-16 combined with 0 (circle), 5 (squares), and 15 (triangles) μmol/L BBGC for the indicated periods. Cellular viability was determined by the MTS method. The means ± SD of triplicate cultures are shown.
Discussion
Methylglyoxal is one of the side products of glycolysis that reacts with such biologic compounds as DNA, RNA, and protein in cells.19 Modifying DNA by means of methylglyoxal induced single-strand breaks and DNA-protein cross-link and cytotoxity.19 GLO1 is ubiquitously distributed in all mammalian cells and plays an important role in catalyzing the formation of S-D-lactoylglutathione from methylglyoxal and glutathione.20 Some groups have reported that tumor tissue expressed higher GLO1 activity than normal tissue. In particular, abnormal expression or activity of GLO1 has been demonstrated in human colon, renal, and prostate cancers.21-24 Thus, the increase of GLO1 expression has been shown to be associated with increased proliferative activity of the tumor. In this study, we found that, in antitumor agent-resistant leukemia cells, GLO1 was frequently overexpressed (Figure 1, Tables 1 and 2). Further, we found that GLO1 activity was also higher in CPT-resistant human colon cancer3 (HT-29/CPT) and CPT-resistant human gastric cancer cells3 (St-4/CPT) than their parental cell lines (data not shown). Because these mutant cell lines were cell populations that survived after treatment with an anticancer agent, it is possible that the development of drug resistance is accompanied by this overexpression of GLO1. These results suggested that GLO1 could be not only a tumor marker but also a drug resistance marker.
We further investigated the effect of GLO1 on antitumor agent-induced apoptosis in the human T-cell leukemia Jurkat cells. We showed that overexpression of GLO1 conferred resistance to ADM and VP-16 (Figure2B). Antitumor agents, such as ADM and VP-16, induce apoptosis through caspase activation in a number of leukemia cells. In GLO1-overexpressed cells, the activation of caspase-3–like protease after drug treatment was significantly suppressed (Figure 2C). Similar results were obtained with the GLO1-transfected HT-1080 human fibrosarcoma cells (data not shown). These results indicated that GLO1 blocked a signaling pathway of antitumor agent-induced apoptosis at a step upstream of caspase activation.
At present, we do not know the target(s) of GLO1 responsible for apoptosis induction. One possibility is that GLO1 inhibits apoptosis by detoxifying methylglyoxal, because methylglyoxal modifies cellular macromolecules and causes damage to the cells. Correspondingly, we found that treating of U937 cells with methylglyoxal induced caspase activation (data not shown). However, we could not exclude the possibility that GLO1 could modify other unidentified factor(s) that play critical roles in apoptosis induction. At least, it is not likely that GLO1 inhibits the accumulation of the drugs because, in the GLO1-overexpressed UK711 cells, initial DNA damage was induced to the same extent as in parental U937 cells.5 Further studies are needed to clarify the role of GLO1 in this process.
GLO1 inhibitor diesters such as BBGC andS-(N-Aryl-N-hydroxycarbamoyl) glutathione diethyl ester have potential antiproliferative activities in vitro and in vivo.12 25 Our results suggest that a GLO1 inhibitor could be an effective modulator of apoptosis resistance to antitumor agents for the following reasons. First, overexpression of GLO1 may frequently occur in apoptosis-resistant leukemia cells. Second, the GLO1 inhibitor selectively sensitized ADM- and VP-16–resistant cells with high GLO1 expression. Because the expression of GLO1 was shown to be lower in normal tissue than in tumors, the inhibitor compounds could be beneficial for clinical reversal of drug resistance.
In summary, our studies indicate that GLO1 is an apoptosis-resistant factor in leukemia cells and that a GLO1 inhibitor is capable of reversing apoptosis resistance. Further studies will elucidate the target of GLO1 that is responsible for the induction of apoptosis.
Acknowledgments
We thank Drs H. Seimiya, N. Fujita, and A. Tomida for helpful discussions.
Supported in part by the Program for Promotion of Fundamental Studies in Health Sciences of the Organization for Pharmaceutical Safety and Research of Japan, Grants-in-Aid for Cancer Research and Scientific Research from the Ministry of Education, Culture and Science of Japan, a grant from the Vehicle Racing Commemorative Foundation, and Public Trust Haraguchi Memorial Cancer Research Fund.
Reprints:Takashi Tsuruo, Institute of Molecular and Cellular Biosciences, The University of Tokyo, Tokyo, 1-1-1 Yayoi, Bunkyo-ku, Tokyo 113-0032, Japan; e-mail: ttsuruo@iam.u-tokyo.ac.jp.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal