Abstract
Mammalian β-globin loci are composed of multiple orthologous genes whose expression is erythroid specific and developmentally regulated. The expression of these genes both from the endogenous locus and from transgenes is strongly influenced by a linked 15-kilobase region of clustered DNaseI hypersensitive sites (HSs) known as the locus control region (LCR). The LCR encompasses 5 major HSs, each of which is highly homologous among humans, mice, and other mammals. To analyze the function of individual HSs in the endogenous murine β-globin LCR, we have used homologous recombination in embryonic stem cells to produce 5 mouse lines, each of which is deficient for 1 of these major HSs. In this report, we demonstrate that deletion of the conserved region of 5′HS 1, 2, 3, 4, or 5/6 abolishes HS formation at the deletion site but has no influence on the formation of the remaining HSs in the LCR. Therefore, in the endogenous murine locus, there is no dominant or initiating site whose formation must precede the formation of the other HSs. This is consistent with the idea that HSs form autonomously. We discuss the implications of these findings for current models of β-globin regulation.
The regulation of the β-like globin genes is of significant interest because it is a paradigm for a complex multigenic and developmentally regulated locus1 and because of the prevalence of mutations causing β-thalassemia and sickle cell anemia. The identification of deletions upstream of the embryonic ε-globin gene that left all of the β-like globin genes intact but that eliminated expression of all of the genes implied the existence of cis-acting regulatory elements in this region.2-4 A series of erythroid-specific DNaseI hypersensitive sites (HSs) were identified in this region,5,6 which subsequently was named the locus control region (LCR). Six 5′HSs have been described in the human and mouse, and homologues to at least 4 have been detected to date in other mammals.7-9 In the mouse, 5′HS 5 forms very weakly, and 5′HS 6 is prominent, while in humans the opposite pattern is observed.
A great number of experiments have been performed in attempts to determine how the LCR influences expression of genes of the β-globin locus located 6 to 50 kilobases (kb) away. The majority of these experiments have used human β-globin transgenes in cell lines or transgenic mice. Interpretation of the transgenic studies have been complicated by variations in integration site, copy number, size of constructs, and the exact deletions made. However, in aggregate, analyses of large transgenes containing sequences from the human β-globin locus have revealed that loci with wild-type LCRs express at the vast majority of integration sites, but not all integration sites, at levels that are within a twofold to fourfold range relative to the number of integrated copies.10-13 In contrast, when 5′HS 1, 2, 3, or 4 is mutated, there is greater variability among lines, and expression levels are often distinctly reduced.14-17
The sequence—the arrangement of gene and regulatory elements—and chromatin structure of the β-globin locus are highly conserved among mammals. This conservation is present within the LCR as well as within the coding regions. Within the LCR, the highest homology is found near the sequences at which the HSs actually form, in what are termed the cores of the HSs. The regions immediately flanking the cores of the HSs have less homology, while regions between HSs have no substantial homology. Sequence and structural conservation extends upstream of the LCR and downstream of the coding regions and includes several orthologous olfactory receptor genes that flank the human and mouse loci.9 The high degree of homology observed between species suggests conservation of function. To gain insights into the regulation of mammalian β-globin loci, we have generated mice with targeted deletions of the individual HSs of the endogenous murine LCR8,18 19 (also in M.A.B., unpublished data). In contrast to the analysis of human β-globin locus transgenes, where one must always consider integration site effects when interpreting results, the HR approach permits analysis of the contribution of the LCR to the function of the endogenous mouse β-globin locus in its native genomic, cellular, and developmental state.
To determine how any single HS influences the formation of other HSs in the native murine locus, we have performed a DNaseI HS formation analysis of mice with each of the individual HS deletions. We show that while the formation of each site is abolished when its core and flanking sequences are deleted, the deletion of any individual HS does not noticeably influence the formation of the remaining HSs. Thus, no single HS is required for the formation of the other HSs in the endogenous locus. These results are most compatible with a model in which multiple elements composing the LCR act independently to alter chromatin structure and to contribute to the level of expression of the linked β-like globin genes.
Materials and methods
Production of mutant mice
Mutant mice were produced by performing homologous recombination in embryonic stem (ES) cells and deriving chimeric mice that transmitted the ES cell genome through the germline. The details of production of the 5′HS 2, 5′HS 3, and 5′HS 5/6 deletion mice were reported previously.8,18 19 The descriptions of production and transcriptional characterization of the 5′HS 1 and 5′HS 4 deletion mice are in preparation. These latter deletions remove nucleotides −4675 to −6999 and −19 849 to −22 561 relative to the Ey cap site, respectively. All selectable markers used were flanked by recombinase target sites for either Flp or Cre site-specific recombinases and were deleted by expression of the recombinases specific to either the Flp (FRT) or the Cre (loxP) site. Correct structure of the targeted alleles was confirmed by Southern blotting.
DNase I hypersensitivity assay
Nuclei were isolated from spleens of mice after phenylhydrazine treatment to induce anemia, at which time more than 50 percent of cells are erythroid. The phenylhydrazine treatment, DNaseI digestion, and Southern blotting were done as described.20 Probes used span the following bases relative to the Ey cap site (−1/+1): probe 1 (−2867 to −3469), probe 2 (−16 779 to −17 656), probe 3 (−20 508 to −222), and probe 4 (−27 981 to −28 567) (Figure1).
Map of the murine β-globin locus.
The top line diagrams the entire locus, including the LCR and the expressed genes. The bottom line is an expansion of the LCR region of the top line showing the restriction fragments and location of probes used in this study and the sites of various deletions. The deleted sequences are specified with negative numbers representing the number of bases from each side of the deletion to the cap site of the Ey messenger RNA, which is designated as −1/+1.
Map of the murine β-globin locus.
The top line diagrams the entire locus, including the LCR and the expressed genes. The bottom line is an expansion of the LCR region of the top line showing the restriction fragments and location of probes used in this study and the sites of various deletions. The deleted sequences are specified with negative numbers representing the number of bases from each side of the deletion to the cap site of the Ey messenger RNA, which is designated as −1/+1.
Results
Formation of 5′HS 1, 2, and 3
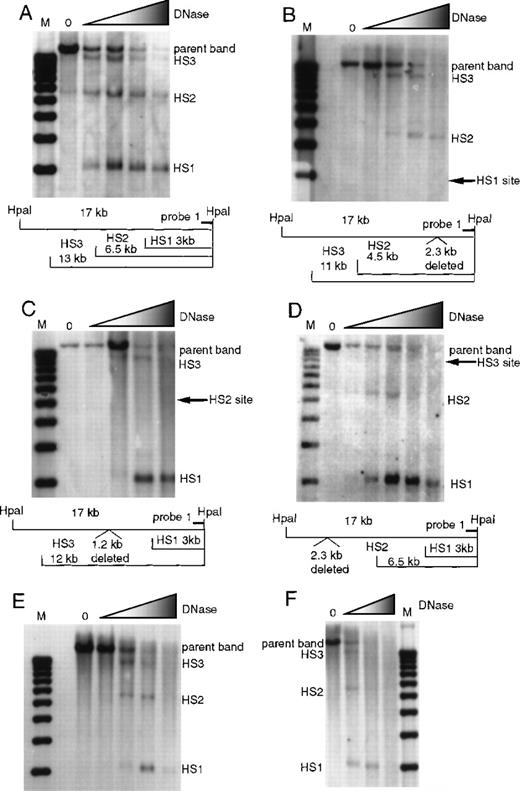
To determine how deletion of a single 5′HS affects the formation of the other 5′HSs, we analyzed the formation of 5′HS 1 through 6 in wild type and HS-knockout mice. Formation of 5′HS 1, 2, and 3 was assayed by digesting nuclei with DNaseI followed by restriction with HpaI and probing Southern blots with probe 1 (Figure 1). Figure 2 presents the data on formation of 5′HS 1, 2, and 3 in mice with deletions of 5′HS 1, 2, 3, 4, or 5/6 and the wild-type control. Figure 2A shows the formation of 5′HS 1, 2, and 3 in the wild-type mice. Figure 2B shows that deletion of 5′HS 1 results in the failure of 5′HS 1 to form; however, 5′HS 2 and 5′HS 3 still form. Figure 2C shows that deletion of the region comprising 5′HS 2 does not impair the formation of 5′HS 1 and 5′HS 3, even though 5′HS 2 does not form. Figure 2D shows that 5′HS 1 and 5′HS 2 form in the absence of the sequences composing 5′HS 3, even though 5′HS 3 does not form. Similarly, 5′HS 1, 2, and 3 form normally in mice with deletions of 5′HS 4 (Figure 2E) or 5′HS 5/6 (Figure 2F), respectively. To confirm the presence and placement of 5′HS 1, 2, and 3, HSs were mapped with the use of probes to the opposite end of the parent HpaI fragment (data not shown). The kinetics of DNaseI digestion varies among different digestion series; thus, quantitative comparisons of sensitivity among experiments cannot be made.
Formation of 5′HS 1, 5′HS 2, and 5′HS 3 in mice with homozygous single-site deletions assayed by HpaI digestion and probe 1.
(A) HS formation in wild-type mice, showing the expected sites and the parent band. M denotes the marker lane; the DNaseI concentration is 0 in the first lane and increases as shown by the shaded triangle. Below the autoradiograph is a restriction map of expected sizes for the HS and parent band. (B) Similar to (A), with the assays done on mice with 5′HS 1 deleted and the site of the deletion shown on the gel and in the map below. (C) Assay done on mice with 5′HS 2 deleted, with the map below. (D) Assay done on mice with 5′HS 3 deleted, with the map below. (E) Assay done on mice with 5′HS 4 deleted. The map is identical to that in (A). (F) Assay done on mice with 5′HS 5/6 deleted. The map is identical to that in (A).
Formation of 5′HS 1, 5′HS 2, and 5′HS 3 in mice with homozygous single-site deletions assayed by HpaI digestion and probe 1.
(A) HS formation in wild-type mice, showing the expected sites and the parent band. M denotes the marker lane; the DNaseI concentration is 0 in the first lane and increases as shown by the shaded triangle. Below the autoradiograph is a restriction map of expected sizes for the HS and parent band. (B) Similar to (A), with the assays done on mice with 5′HS 1 deleted and the site of the deletion shown on the gel and in the map below. (C) Assay done on mice with 5′HS 2 deleted, with the map below. (D) Assay done on mice with 5′HS 3 deleted, with the map below. (E) Assay done on mice with 5′HS 4 deleted. The map is identical to that in (A). (F) Assay done on mice with 5′HS 5/6 deleted. The map is identical to that in (A).
Formation of 5′HS 4, 5, and 6
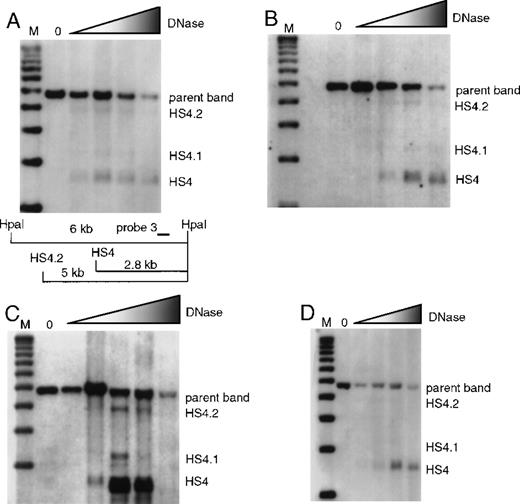
Formation of 5′HS 4 and minor bands 5′HS 4.1 and 4.2 were analyzed with the use of HpaI-digested blots and probe 3. Unlike the major HS bands 5′HS 1, 2, 3, 4, and 6, which always form strong sites in erythroid tissues, 5′HS 4.1 and 4.2 are more variable in intensity. There is formation of 5′HS 4 in wild-type mice and in mice with individual deletions of 5′HS 1, 2, or 3 (Figure 3); 5′HS 4 also forms normally in mice with deletion of 5′HS 5/6, as previously published8 (data not shown).
Formation of 5′HS 4 and minor sites 5′HS 4.1 and 5′HS 4.2 in mice with single-site deletions assayed by HpaI digestion and probed with probe 3.
The map appearing in (A) is appropriate for all parts of this figure, and is explained in the legend describing (A). (A) HS formation in wild-type mice with a restriction map below the film showing the expected sizes for the HS and parent band. M denotes the marker lane; the DNaseI concentration is 0 in the first lane and increases as shown by the shaded triangle. (B) Similar to (A), with the assay done on mice with 5′HS 1 deleted. (C) Similar to (A), with the assay done on mice with 5′HS 2 deleted. (D) Similar to A, with the assay done on mice with 5′HS 3 deleted.
Formation of 5′HS 4 and minor sites 5′HS 4.1 and 5′HS 4.2 in mice with single-site deletions assayed by HpaI digestion and probed with probe 3.
The map appearing in (A) is appropriate for all parts of this figure, and is explained in the legend describing (A). (A) HS formation in wild-type mice with a restriction map below the film showing the expected sizes for the HS and parent band. M denotes the marker lane; the DNaseI concentration is 0 in the first lane and increases as shown by the shaded triangle. (B) Similar to (A), with the assay done on mice with 5′HS 1 deleted. (C) Similar to (A), with the assay done on mice with 5′HS 2 deleted. (D) Similar to A, with the assay done on mice with 5′HS 3 deleted.
HSs in the region of 5′HS 4 were mapped in the 5′HS 4 deletion mice with the use of an SphI digest and probe 4 (Figure4). HpaI was not used because the targeted deletion of 5′HS 4 alters the HpaI restriction map. Analysis of the wild-type samples on the left of Figure 4 reveals the presence of 5′HS 4, 4.1, and 4.2. In this assay, 5′HS 3 is difficult to discern since the gel did not clearly resolve the 14-kb 5′HS 3 sub-band from the 16.2-kb parent band. The targeted deletion of 5′HS 4 removes the region encompassing 5′HS 4, but leaves the region of 5′HS 4.1 and 4.2 intact. HS mapping of the 5′HS 4 deletion mice reveals that 5′HS 4.1 and 4.2 form, but that 5′HS 4 is not present (Figure 4). Two additional minor bands are visible but do not map to the region of the 5′HS 4 deletion. The band just below the parent band is 5′HS 3, which is resolved more easily from the parent band owing to the 2.8-kb 5′HS 4 deletion and which is also more easily visualized in the absence of 5′HS 4. The smaller of the 2 bands, 5′HS 3.5, is seen sporadically in these assays and is more prominent in the absence of 5′HS 4 since the fragment is more frequently available to be cut by DNaseI at 5′HS 3.5. In both series, the major sites, 5′HS 3 and 5′HS 4, first appear at lower DNaseI concentration than the minor sites (5′HS 4.1, 4.2, and 3.5), suggesting their greater accessibility to DNaseI. As with the mapping of 5′HS 1, 2, and 3, these results were confirmed by mapping sites from the opposite end of the parent fragment.
Formation of 5′HS 3, 5′HS 4, and minor associated sites in mice with the 5′HS 4 region deleted.
The restriction digest is with SphI, and the probe is probe 4 from Figure 1. M denotes the marker lane; the DNaseI concentration is 0 in the first lane; and increases as shown by the shaded triangle. The portion of the film to the left of the marker shows an assay done on wild-type mice, and below this is the restriction map of expected bands. The portion of the autoradiograph to the right of the marker shows an assay done on mice with 5′HS 4 deleted, and below this is the restriction map of expected bands.
Formation of 5′HS 3, 5′HS 4, and minor associated sites in mice with the 5′HS 4 region deleted.
The restriction digest is with SphI, and the probe is probe 4 from Figure 1. M denotes the marker lane; the DNaseI concentration is 0 in the first lane; and increases as shown by the shaded triangle. The portion of the film to the left of the marker shows an assay done on wild-type mice, and below this is the restriction map of expected bands. The portion of the autoradiograph to the right of the marker shows an assay done on mice with 5′HS 4 deleted, and below this is the restriction map of expected bands.
Previously, we showed that deletion of the region that spans 5′HS 5 and 6 results in no formation of HSs at the site of the deletion, while 5′HS 4, 4.1, and 4.2 form normally.8 We extend these findings to show that deletion of 5′HS 5/6 does not affect formation of 5′HS 1, 2, or 3 (Figure 2F). Thus, our analyses reveal that deletion of any LCR 5′HSs (5′HS 1, 2, 3, 4, or 5/6) does not affect the formation of the other LCR HSs.
Discussion
A great deal of experimental effort has been devoted to understanding the role of the LCR in regulating the β-globin locus; however, many of the fundamental questions remain unresolved. We have taken the approach of investigating the LCR by using homologous recombination in ES cells and deriving mouse lines with individual deletions of each of the major HSs in the murine LCR8,18 19(also in M.A.B., unpublished data). None of these deletions in the endogenous murine β-globin LCR disrupts the normal pattern of β-like–globin switching. Deletion of HS 2, HS 3, or HS 4 reduces expression of β-globin in adults by roughly 30%, 40%, and 20% respectively, whereas deletion of 5′HS 1 or 5′HS 5/6 results in more minor reductions in β-globin gene expression. In this report, we have investigated the formation of HSs in adult mice carrying individual HS deletions. Our results, in combination with previous studies, lead to 3 major conclusions regarding the murine LCR HSs: (1) Although each HS contributes to the overall expression of the locus, no individual HS is necessary for the expression of a specific gene or for expression at any specific stage of development. (2) Deletion of the DNA region that forms an HS results in the elimination of that HS. (3) Deletion of a given HS has little or no effect on formation of the other HSs. Our data support the notion that the HSs form and act independently and, in an additive or synergistic manner, contribute to overall LCR activity.
On the basis of results obtained with single-copy transgenes containing a single 5′ HS, it was suggested that 5′HS 3 possessed the dominant chromatin opening activity in the LCR; thus, the chromatin overlaying 5′HS 3 would be altered initially, and subsequently an open chromatin state could spread to the rest of the LCR and β-globin locus.21 Although, compared with the other HSs in the LCR, HS 3 may have a greater chromatin opening activity when linked by itself to globin genes in transgenic mice, it is clearly not required for establishment of normal chromatin structure of the other HSs in the endogenous mouse LCR. Similarly, during differentiation of a multipotent murine hematopoietic cell line, the endogenous 5′HS2 formed prior to the other HSs,22 suggesting that formation of 5′HS2 might be required to precede the formation of the other HSs. We now demonstrate that this is not the case.
A current model for LCR function postulates that the HSs and associated bound proteins form a single large “holocomplex” that is required for expression of the β-globin locus.21,23 The observations that HSs act additively or synergistically to stimulate expression, both in the endogenous locus and in transgenes, have been cited in support of this model. Clearly, however, these data are equally compatible with an additive effect of the HSs on modifying chromatin, directing subnuclear localization, fostering tracking of factors from the LCR to the genes, or other plausible models of LCR function. We have shown that in the endogenous murine locus, HSs form independently, and no particular HS is necessary for near-normal expression of any of the genes. Moreover, synthetic LCRs, which include various combinations of HS sites and intervening DNA, generally confer high-level expression in cell lines and in mice.21,24-26 In fact, HS 2, 3, or 4 stimulates expression of linked globin transgenes when it is the only HS present.23,27,28 Thus, if the postulated holocomplex does exist and is required for LCR function, then no specific HS is required to participate in such a complex, and formation of such a holocomplex would not be dependent on the presence or arrangement of any of the individual LCR HSs or associated binding proteins. Such a complex, with little or no structural constraints, would be unique among present examples of multiprotein complexes regulating transcription in higher eukaryotes.29Ultimately, the holocomplex model can be proved or disproved only by biochemical and structural analyses of the protein-bound LCR in a normal erythroid cell.
Our failure to observe any influence of the deletion of 1 HS on the formation of others in the endogenous murine locus contrasts with the more variable results obtained with human β-globin locus transgenes in mice. Comparison of results between homologous-recombination–mediated HS deletion at the endogenous locus and the deletion of HSs in human transgenes is difficult owing to several factors, including position effects due to variable integration sites of the transgenes, differences in the sizes of the deletions, and cross-species differences.
One unavoidable variable in the transgenic studies is that each of the transgenes is integrated at a different position in the genome, and position effects on expression of transgenes are well documented. Although the full LCR significantly suppresses position effects, this suppression is incomplete; the same large, apparently unrearranged, single-copy and wild-type transgenes consistently show twofold to fourfold variability in expression level in different mouse lines. In addition, a 150-kb YAC transgene containing the human β-globin locus and an intact LCR demonstrates position effect variegation in mice: the transgene is not expressed in 58% of the red cells in these animals.30 Thus, LCRs containing transgenes are sensitive to position effects. Moreover, deletion of any of the HSs further sensitizes the transgene to position effects.14 Depending on the integration site, position effects may influence the chromatin structure of the transgene, which may be manifested in part as variable HS formation. Examination of each construct at a larger number of integration sites or examination of multiple constructs at a single integration site will be required to address this issue.
In general, studies of transgenes with relatively large HS deletions encompassing the HS core and flanking regions revealed minor effects on transcription, similar to the effects of our studies of targeted deletions in the endogenous LCR. However, chromatin structure has not been evaluated in transgenic mice with these large HS deletions. In contrast, small deletions of the HS cores have been reported to result in dramatic decreases in transcription and in significant diminution of HS formation.17,31 As discussed previously,32these core deletions may remove binding sites for activators and leave intact sites for the binding of repressors that may nucleate formation of a chromatin structure incompatible with transcription. It is possible that such core deletions in the endogenous mouse locus may reveal suppressive effects on the formation of the remaining HSs in the mouse locus; these experiments are currently underway.
It is also possible that species differences may account for the different results obtained in the analysis of the human β-globin locus in the transgenes compared with studies of the murine locus. Although we cannot completely rule out any cross-species effects, the high degree of sequence and structural homology between the mouse and human loci, and the fact that the human locus functions in a transgenic mouse, further support the idea that the loci are regulated similarly. Studies of the endogenous human β-globin locus in mouse erythroleukemia somatic cell hybrids carrying the entire human chromosome 11 revealed that deletion of human 5′ HS 2 through 5′ HS 5 eliminates β-globin gene expression without affecting the formation of the remaining site, 5′HS 1.33Similarly, deletion of 5′HS 2 through 5′HS 4 leads to complete loss of expression but normal formation of 5′HS 1 and 5. The HSs at either the murine or the human endogenous β-globin loci form independently of each other. Thus, the endogenous human locus in a murine environment has a chromatin phenotype similar to that observed in our knockout mouse studies.
Further insight into the differences in results obtained in studies of LCR function at ectopic genomic sites and those at the endogenous locus may derive from our recent demonstration that 5′HS 2 confers stability of transgene expression at repressive genomic sites by influencing the subnuclear localization of the transgene locus.34 By extrapolation, we suggest that the deletion of individual or multiple HSs from β-globin locus transgenes increases the probability that the transgene associates with heterochromatic nuclear compartments, resulting in the lack of transcription factor accessibility, the loss of HS formation and the failure to activate transcription. In this scenario, transgenes with mutated LCRs would be expressed only at genomic sites that are normally in a subnuclear compartment permissive for expression in erythroid cells, whereas these mutated transgenes would not be expressed at genomic sites that are normally heterochromatized in erythroid cells. In contrast, multiple, redundant elements in the endogenous globin locus prevent its localization in such repressive subnuclear compartments, resulting in the maintenance of the locus in an “open” DNaseI sensitive conformation upon deletion of the LCR. Clearly, elucidation of the molecular mechanism(s) by which the LCR influences the β-globin locus will require additional experimentation.
Acknowledgments
We are grateful to Jessica Halow, Joan Hamilton, and Jennie Close for expert technical assistance.
Supported by the National Institutes of Health grants DK54071 (S.N.F., M.G.), DK44746 (M.G.), and P30 HD28834 through the University of Washington Child Health Research Center (M.A.B.); and by a Burroughs-Wellcome Fund Career Development Award (S.N.F.). M.A.B. is a Howard Hughes Medical Institute Physician Postdoctoral Fellow and a J. S. McDonnell Foundation Scholar.
Reprints:Steven Fiering, 6W Borwell, DHMC, Lebanon, NH, 03756; e-mail, fiering@dartmouth.edu; or Mark Groudine, FHCRC, 1100 Fairview Ave N, Seattle, WA 98109; e-mail, markg@fhcrc.org.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal