Abstract
Genes of the MAGE-A family are expressed in several types of solid tumors but are silent in normal tissues with the exception of male germline cells, which do not carry HLA molecules.Therefore, peptides encoded by MAGE-A genes are strictly tumor-specific antigens that can be recognized by CTL and constitute promising targets for immunotherapy. The expression of 6 genes of the MAGE-A family was tested with reverse transcriptase-polymerase chain reaction in lymphoma samples. Among 38 samples of non-Hodgkin lymphoma, 1 anaplastic large cell lymphoma expressed genes MAGE-A1, -A2, -A3, -A4, and -A12, and 1 lymphoepithelioid T-cell lymphoma expressed geneMAGE-A4. Five of 18 samples (28%) from patients with Hodgkin disease expressed gene MAGE-A4. In tissue sections, staining by a monoclonal antibody that recognizes the MAGE-A4 protein was observed in 11 of 53 samples (21%) from patients with Hodgkin disease. In the positive samples, the Reed-Sternberg cells were strongly stained whereas the surrounding cells were not. These results indicate that Hodgkin disease may be a target for specific immunotherapy involving MAGE-A4 antigens.
Intensive chemotherapy and bone marrow transplantation have improved the disease-free survival of patients with non-Hodgkin lymphoma (NHL) and Hodgkin disease (HD). However, the relapse rate remains important, particularly in NHL. Clinical experience with allogeneic bone marrow transplantation demonstrated that immune-mediated graft-versus-lymphoma reactions may be induced in NHL patients, a situation that may be analogous to the graft-versus-leukemia (GVL) effect that participates in the cure of leukemia patients.1-6 Immunotherapy involving the administration of immunostimulatory cytokines or donor lymphocytes seemed to improve the clinical outcome of a few lymphoma patients.7 8 However, optimizing the antilymphoma effect of allogeneic reactions is clearly difficult, considering the mortality and morbidity associated with graft-versus-host disease. New immunotherapeutic tools would be necessary, at least as adjuvant treatment, to control residual disease.
Tumor-specific antigens consisting of peptides presented to autologous cytotolytic T lymphocytes by class I human leukocyte antigens (HLAs) may constitute targets for specific immunotherapy.9 Several types of antigens present on leukemia, lymphoma, or melanoma cells have been described: chimeric proteins resulting from gene translocations (for example, PML-RARα in acute promyelocytic leukemia and p210 [BCR-ABL] in chronic myelocytic leukemia); proteinase 3, which is present in leukemia cells but also in normal promyelocytes; the mucin MUC-1 in myeloma cells; the PRAME protein that appears to be overexpressed in many acute leukemias; or viral proteins such as LMP1 and LMP2 in lymphomas associated with Epstein-Barr virus infection.10-16
An interesting category of tumor antigens is encoded by genes such asMAGE, BAGE, GAGE, or SSX, which are expressed in many types of solid tumors but are silent in normal tissues with the exception of male germinal cells.9,17-19Because these cells do not carry HLA molecules, they cannot present antigenic peptides. Antigens encoded by such “cancer/testis” genes are therefore strictly tumor-specific and can be used safely in immunotherapy trials. Regressions of metastatic melanoma lesions were reported after immunization with an antigenic peptide encoded by geneMAGE-A3 and presented by HLA-A1 molecules.20
MAGE-A is 1 of the 4 known MAGE gene families, and it comprises 12 genes. We reported that gene MAGE-A1 was silent in bone marrow or blood samples from 48 leukemia patients.21The only hematologic malignancies in which MAGE gene expression has been detected are adult T-cell acute lymphoblastic leukemia and multiple myeloma.22-24 Among the members of the SSX genes family, SSX-2 was shown to be expressed in 4 of 11 NHL samples. Here we report on the analysis ofMAGE-A gene expression in lymph node biopsies from lymphoma patients.
Patients, materials, and methods
Control cell lines
Melanoma cell line MZ2-MEL-3.0 expresses genes MAGE-A1, -A2, -A3, and -A6 but not MAGE-A4. Sarcoma cell line LB23-SARC expresses MAGE-A4 but not MAGE-A1, -A2, -A3, -A6, or -A12. Melanoma cell line LB373-MEL expresses geneMAGE-A12. These cell lines were used as positive controls in the reverse transcriptase-polymerase chain reaction (RT-PCR) assays ofMAGE-A genes expression.
Normal lymphoid cells
Samples of peripheral blood mononuclear cells (PBMCs) (n = 8), phytohemaglutanin (PHA)-activated blood T cells (n = 3), normal bone marrow (n = 5), normal lymph nodes (n = 5), and cord blood cells (n = 4) from 25 patients without leukemia or lymphoma were used as controls in RT-PCR assays.
Patients
Fifty-six lymphoma patients were included in the study: 38 with NHL and 18 with HD. Diagnosis was based on conventional morphologic examination of paraffin-embedded material, fixed with formalin or Bouin's fluid. The slides were examined by 2 pathologists (L.X. and I.T.) and, when required, diagnoses were refined with immunohistochemistry using monoclonal antibodies (MAbs) recognizing B cells, T cells or Reed-Sternberg cells. According to the REAL classification,25 these lymphoid neoplasms were distributed as follows: 29 B-cell NHLs (2 small lymphocytic lymphomas, 5 mantle cell lymphomas, 14 follicle center lymphomas, 1 marginal zone B-cell lymphoma, 6 diffuse large B-cell lymphomas, and 1 plasmacytoma/plasma cell lymphoma), 9 T-cell NHLs (3 precursor T-lymphoblastic lymphomas, 5 anaplastic large cell lymphomas, and 1 lymphoepithelioid peripheral T-cell lymphoma), and 18 HD samples: 2 (HD1 and HD2) of type I (lymphocytic predominance), 8 (HD3-10) of type II (nodular sclerosis), 7 (HD11-17) of type III (mixed cellularity), and 1 (HD18) of type IV (lymphocyte depletion). Fifty-three HD samples, including 3 of type I, 34 of type II, 14 of type III, and 2 of type IV, were used for the immunohistochemical analysis. HD samples from 2 HD patients (HD3 and HD4) belonged to both series (RT-PCR analysis and immunohistochemistry).
Lymph node biopsies
Lymph nodes were either frozen in liquid nitrogen immediately after surgical excision, or they were minced to obtain a suspension of cells that were frozen in RPMI (GIBCO BRL, Gaithersburg, MD) with 37.5% (vol/vol) fetal calf serum (GIBCO BRL) and 10% dimethyl sulfoxide and stored in liquid nitrogen. For patient HD3, cryopreserved lymph node cells were thawed, treated by trypsin ethylenediaminetetraacetic acid, and separated by positive immunoselection with CD30 MAb and goat antimouse magnetic beads to obtain fractions enriched or depleted in CD30+ cells before RNA extraction.
RT-PCR assay
Total cellular RNA was isolated by the guanidine-isothiocyanate/cesium chloride procedure26 or with TRIzol (GIBCO BRL). Complementary DNA (cDNA) from 2 μg of total RNA was obtained by incubation at 42°C for 90 minutes with oligo(dT) primer and 200 units of Moloney murine leukemia virus RT (GIBCO BRL). PCR amplification was performed on 2.5% of the cDNA with 0.625 units of Taq DNA polymerase in a final volume of 25 μL. Reaction mixtures were heated at 94°C for 4 minutes and subjected to amplification for 30 (MAGE-A1 to -A6) or 32 cycles (MAGE-A12) consisting of 1 minute at 94°C; 1 minute (2 minutes for MAGE-A6 and -A12) at 72°C (MAGE-A1 and -A3), 70°C (MAGE-A6), 68°C (MAGE-A4), 67°C (MAGE-A2), or 62°C (MAGE-A12); and 1 minute (2 minutes for MAGE-A6 or 3 minutes for MAGE-A12) at 72°C. Final extension was obtained with 15 minutes at 72°C. TheMAGE-specific primers were described previously,18,27 except for MAGE-A12: 5′-GCCCTCCACTGATCTTTAGCAA-3′ (exon 3)18 and 5′-CGTTGGAGGTCAGAGAACAG-3′ (exon 1). Each primer was chosen in a different exon to avoid false positives caused by DNA contamination of the RNA preparation. Samples with degraded RNA were excluded by testing the expression of the β-actin gene. The amounts of amplified products were evaluated visually on ethidium bromide–stained agarose gels and identified by comparing the size of the band with the molecular weight marker ΦX174 (GIBCO BRL). Bands of test samples were compared with those resulting from RT-PCR performed on serial dilutions (1:1, 1:3, 1:9, 1:27) of RNA from a positive control cell line: MZ2-MEL-3.0 for MAGE-A1, -A2, -A3, -A6; LB23-SARC for MAGE-A4; and LB373-MEL for MAGE-A12.
Immunohistochemistry
Lymph node biopsy specimens from HD were formalin-fixed and embedded in paraffin. Immunohistochemical detection of the MAGE-A4 protein was performed on paraffin sections previously heated for 10 minutes in the ChemMate Buffer for Antigen Retrieval (DAKO, Trappes, France) with the mouse MAb 57B.28 The positive control was a sample of normal testis, whereas negative control consisted of replacing antibody 57B by an irrelevant isotype-matched MAb. The slides were processed using the alkaline phosphatase LSAB+ detection kit (DAKO) and the New Fuschin Substrate System (DAKO), according to the manufacturer's recommendations, and counterstained with hematoxylin.
Results
RT-PCR analysis of MAGE-A expression in lymph nodes from NHL patients
We tested 38 samples corresponding to various types of NHL. None of the 29 B-cell lymphomas expressed any of the 6 MAGE-A genes that were tested, whereas 2 of the 9 T-cell lymphomas were positive for at least 1 MAGE-A gene (Table 1). One anaplastic large-cell lymphoma expressed genes MAGE-A1, -A2, -A3, -A4, and -A12 at a significant level, whereas expression of gene MAGE-A6 was not detected. A low amount ofMAGE-A4 transcript was detected in 1 sample of lymphoepithelioid peripheral T-cell lymphoma (Figure1).
Expression of MAGE-A genes
| Patients . | Pathology (according to the REAL classification) . | Fraction of MAGE-positive samples . |
|---|---|---|
| B-cell NHL (n = 29) | Small lymphocytic lymphoma | 0/2 |
| Mantle cell lymphoma | 0/5 | |
| Follicle center lymphoma | 0/14 | |
| Marginal zone B-cell lymphoma | 0/1 | |
| Diffuse large B-cell lymphoma | 0/6 | |
| Plasmacytoma/plasma cell lymphoma | 0/1 | |
| Total B-cell NHL | 0/29 | |
| T-cell NHL (n = 9) | Precursor T-lymphoblastic lymphoma | 0/3 |
| Anaplastic large cell lymphomas | 1/5* | |
| Lymphoepithelioid peripheral T-cell lymphoma | 1/1* | |
| Total T-cell NHL | 2/9 (22.2%) | |
| HD (n = 18) | Type I (lymphocyte predominance) | 0/2 |
| Type II (nodular sclerosis) | 4/8* | |
| Type III (mixed cellularity) | 1/7* | |
| Type IV (lymphocyte depletion) | 0/1 | |
| Total HD | 5/18 (27.8%) |
| Patients . | Pathology (according to the REAL classification) . | Fraction of MAGE-positive samples . |
|---|---|---|
| B-cell NHL (n = 29) | Small lymphocytic lymphoma | 0/2 |
| Mantle cell lymphoma | 0/5 | |
| Follicle center lymphoma | 0/14 | |
| Marginal zone B-cell lymphoma | 0/1 | |
| Diffuse large B-cell lymphoma | 0/6 | |
| Plasmacytoma/plasma cell lymphoma | 0/1 | |
| Total B-cell NHL | 0/29 | |
| T-cell NHL (n = 9) | Precursor T-lymphoblastic lymphoma | 0/3 |
| Anaplastic large cell lymphomas | 1/5* | |
| Lymphoepithelioid peripheral T-cell lymphoma | 1/1* | |
| Total T-cell NHL | 2/9 (22.2%) | |
| HD (n = 18) | Type I (lymphocyte predominance) | 0/2 |
| Type II (nodular sclerosis) | 4/8* | |
| Type III (mixed cellularity) | 1/7* | |
| Type IV (lymphocyte depletion) | 0/1 | |
| Total HD | 5/18 (27.8%) |
Expression of MAGE-A genes was determined semiquantitatively with RT-PCR as explained in the legend to Figure 1.
Levels of expression of the individual MAGE-A genes in the positive samples were as follows: 1 anaplastic large cell lymphoma:MAGE-A1+, A2+, A3+, A4+, andA12+; 1 lymphoepithelioid cell lymphoma: MAGE-A4±; 4 HD type II: 1 MAGE-A4++ (HD4), 1 MAGE-A4+ (HD5), and 2MAGE-A4± (HD3, HD6); and 1 HD type III: MAGE-A4+ (HD17).
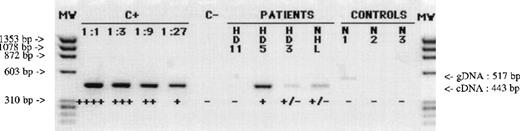
RT-PCR amplification products of gene MAGE-A4.
Products were obtained with lymph node samples from patients with HD (HD11, HD5, HD3) or NHL and obtained with samples of normal lymphoid cells (N1, lymph node; N2, PBMC; N3, PHA-activated peripheral T cells). C+, positive control sarcoma line LB23-SARC. C−, negative control (no RNA present in the RT reaction). Amplification of MAGE-A4transcripts gives a band of 443 base pairs, distinct from that of the amplified genomic sequence of 517 base pairs (faintly visible in line N1). The levels of gene expression were evaluated by comparing band intensities with those obtained for serial dilutions (1:1, 1:3, 1:9, 1:27) from the positive and scored as follows : ++++, 100%; +++, 30% to 100%; ++, 10% to 30%; +, 3% to 10%; or ±, less than 3%. Absence of RT-PCR product is indicated as −. RNA integrity was assessed by RT-PCR amplification of the β-actin mRNA (data not shown).
RT-PCR amplification products of gene MAGE-A4.
Products were obtained with lymph node samples from patients with HD (HD11, HD5, HD3) or NHL and obtained with samples of normal lymphoid cells (N1, lymph node; N2, PBMC; N3, PHA-activated peripheral T cells). C+, positive control sarcoma line LB23-SARC. C−, negative control (no RNA present in the RT reaction). Amplification of MAGE-A4transcripts gives a band of 443 base pairs, distinct from that of the amplified genomic sequence of 517 base pairs (faintly visible in line N1). The levels of gene expression were evaluated by comparing band intensities with those obtained for serial dilutions (1:1, 1:3, 1:9, 1:27) from the positive and scored as follows : ++++, 100%; +++, 30% to 100%; ++, 10% to 30%; +, 3% to 10%; or ±, less than 3%. Absence of RT-PCR product is indicated as −. RNA integrity was assessed by RT-PCR amplification of the β-actin mRNA (data not shown).
RT-PCR analysis of MAGE-A expression in lymph nodes from HD patients
Eighteen samples were tested. Gene MAGE-A4 was found to be expressed in 5 of them, whereas the other MAGE-A genes were never detected (Table 1). The levels of MAGE-A4 gene expression were assessed semiquantitatively as indicated in Table 1 and Figure 1. Among the 4 samples of HD type II that were tested, 1 (HD4) expressed gene MAGE-A4 at a level that corresponded to 10% to 30% of that found in the reference sarcoma cell line, whereas the other 3 expressed 3% to 10% (HD5) or less than 3% of that level (HD3 and HD6). The positive sample of HD type III (HD17) expressed 3% to 10% of that level. Representative results are shown in Figure 1 for patient HD11 (−), HD5 (+), and HD3 (±). Expression of genesMAGE-A1, -A2, -A3, -A4, -A6, and -A12 was not detected in any of the 25 samples of normal hematopoietic cells (PBMCs, PHA-activated peripheral T cells, bone marrow, benign lymph node, cord blood cells) (Figure 1).
We used a monoclonal antibody against CD30/Ki-1, which is present on Reed-Sternberg cells, to sort CD30+ and CD30−cells from a lymph node of patient HD3. MAGE-A4expression, analyzed with RT-PCR on the 2 cell populations, was restricted to the CD30+ cells (data not shown).
Immunohistochemical detection of the MAGE-A4 protein in HD lymph nodes
The low level of expression of MAGE-A4 detected in 4 of 5 lymph nodes from HD patients could result either from a low level of transcription of the gene in most of the cells present in the sample or from a higher expression by a minority of the cells. The latter possibility was obviously relevant, considering that Reed-Sternberg cells represent usually less than 5% to 10% of the total cell population in HD lymph nodes. We therefore analyzed paraffin sections of lymph nodes from 53 HD patients with MAb 57B. This antibody recognizes cells transfected with cDNAs encoding MAGE-A1, -A2, -A3, -A4, -A6, and -A12 in tissue sections, and it stains the tumors that express gene MAGE-A4 and only these tumors, regardless of the expression of the other MAGE-Agenes.28
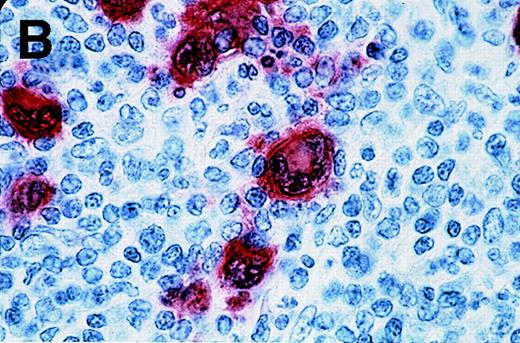
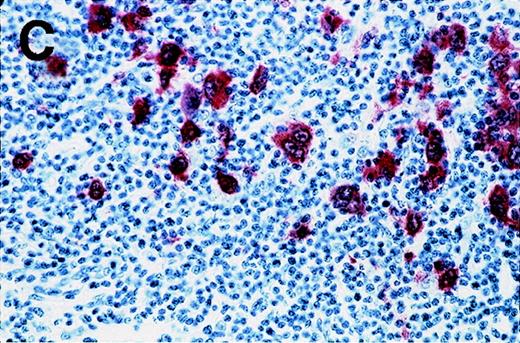
Antibody 57B stained 11 of 53 HD samples (21%), whereas no sample was stained by an isotype-matched control antibody. The positive samples included 1 of 3 HD type I, 8 of 34 HD type II, 2 of 14 HD type III, and 0 of 2 HD type IV. Representative results from patients HD3 and HD4 are shown in Figure 2. Positive cells are distributed in the different nodules (Figure 2A). At higher magnification, the staining is clearly observed in Reed-Sternberg cells (Figure 2B and 2C). Even though a weak background staining is present around some Reed-Sternberg cells, the reactive small lymphocytes and histiocytes are not stained.
Labeling of Reed-Sternberg cells with anti–MAGE-A4 MAb 57B in nodular sclerosing lymph nodes.
Patients include HD3 (A and B) and HD4 (C) at low (A), high (B) and intermediate (C) magnification. (B) and (C) correspond to MAGE-A4expression in HD3. Formalin-fixed sections were deparaffinized, heated in a citrate buffer, and incubated with MAb 57B. The antibody was detected with biotinylated secondary antibodies and avidin-coupled alkaline phosphatase. The chromogenic substrate was New Fuschin, and hematoxylin was used to counterstain the tissue.
Labeling of Reed-Sternberg cells with anti–MAGE-A4 MAb 57B in nodular sclerosing lymph nodes.
Patients include HD3 (A and B) and HD4 (C) at low (A), high (B) and intermediate (C) magnification. (B) and (C) correspond to MAGE-A4expression in HD3. Formalin-fixed sections were deparaffinized, heated in a citrate buffer, and incubated with MAb 57B. The antibody was detected with biotinylated secondary antibodies and avidin-coupled alkaline phosphatase. The chromogenic substrate was New Fuschin, and hematoxylin was used to counterstain the tissue.
Discussion
We report the expression of MAGE-A genes in 7 of 56 lymph nodes from lymphoma patients. Wide differences were observed according to the type of pathology. Whereas B-cell lymphomas did not express any of the 6 MAGE-A genes that were tested, expression was found in 2 of 9 T-cell lymphomas. A higher proportion of positive samples has been reported previously in T-cell leukemias, which expressed geneMAGE-A1.23 A notable difference between the 2 diseases is the pathogenesis of the T-cell leukemias, namely its frequent association with human T cell leukemia-lymphoma virus (HTLV-1) in Japan and the Philippines but not in European countries.
In 20% to 25% of lymph nodes from HD patients, we found expression of gene MAGE-A4 but not of any of the other members of theMAGE-A gene family that were tested. The level ofMAGE-A4 expression was at least 10 times lower than that found in a control sarcoma cell line. These results matched with the observation that the MAGE-A4 protein was detected only in Reed-Sternberg cells, which represent only 5% to 10% of the total cell population in most samples of HD lymph nodes. The proportion of samples that expressed gene MAGE-A4 (28%), detected with RT-PCR, was similar to the proportion of samples in which a specific staining was observed with antibody 57B (21%). In addition, in the 2 positive samples that were analyzed with the 2 techniques,MAGE-A4 expression was associated with the staining of Reed-Sternberg cells by the anti–MAGE-A4 antibody. This MAGE-A4 positivity restricted to Reed-Sternberg cells validates the concept that HD patients whose lymph node samples display low levels ofMAGE-A4 gene expression (less than 3% of the level found in the reference cell line) should also be considered eligible forMAGE-A4–based immunotherapy. Our results are in line with the recent observation, based on a global gene expression analysis, thatMAGE-A4 is expressed in Reed-Sternberg cells.29
Interestingly, all 7 MAGE-A–positive lymphoma samples identified in this study expressed MAGE-A4, but only 1 of them expressed other MAGE-A genes in addition to MAGE-A4. All 5 MAGE-A–positive HD samples expressed onlyMAGE-A4. This observation contrasts with the frequent coexpression of several MAGE-A genes in solid tumors.18 Unlike the other MAGE-A genes,MAGE-A4 contains several alternative promoters.30It is therefore possible that 1 of them is specifically activated in some lymphoma cells, possibly through lymphoma-specific transcription factors.
Reed-Sternberg cells carry HLA class I molecules, have functional TAP1 (transporter-associated protein) and TAP2 transporters, and bear the B7.1 and B7.2 costimulatory molecules.31-34 Therefore, they may be targets of anti–MAGE-A4 T-cell responses. Reed-Sternberg cells from Epstein-Barr virus–positive patients were shown to be lysed by cytolytic T lymphocytes recognizing peptides encoded by the LMP1 orLMP2 genes of Epstein-Barr virus, indicating that these cells were indeed capable to process and present antigenic peptides derived from endogenous proteins.16 35 It is therefore worth considering the possibility of vaccinating Hodgkin patients, selected on the basis of the expression of gene MAGE-A4 by their tumor cells, with MAGE-A4–encoded antigens.
Acknowledgments
We are particularly grateful to Mrs. M. Swinarska and M. Van Malderen for excellent technical assistance. We thank Dr R. Costello for helpful discussions.
Supported in part by a Swiss National Fonds grant (no. 31-57′473.99) and by grants from the Association contre le Cancer, Brussels, Belgium; from CGER-Assurances and VIVA, Brussels, Belgium; from the Fonds National de la Recherche Scientifique (TELEVIE grants), Brussels, Belgium; and from the Ligue Nationale contre le Cancer, France.
Submitted October 4, 1999; accepted January 30, 2000.
Reprints:Daniel Olive, INSERM U119 and Institut Paoli Calmettes, 27 Boulevard Leı̈ Roure, Marseille, France; e-mail:olive@marseille.inserm.fr.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal