Shear stress causes the platelet glycoprotein (Gp) Ib/IX/V to bind to von Willebrand factor, resulting in platelet adhesion. GpIb/IX/V also functions to stimulate transmembranous signaling, leading to platelet activation and the expression of a ligand-receptive GpIIb-IIIa complex. The highly conserved cytoplasmic domain of GpIb binds directly to a dimeric 14-3-3 adapter protein ζ isoform. To explore structural determinants of GpIb/IX/V binding to 14-3-3ζ, the authors examined 14-3-3ζ interactions with GpIb and GpIbβ in heterologous cells and platelets. Truncations of GpIb at amino acid 542 or 594, or deletions of residues 542 through 590, inhibited binding of 14-3-3ζ. Deletion of GpIb from Trp570 to Ser590 eliminated 14-3-3ζ binding, and deletion of the sequence from Arg542-Trp570 enhanced binding of 14-3-3ζ to GpIb. All GpIb mutations that eliminated GpIb binding to the GST-14-3-3ζ fusion protein also eliminated GpIbβ binding to the fusion protein. Forskolin treatment of Chinese hamster ovary cells expressing wild-type GpIb/β/IX resulted in the phosphorylation of GpIbβ associated with enhanced binding of GpIbβ to GST-14-3-3ζ fusion protein and increased 14-3-3ζ coimmunoprecipitated with GpIb. When intact human platelets aggregated in response to 90 dynes/cm2 shear stress, 14-3-3ζ disassociated from GpIb. Prostacyclin treatment of platelets inhibited shear stress-induced aggregation and the release of 14-3-3ζ from GpIb. These data demonstrate that amino acid residues in the cytoskeletal interaction domains of GpIb regulate 14-3-3ζ binding to GpIb/β/IX, and suggest that protein kinase A-dependent phosphorylation of GpIbβ enhances 14-3-3ζ binding to the GpIb/IX/V complex in human platelets.
The platelet glycoprotein (Gp) Ib/V/IX complex is a molecular trigger of arterial thrombosis.1 When rheological conditions, such as those resulting from a ruptured atherosclerotic plaque, cause elevated wall shearing stress to develop, GpIb/V/IX binds to plasma and vessel wall von Willebrand factor (vWF). GpIb/V/IX binding to vWF tethers platelets to the damaged arterial wall (adhesion) and to each other (aggregation). It also stimulates platelets, causing the secretion of stored proaggregatory and vasoconstricting substances and in the activation of the integrin receptor αIIbβ3 (GpIIb-IIIa). These responses culminate in a platelet-rich thrombus that occludes blood flow and results in tissue ischemia and infarction.
Gp Ib/V/IX complex is comprised of 4 protein subunits.2GpIbα is the largest member of this complex. It is a heavily glycosylated polypeptide of 610 amino acids with a globular extracellular domain that contains the major ligand-recognition site for the entire complex. A 96-amino acid carboxyl-terminal tail follows a single transmembranous domain. GpIbα is disulfide linked through its perimembranous extracellular domain to GpIbβ, which is a 181-amino acid protein with a single transmembranous domain and a carboxyl-terminal cytoplasmic tail of ∼36 amino acids. GpIX is a 160-amino acid polypeptide with a single transmembranous domain and an ∼6 amino acid cytoplasmic tail. It is noncovalently complexed with GpIbα and GpIbβ in a 1:1:1 molar ratio. GpIbα/GpIbβ/GpIX is complexed noncovalently with GpV in a 2:1 molar ratio. GpV is a 560-amino acid protein with a single transmembranous domain and a 16-amino acid carboxyl-terminal tail.
The 4 proteins in the human complex share a high degree of homology in their extracellular domains, primarily because they all contain leucine-rich repeats, from 15 in GpV to 7 in GpIbα to a single repeat in GpIbβ and GpIX. For those with available data, it appears that the interspecies homology of the extracellular domains of the 4 proteins is lower. For example, the extracellular domains of canine and mouse GpIbα are only ∼60% to 70% homologous with human GpIbα. This structural divergence is functionally important because several antibodies that recognize human GpIbα do not bind to canine GpIbα, and canine GpIbα does not support ristocetin-induced vWF binding. In contrast, there is a very high degree of homology in the primary structure in the cytoplasmic domain of GpIbα from humans, dogs, and mice, with sequence identity in several large regions (Figure1).
Interspecies homology of the cytoplasmic domain of GpIb.
Amino acid identity is reported by bold type, and identity among the 3 species is designated by the bold underline. The actin-binding protein (ABP)/filamen α and 14-3-3ζ interaction domains are designated by the double underline bordered by asterisked residue numbers.
Interspecies homology of the cytoplasmic domain of GpIb.
Amino acid identity is reported by bold type, and identity among the 3 species is designated by the bold underline. The actin-binding protein (ABP)/filamen α and 14-3-3ζ interaction domains are designated by the double underline bordered by asterisked residue numbers.
The conserved amino acid sequence in the cytoplasmic domain of GpIbα suggests that it may be functionally important. The functions that it serves, however, are not clearly elucidated. GpIbα contains at least 2 adjacent filamin α-binding domains (actin-binding protein 280). Residues ∼540-570 are probably the primary filamen α-binding site,3 whereas residues 570-590 contribute to filamen α binding and are absolutely required for GpIb/V/IX attachment to the cytoskeleton.4 GpIbα also binds directly to the 14-3-3-adapter protein ζ isoform found in human platelets.Residues 605-610 at the extreme carboxyl terminus are reportedly absolutely required for GpIbα binding to 14-3-3ζ,5 though peptide inhibition studies indicate that more proximal amino acid sequences enhance or stabilize binding between GpIbα and 14-3-3ζ.6
The consequences of 14-3-3ζ binding to platelet GpIbα in the physiologic or pathologic development of platelet function in vivo are obscure. Although there is ample evidence that shear-induced vWF binding to GpIbα causes activating platelet-signaling responses, there is no evidence that 14-3-3ζ is involved in these responses.7-10 Recent data from heterologous cell experiments, however, indicate that the primary 14-3-3ζ interaction domain of GpIbα is essential for signaling growth arrest in Chinese hamster ovary (CHO) cells.11 Platelets are anucleate, but, because there may be conserved protein function among platelets, megakaryocytes, and heterologous cells, these results suggest that the interaction between GpIb/IX/V and 14-3-3ζ could also direct functional responses in platelets. To begin to investigate the hypothesis that shear stress activates platelets through a pathway that involves GpIbα binding to 14-3-3ζ, we examined 14-3-3ζ interactions with GpIbα and GpIbβ in genetically engineered heterologous cells and platelets.
Methods
Platelet aggregation studies
Venous blood was obtained from healthy volunteer donors and collected in 15% (vol/vol) acid-citrate-dextrose. Blood was centrifuged at 270g for 14 minutes at 24°C, and the platelet-rich plasma was acidified to pH 6.5 with acid-citrate-dextrose and treated with phosphocreatine (5 mmol/L) and creatine phosphokinase (25 U/mL). The platelets were separated from the platelet-rich plasma by a second centrifugation at 1600g for 15 minutes at 24°C. The platelet pellet was then washed in 10 mL Tyrode's buffer (10 mmol/L HEPES, 12 mmol/L NaHCO3, 137 mmol/L NaCl, 2.7 mmol/L KCl, 1 mmol/L CaCl2, and 5 mmol/L glucose, pH 6.5) (supplemented with phosphocreatine and creatine phosphokinase at the same final concentrations as above) at 1200g for 10 minutes at 24°C. The resultant platelet pellet was finally suspended in buffer containing 6 mmol/L glucose, 130 mmol/L NaCl, 9 mmol/L NaHCO3, 10 mmol/L sodium citrate, 10 mmol/L Tris base, 3 mmol/L KCl, 2 mmol/L HEPES, and 0.9 mmol/L MgCl2, with 1 mmol/L CaCl2 at pH 7.35 at a concentration of 2.5 × 108 platelets/mL.
GpIb cloning and mutagenesis
Wild-type and recombinant constructs of GpIbα are shown in Figure2. The full-length human GpIbα cDNA (from −42 to 2420 bp), obtained from Dr José López, was cloned to the pBluescript SK vector (Stratagene, La Jolla, CA) at its EcoRI insertion site and then subcloned to the mammalian expression vector pcDNA3.1/Zeo (Invitrogen, San Diego, CA) at BamHI and XhoI sites. A truncated GpIbα (amino acid residues Met1-Gln541) was generated by the ligation of 2 fragments of GpIbα cDNA in the SK vector from theBamHI to XbaI and XbaI to PstI sites and cloned to the pcDNA3.1/Zeo expression vector at the BamHI and XhoI sites. A second truncated GpIbα (amino acid residues Met1-Gly594) was constructed by making a fragment of wild-type GpIbα in pBluescript SK with an excision from its HindIII to its SmaI restriction site. This fragment was ligated to a HindIII plus SmaI digested product of wild-type GpIbα cDNA after amplification by polymerase chain reaction (PCR) using primers spanning codons 547 through 595 (5′ACAGTGCCCCGGGCCTGGCTGCTC3′ and 5′CAGGTCCTGACCTCGAGCCTGACTCAG3′, respectively). A cDNA for GpIbα, deleted of its actin-binding domain (Arg542-Ser590), was constructed by ligating 3 fragments of GpIbα cDNA in pBluescript SK. The first fragment was wild-type GpIbα with an excision from its HindIII to itsXbaI restriction site. The second was wild-type GpIbα with an excision from its XbaI to its PstI restriction site. The third fragment was generated by PCR amplification of wild-type GpIbα using primers that span codon 587 (5′TCAGCTCTGCTGCAGGGTCGTGGTCAG3′) with a sequence for the 3′ untranslated region of the cDNA (5′ATGCAGCATCTCGAGCTTTGTCTTGTC3′). After ligation, the mutant GpIbα species were cloned to pcDNA3.1/Zeo at theHindIII and XhoI sites. Another deleted form of GpIbα (Arg542-Trp570) was generated by the ligation of 3 fragments of human GpIbα cDNA, derived from the SK vector, to pcDNA3.1/Zeo at HindIII and XhoI sites. The first fragment, coding 1041 bp, was excised from the SK vector byHindIII and XbaI digestion. The second fragment, coding 671 bp, was generated by digestion with XbaI and PstI. The third piece, coding 370 bp, was synthesized by PCR using GpIbα cDNA as the template and primers 5′AGCCTCTTCCTGCAGGTACGGCCTAAT3′ and 5′ATGCAGCATCTCGAGCTTTGTCTTGTC3′. Polymerase chain reaction products were digested with PstI and XhoI at 235 bp downstream of the translation stop codon. The deleted form (Trp570-Ser590) was constructed by the ligation of 3 fragments of human GpIbα cDNA to pcDNA3.1/Zeo at HindIII and XhoI sites. The first fragment, coding 1738 bp, was generated by the digestion of the full-length GpIbα cDNA atHindIII and SmaI sites. The second fragment, coding 100 bp, was produced by PCR with primers 5′ATTAGGCCGTACCTGCAGGAAGAGGCT3′ and 5′ACAGTGCCCCGGGCCTGGCTGCTC3′. These PCR fragments were digested at SmaI and PstI sites. The third fragment, coding 335 bp, was generated by PCR with primers 5′TCAGCTCTGCTGCAGGGTCGTGGTCAG3′ and 5′ATGCAGCATCTCGAGCTTTGTCTTGTC3′. The PCR fragment was digested at PstI and XhoI sites. In all cases, the integrity of the mutant cDNA was verified by sequence analysis.
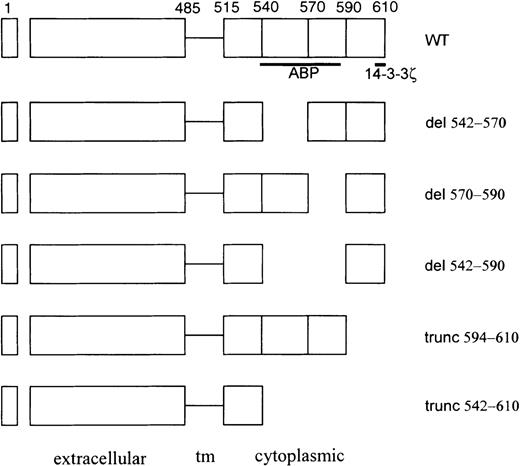
Schematic of GpIbα cytoplasmic domain mutants used in these studies.
The actin-binding protein (ABP)/filamen α and 14-3-3ζ binding domains are designated schematically by the bold lines beneath the wild-type (WT) construct. tm, transmembrane.
Schematic of GpIbα cytoplasmic domain mutants used in these studies.
The actin-binding protein (ABP)/filamen α and 14-3-3ζ binding domains are designated schematically by the bold lines beneath the wild-type (WT) construct. tm, transmembrane.
Transfections
CHOβ/IX cells (CHO cells expressing GpIbβ and GpIX) were a gift of Dr J. A. López and Dr J. F. Dong (Baylor College of Medicine and Veterans Affairs Medical Center, Houston, TX). The cells were grown in α-minimum essential medium (Life Technologies, Grand Island, NY) containing 5% fetal bovine serum (Life Technologies). All cells were maintained in an atmosphere of 5% CO2 and 99% humidity at 37°C. CHOβ/IX cells (5 × 105) in 25-cm2 culture flasks were washed twice with phosphate-buffered saline (PBS) and maintained in 1.5 mL serum-free minimum essential medium. A mixture of 15 μL lipofectamine (Life Technologies) with 5 μg plasmid DNA (full-length GpIbα, mutant GpIbα, or control empty vector) was warmed to room temperature for 15 minutes in 100 μL PBS before it was added to each flask. The transfection mixtures were incubated at 37°C for 10 hours. After transfection, the cells were selected in minimum essential medium supplemented with 10% fetal bovine serum, 500 μg/mL Zeocin (Invitrogen), 500 μg/mL G418, and 80 μmol/L methotrexate.
Flow cytometry
The expression of the GpIb/IX complex on the cell surface was analyzed by flow cytometry. Cells were washed with PBS, detached with EDTA, and incubated with 1 μg/mL fluorescein isothiocyanate-conjugated AN51 (DAKO, Carpinteria, CA) to identify GpIbα. The other members of the GpIb/IX complex were identified with anti-GpIX antibody SZ1 (provided by Dr J. A. López) and an anti-GpIbβ antibody (Santa Cruz Biotechnology, Santa Cruz, CA). For GpIbα, samples were washed twice with PBS, resuspended in 0.5 mL PBS, and directly analyzed for emission at 520 nm in a FACStar flow cytometer (Becton Dickinson, Mountain View, CA) after stimulation with an argon ion laser at a wavelength of 488 nm. For GpIbβ and GpIX, samples were immunostained with a second antibody (a goat antimouse antibody conjugated with fluorescein isothiocyanate), washed twice with PBS, and analyzed by flow cytometry. In some cases, fluorescence-activated cell sorting and single-cell cloning were performed as part of the selection process.
Immunoprecipitation procedure
Cell samples were collected and lysed in an equal volume of ice-cold PBS containing 1% Nonidet P-40 (NP-40), 100 mmol/L Na3VO4, 10 mmol/L Na4P2O7, 5 mmol/L EGTA, 1 mmol/L phenylmethylsulfonyl fluoride, and 1 μg/mL each of aprotinin, pepstatin, and leupeptin. The samples were then sonicated briefly (5 seconds) and incubated on ice for 30 minutes. Lysates were cleared of insoluble debris by centrifugation at 13,000g for 15 minutes at 4°C and diluted with PBS to bring the final NP-40 concentration to 0.5%. The GpIbα protein was immunoprecipitated using either mAb AN51 (which recognizes the N-terminal vWF-binding domain; DAKO) or WM23 (which recognizes the macroglycopeptide repeat domain). Immunoprecipitation was carried out by incubating lysates with antibody overnight at 4°C, followed by incubation with 40 μL Protein A-Sepharose (Sigma) for 1 hour at 4°C. After 3 washes with ice-cold PBS buffer containing protease inhibitors, precipitated protein was separated by sodium dodecyl sulphate–polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes. Western blotting was then performed using primary antibodies for GpIbα (WM23), GpIbβ (Santa Cruz Biotechnology), or 14-3-3ζ (Santa Cruz). Polyvinylidene difluoride membranes were blotted with the appropriate horseradish peroxidase-conjugated secondary antibody, and reactive bands were visualized by chemiluminescence (ECL kit; Amersham).
GST-14-3-3ζ fusion protein-binding assay
Full-length human 14-3-3ζ fused to glutathione-S-transferase (GST) was cloned to pGEX-2T-expressing vector at BamHI andEcoRI sites (Pharmacia, Piscataway, NJ) and transfected to DH5α Escherichia coli. The expression of GST-14-3-3ζ (or just GST in pGEX-2T as a control) was induced with 0.5 mmol/L isopropyl b-D-thiogalactopyranoside for 4 hours, the fusion protein was released by bacterial lysis using freezing/thawing in PBS buffer, and GST-14-3-3ζ was separated by centrifugation at 40,000g for 2 hours. CHO β/IX cells transfected with wild-type or mutant GpIbα or empty vector were lysed in 1% NP-40 in PBS buffer supplemented with 5 mmol/L EGTA; 1 mmol/L phenylmethylsulfonyl fluoride; 1 μg/mL each of aprotinin, leupeptin, and pepstatin; 1 mmol/L Na3VO4; and 100 mmol/L NaF. After centrifugation at 14,000g for 20 minutes, cell lysates were incubated with GST-14-3-3ζ or GST overnight at 4°C and then by a 30-minute incubation with glutathione-Sepharose beads (Pharmacia) at room temperature. Bound proteins were washed extensively with lysis buffer and were analyzed by immunoblotting.
Densitometry
To quantify proteins binding to GST-14-3-3ζ or coimmunoprecipitating with GpIbα, immunoblots were scanned with a laser densitometer (LKB Bromma Ultra Scan XL Enhanced Laser Densitometer; Pharmacia). Densitometry data were combined to generate mean and SEM, and differences were analyzed by the Student ttest.
Results
GpIb binding to 14-3-3ζ is inhibited by truncating the cytoplasmic tail, or by deleting residues 542-590, of GpIb
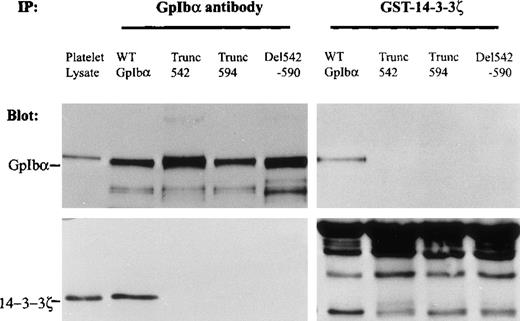
To identify changes in the primary structure of the cytoplasmic domain of GpIbα that affect 14-3-3ζ binding to GpIbα, several GpIbα mutants were expressed with wild-type GpIbβ and GpIX. The capacity of these mutants to bind to 14-3-3ζ was examined in cell lysates precipitated with the GST-14-3-3ζ fusion protein or a monoclonal antibody that recognizes the extracellular domain of GpIbα (AN51). Figure 3 shows that truncation of the cytoplasmic domain of GpIbα at residue 542 eliminates binding to the 14-3-3ζ fusion protein and eliminates endogenous 14-3-3ζ coimmunoprecipitating with GpIbα. Deletion of the large cytoskeletal interaction domain(s) between residues 542 and 590 (ABP [actin-binding protein] in Figure 2, which maintains the C-terminal 14-3-3ζ binding site) also eliminates the capacity of recombinant GpIbα to bind to the GST-14-3-3ζ fusion protein and eliminates the interaction of endogenous 14-3-3ζ with recombinant GpIbα in CHO cells. Truncation of GpIbα at amino acid 594 eliminates 14-3-3ζ coimmunoprecipitating with GpIbα, and consistent with a previous report,5 no immunodetectable GpIbα truncated at residue 594 binds to recombinant GST-14-3-3ζ. The empty GST fusion protein does not bind any GpIbα, and an irrelevant isotype-specific IgG does not immunoprecipitate any GpIbα or 14-3-3ζ (data not shown).
GpIb binding to 14-3-3ζ inhibited by truncations or deletions in its cytoplasmic domain.
GpIbα mutants were expressed with wild-type GpIbβ and GpIX. The capacity of these mutants to bind to 14-3-3ζ was examined in cell lysates precipitated with a GST-14-3-3ζ fusion protein (right) or with mAb AN51 that recognizes the extracellular domain of GpIbα (left). Truncation of the cytoplasmic domain of GpIbα at residue 542 eliminated binding to the 14-3-3ζ fusion protein and eliminated endogenous 14-3-3ζ coimmunoprecipitating with GpIbα. Deletion of the large cytoskeletal interaction domain(s) between residues 542 and 590 (ABP in Figure 2, which maintains the C-terminal 14-3-3ζ binding site) also eliminated the capacity of recombinant GpIbα to bind to the GST-14-3-3ζ fusion protein and eliminated the interaction of endogenous 14-3-3ζ with recombinant GpIbα in CHO cells. Truncation of GpIbα at amino acid 594 eliminated 14-3-3ζ coimmunoprecipitating with GpIbα. No immunodetectable GpIbα truncated at residue 594 bound GST-14-3-3ζ (IP, immunoprecipitation; n = 3).
GpIb binding to 14-3-3ζ inhibited by truncations or deletions in its cytoplasmic domain.
GpIbα mutants were expressed with wild-type GpIbβ and GpIX. The capacity of these mutants to bind to 14-3-3ζ was examined in cell lysates precipitated with a GST-14-3-3ζ fusion protein (right) or with mAb AN51 that recognizes the extracellular domain of GpIbα (left). Truncation of the cytoplasmic domain of GpIbα at residue 542 eliminated binding to the 14-3-3ζ fusion protein and eliminated endogenous 14-3-3ζ coimmunoprecipitating with GpIbα. Deletion of the large cytoskeletal interaction domain(s) between residues 542 and 590 (ABP in Figure 2, which maintains the C-terminal 14-3-3ζ binding site) also eliminated the capacity of recombinant GpIbα to bind to the GST-14-3-3ζ fusion protein and eliminated the interaction of endogenous 14-3-3ζ with recombinant GpIbα in CHO cells. Truncation of GpIbα at amino acid 594 eliminated 14-3-3ζ coimmunoprecipitating with GpIbα. No immunodetectable GpIbα truncated at residue 594 bound GST-14-3-3ζ (IP, immunoprecipitation; n = 3).
14-3-3ζ binding to GpIb and GpIbβ is affected by partial deletions within the cytoskeletal interaction domain of GpIb
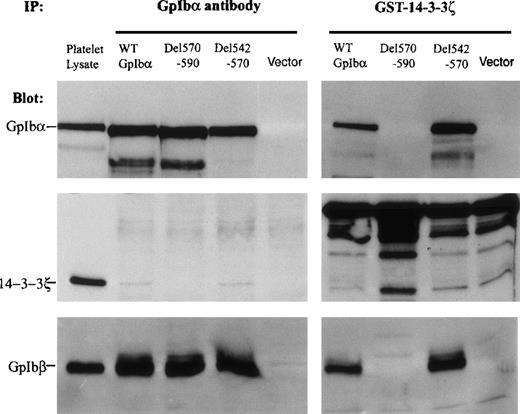
Figure 3 demonstrates that the ABP/cytoskeletal interaction domain of GpIbα influences 14-3-3ζ binding to the C-terminal domain of GpIbα. To begin to elucidate how these proximal amino acid residues regulate 14-3-3ζ binding to GpIbα, 2 deletions were made within the large ABP/cytoskeletal interaction domain(s) (see Figure 2). One deletion encompasses the primary ABP binding domain, as determined by peptide inhibition assays: residues 542-570.3 The second deletion encompasses a second, perhaps regulatory, cytoskeletal interaction domain identified by mutagenesis assays: residues 570-590.4 Figure 4 shows that the deletion of residues 542-570 does not inhibit the binding of GpIbα to a GST-14-3-3ζ fusion protein and does not inhibit the quantity of 14-3-3ζ that coimmunoprecipitates with GpIbα. This figure suggests that the 542-570 deletion enhances the interaction between 14-3-3ζ and GpIbα, and this enhancement was seen consistently with the GST-14-3-3ζ binding assay in 5 separate experiments. Enhancement is less obvious in 5 coimmunoprecipitation experiments. In contrast to the 542-570 deletion, deletion of the sequence from 570-590 of GpIbα eliminates all binding of 14-3-3ζ to GpIbα. This result indicates that the 570-590 domain, which has been reported to regulate cytoskeletal interactions with GpIbα,4 may regulate 14-3-3ζ binding to the C-terminus of GpIbα.
14-3-3ζ binding to GpIb and GpIbβ regulated by partial deletions within the cytoskeletal interaction domain of GpIb.
Two deletions were made within the large actin-binding protein (ABP)/cytoskeletal interaction domain(s) (see Figure 2). One encompassed the primary ABP domain as determined by peptide inhibition assays: residues 542-570. The second encompassed a second, perhaps regulatory, cytoskeletal interaction domain identified by mutagenesis assays: residues 570-590. Deletion of residues 542-570 enhanced the binding of GpIbα to a GST-14-3-3ζ fusion protein (right) and did not affect the quantity of 14-3-3ζ that coimmunoprecipitated with GpIbα (left). Deletion of the sequence 570-590 of GpIbα eliminated all binding of 14-3-3ζ to GpIbα. The absence of GpIbα (vector) or the deletion of residues 570-590 prevented GpIbβ from binding to the GST-14-3-3ζ protein (bottom panels) (n = 5).
14-3-3ζ binding to GpIb and GpIbβ regulated by partial deletions within the cytoskeletal interaction domain of GpIb.
Two deletions were made within the large actin-binding protein (ABP)/cytoskeletal interaction domain(s) (see Figure 2). One encompassed the primary ABP domain as determined by peptide inhibition assays: residues 542-570. The second encompassed a second, perhaps regulatory, cytoskeletal interaction domain identified by mutagenesis assays: residues 570-590. Deletion of residues 542-570 enhanced the binding of GpIbα to a GST-14-3-3ζ fusion protein (right) and did not affect the quantity of 14-3-3ζ that coimmunoprecipitated with GpIbα (left). Deletion of the sequence 570-590 of GpIbα eliminated all binding of 14-3-3ζ to GpIbα. The absence of GpIbα (vector) or the deletion of residues 570-590 prevented GpIbβ from binding to the GST-14-3-3ζ protein (bottom panels) (n = 5).
The bottom panels in Figure 4 represent the quantity of immunodetectable GpIbβ that coimmunoprecipitates with GpIbα (left side) or that binds to the 14-3-3ζ-GST fusion protein (right side). Results with the fusion protein indicate that the absence of GpIbα (vector) or the deletion of residues 570-590 (which eliminates GpIbα binding) prevents GpIbβ from binding to the GST-14-3-3ζ protein. Similarly, truncation of the cytoplasmic domain of GpIbα at amino acids 542 or 594, both of which prevented GpIbα binding to 14-3-3ζ-GST (Figure 3), also eliminates all GpIbβ binding to the fusion protein (data not shown). Taken together, these data suggest that the primary interaction of dimeric 14-3-3ζ with the GpIb/IX complex is between 14-3-3ζ and GpIbα. When the GpIbα/14-3-3ζ interaction is lost, GpIbβ cannot bind to GST-14-3-3ζ.
Forskolin-treatment of CHO cells expressing wild-type GpIb/β/IX enhances the interaction between 14-3-3ζ and the GpIb complex
Figure 4 shows that GST-14-3-3ζ must bind to GpIbα to bind to GpIbβ, suggesting a bivalent interaction modulated primarily by the structure of GpIbα. Using a different method (the yeast 2-hybrid system), others have shown that 14-3-3ζ interacts with both GpIbα and GpIbβ and that this interaction is in part mediated by the phosphorylation of Ser166 of GpIbβ.12 The phosphorylation of Ser166 of GpIbβ is directed by platelet cyclic adenosine monophosphate-dependent protein kinase, designated protein kinase A (PKA).13 14 To determine whether 14-3-3ζ binding to the GpIb/IX/V complex is regulated by the phosphoserine at residue 166 of GpIbβ, CHO β/IX cells expressing either vector, wild-type GpIbα, or GpIbα deleted of residues 570-590 were treated for 1 hour with 100 μmol/L forskolin, which is membrane permeable and directly activates the catalytic subunit of PKA. This treatment results in increased phosphorylation of recombinant GpIbβ coimmunoprecipitated with GpIbα from the CHO cells (Figure5A). Figure 5B shows that forskolin increases the amount of recombinant GpIbβ that binds to the GST-14-3-3ζ fusion protein in cells expressing wild-type GpIbα. In cells expressing no (vector) or deleted GpIbα (Del 570-590), phosphorylation of GpIbβ is insufficient to direct the binding of GpIbβ to GST-14-3-3ζ. Figure 5C shows that forskolin increases the quantity of 14-3-3ζ that coimmunoprecipitates with recombinant GpIbα from CHO β/IX cells transduced with wild-type GpIbα. GpIbβ phosphorylation is not, however, sufficient to permit the binding of endogenous 14-3-3ζ to the GpIb complex when GpIbα is deleted from residues 570-590. Figure 5D presents quantitative data demonstrating that PKA activation enhances the interactions between 14-3-3ζ, GpIbβ, and GpIbα in CHO cells expressing wild-type GpIbα, GpIbβ, and GpIX.
Forskolin-treatment of Chinese hamster ovary cells expressing wild-type GpIb/β/IX enhanced the interaction between 14-3-3ζ and the GpIb complex.
CHO β/IX cells expressing vector, wild-type GpIbα or GpIbα deleted from residues 570-590 were treated for 1 hour with 100 μmol/L forskolin, which stimulated cyclic adenosine monophosphate-dependent protein kinase. (A) Forskolin caused increased phosphorylation of recombinant GpIbβ in CHO cells radiolabeled with 32P] orthophosphate. (B) Forskolin increased the amount of recombinant GpIbβ that bound to the GST-14-3-3ζ fusion protein only in cells expressing wild-type GpIbα. (C) Forskolin increased the quantity of 14-3-3ζ that coimmunoprecipitated with recombinant GpIbα from CHO cells transduced with wild-type GpIbα, GpIbβ, and GpIX. (D) Quantitative analysis of the effect of forskolin-induced PKA activation on the amount of 14-3-3ζ that coimmunoprecipitated with GpIbα and the amount of GpIbβ and GpIbα that bound to the GST-14-3-3ζ fusion protein, in CHO cells stably expressing wild-type GpIbα, GpIbβ, and GpIX. The amount of protein is reported as arbitrary densitometry units. (n = 3; *P < .001, Student ttest).
Forskolin-treatment of Chinese hamster ovary cells expressing wild-type GpIb/β/IX enhanced the interaction between 14-3-3ζ and the GpIb complex.
CHO β/IX cells expressing vector, wild-type GpIbα or GpIbα deleted from residues 570-590 were treated for 1 hour with 100 μmol/L forskolin, which stimulated cyclic adenosine monophosphate-dependent protein kinase. (A) Forskolin caused increased phosphorylation of recombinant GpIbβ in CHO cells radiolabeled with 32P] orthophosphate. (B) Forskolin increased the amount of recombinant GpIbβ that bound to the GST-14-3-3ζ fusion protein only in cells expressing wild-type GpIbα. (C) Forskolin increased the quantity of 14-3-3ζ that coimmunoprecipitated with recombinant GpIbα from CHO cells transduced with wild-type GpIbα, GpIbβ, and GpIX. (D) Quantitative analysis of the effect of forskolin-induced PKA activation on the amount of 14-3-3ζ that coimmunoprecipitated with GpIbα and the amount of GpIbβ and GpIbα that bound to the GST-14-3-3ζ fusion protein, in CHO cells stably expressing wild-type GpIbα, GpIbβ, and GpIX. The amount of protein is reported as arbitrary densitometry units. (n = 3; *P < .001, Student ttest).
14-3-3ζ dissociates from GpIb during platelet aggregation in response to elevated shear stress, and this release is inhibited by prostacyclin
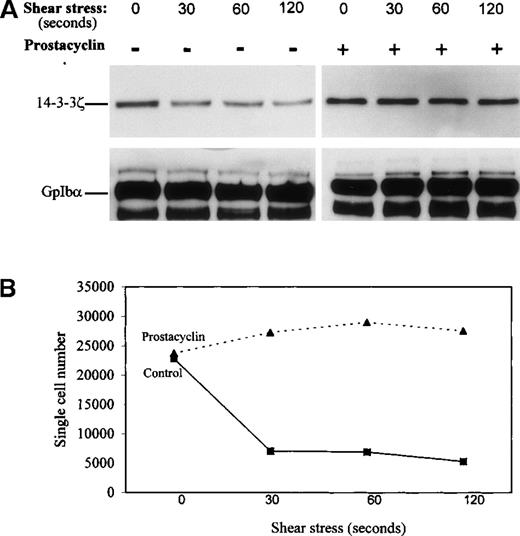
Figure 5 shows that PKA-dependent phosphorylation of CHO cells increased the amount of immunodetectable 14-3-3ζ that bound to the recombinant wild-type GpIbα/β/IX complex. PKA is well known to inhibit platelet activation, including shear stress-induced aggregation,15 and PKA-mediated phosphorylation of GpIbβ has been reported to inhibit collagen-induced actin polymerization in platelets.16 Our observations in CHO cells suggested that PKA-mediated inhibition of shear-induced platelet aggregation could have involved changes in the association of 14-3-3ζ with GpIb/IX/V. To investigate whether changes in GpIbα/14-3-3ζ interactions occur in intact human platelets stimulated by pathologic shear stress, washed platelets were sheared in a cone-plate viscometer at a force of 90 dynes/cm2. These conditions are associated with platelet activation and aggregation resulting from vWF released by the sheared platelets.7 8 Platelet lysates collected at several time points after shear were immunoprecipitated for GpIbα, and immunoprecipitates were immunoblotted for 14-3-3ζ. Figure6A shows that 90 dynes/cm2shear stress caused the dissociation of 14-3-3ζ from platelet GpIbα. To determine whether shear stress-induced loss of 14-3-3ζ from the GpIbα immunoprecipitates was caused by proteolytic cleavage of GpIbα, the immunoprecipitates were blotted with an antibody that recognized the C-terminal 15 amino acids of GpIbα (antibody IbαC, kindly provided by Dr Xiaoping Du, University of Illinois at Chicago; 5). No decrease in the amount of IbαC binding to GpIbα immunoprecipitated with monoclonal antibody AN51 was observed (data not shown).
14-3-3ζ dissociates from GpIb during platelet aggregation in response to pathologic shear stress, and disassociation is inhibited by prostacyclin.
Intact washed human platelets were sheared in a cone-plate viscometer at a force of 90 dynes/cm2. (A) Shear stress caused the dissociation of 14-3-3ζ from platelet GpIbα immunoprecipitated with mAb AN51, and that platelet treatment with 100 ng/mL prostacyclin, which stimulated PKA, inhibited the decrease of immunodetectable 14-3-3ζ from the immunoprecipitated GpIbα. (B) Platelet aggregation in response to 90 dynes/cm2 shear stress and its inhibition by prostacyclin. Aggregation is reported as a decrease in the number of single particles in the sheared platelet suspension (n = 3).
14-3-3ζ dissociates from GpIb during platelet aggregation in response to pathologic shear stress, and disassociation is inhibited by prostacyclin.
Intact washed human platelets were sheared in a cone-plate viscometer at a force of 90 dynes/cm2. (A) Shear stress caused the dissociation of 14-3-3ζ from platelet GpIbα immunoprecipitated with mAb AN51, and that platelet treatment with 100 ng/mL prostacyclin, which stimulated PKA, inhibited the decrease of immunodetectable 14-3-3ζ from the immunoprecipitated GpIbα. (B) Platelet aggregation in response to 90 dynes/cm2 shear stress and its inhibition by prostacyclin. Aggregation is reported as a decrease in the number of single particles in the sheared platelet suspension (n = 3).
Figure 6A also shows that pretreatment of platelets for 5 minutes with 100 ng/mL prostacyclin (prostaglandin I2), which causes receptor-mediated stimulation of adenylyl cyclase leading to the activation of platelet PKA, inhibits shear stress-induced dissociation of 14-3-3ζ from GpIbα. Figure 6B shows platelet aggregation in response to 90 dynes/cm2 shear stress and its inhibition by prostacyclin.
Discussion
The highly conserved cytoplasmic tail of platelet GpIbα contains at least 2 functional binding domains. These 2 domains directed interactions with the cytoskeleton (residues approximately 540-590) and a 14-3-3ζ adapter protein (residues ∼605-610 at the extreme carboxyl terminus). The role of these interactions in modulating platelet activation is not well defined. Because platelet activation in response to shear stress depends in large part on GpIbα engagement by vWF, we hypothesized that the highly conserved cytoplasmic tail of GpIbα regulates functional platelet responses to pathologic shear stress. Results of the experiments presented in this report, using heterologous cells under static conditions and platelets subjected to 90 dynes/cm2 shear stress, were consistent with the hypothesis that GpIbα transduced aggregation signals through its 14-3-3ζ interaction domains. We showed that 14-3-3ζ binding to the GpIb/IX/V complex was regulated by the cytoskeletal interaction domains of GpIbα and the phosphorylation of GpIbβ. We also presented data suggesting that dimeric 14-3-3ζ binds to GpIbα and GpIbβ only when a primary interaction with GpIbα is established. Finally, we demonstrated that 14-3-3ζ dissociated from the GpIb/IX/V complex in platelets stimulated by shear stress. These observations support the concept that 14-3-3ζ is involved in platelet signaling and lay the foundation for investigations of its mechanisms of action.
The regulation of 14-3-3ζ binding by the cytoskeletal interaction domain of GpIbα is possibly related to functional domains within the amino acid 542-590 sequence. Deletion of the entire sequence eliminated all 14-3-3ζ binding. Elimination of the more distal cytoskeletal interaction regulatory domain (residues 570-590)4 similarly eliminated 14-3-3ζ binding. In contrast, and perhaps paradoxically, deletion of the primary ABP domain (residues 542-570)3 did not inhibit (and may have enhanced) 14-4-3ζ binding. At the least, this result suggests that tail length alone does not modulate 14-3-3ζ binding. More importantly, perhaps, these data suggest that 14-3-3ζ binding to the GpIb/IX/V complex may be regulated by dynamic interactions between GpIbα and the platelet cytoskeleton. Reportedly, CHO cells contain an ABP closely related to that found in platelets (ABP-280 or filamen α).17 Thus, our data could be interpreted as indicating that GpIbα binding to ABP-280/filamin α inhibits 14-3-3ζ binding. Furthermore, the data indicate that the domain in GpIbα encompassed by residues 570-590 may regulate both ABP/filamen and 14-3-3ζ interactions.
Recent data from this laboratory indicate that ABP-280/filamin α associates rapidly with GpIbα in platelets subjected to elevated shear stress.18 The molecular interactions that develop under pathologic shear stress to trigger this response are largely unknown. ABP-280/filamin α binding to GpIbα occurs in the absence of vWF binding to GpIbα, suggesting that it is an early direct effect of shear that could precede vWF engagement by GpIbα. After vWF binding, increasing amounts of filamentous actin associate with GpIbα. Thus a model emerges in which increased ABP-280/filamin α binding, possibly in conjunction with increasing tethering of GpIbα to the actin cytoskeleton, causes the dissociation of 14-3-3ζ from the C-terminus of GpIbα. Whether this release comes from a conformational change or a steric effect is unknown, but 14-3-3ζ translocates to the cytoskeleton of platelets stimulated by vWF and ristocetin.12 This observation is consistent with the idea that 14-3-3ζ is released from GpIbα by steric effects resulting from an enlarging nexus of cytoskeletal filaments that trap 14-3-3ζ as they nucleate around and extend beyond residues 542-570 of GpIbα's cytoplasmic tail.
Truncations 542 and 594 and deletions 542-590 and 570-590 that eliminate GpIbα association with GST-14-3-3ζ also eliminate GpIbβ association with GST-14-3-3ζ (Figure 4). Because these experiments were conducted in nonreduced specimens, such results indicate that GpIbβ associates with GST-14-3-3ζ only when GpIbα associates with 14-3-3ζ. This conclusion is further supported by the absent binding of GST-14-3-3ζ to GpIbβ from CHO β/IX cell lysates. Conversely, whenever GpIbα binds to GST-14-3-3ζ, GpIbβ is present in the fusion protein bead eluate. This reflects either passive attachment through GpIbα or a specific noncovalent interaction between GpIbβ and 14-3-3ζ. Several previous studies using different experimental approaches support the latter interpretation that dimeric 14-3-3ζ bridges GpIbα and GpIbβ, and that 14-3-3ζ binding to GpIbβ involves a phosphorylation recognition domain.6,12 19Additional support for the idea that 14-3-3ζ binds both GpIbα and GpIbβ simultaneously, and that this bridging is regulated by the phosphorylation of the β subunit of GpIb, is shown in Figure 5. This figure shows that PKA-dependent phosphorylation of GpIbβ increases both the amounts of immunodetectable GpIbα and GpIbβ that bind to GST-14-3-3ζ, and the amount of immunodetectable 14-3-3ζ that coimmunoprecipitates with wild-type GpIbα. Phosphorylation of GpIbβ cannot induce GpIbβ binding to 14-3-3ζ in cells expressing mutations of GpIbα that inhibit 14-3-3ζ interactions.
The functional importance of GpIbα binding to 14-3-3ζ can only be theorized based on experiments is which 14-3-3ζ release is associated with shear-induced platelet aggregation, and the inhibition of 14-3-3ζ release is associated with the inhibition of aggregation. Furthermore, no downstream effectors of platelet activation stimulated by released 14-3-3ζ have yet been identified. Nonetheless, data presented in this article begin to focus on specific molecular responses that are inextricably coupled to 2 well-known and prominent functions of the cytoplasmic domains of the GpIb/IX/V complex. Additional studies should establish, or refute, the validity of this novel model of the molecular mechanism of shear stress-induced platelet aggregation.
Acknowledgments
The authors thank Xiaoping Du and José López for generously providing reagents, and they thank Fay Houston and June Osterholm for assisting with manuscript preparation.
Supported by the Research Service of the Department of Veterans Affairs; the National Heart, Lung and Blood Institute (HL18584); the American Heart Association; and the National Heart Foundation of Australia.
Reprints:Michael H. Kroll, Section of Hematology-Oncology, 111H, Veterans Administration Medical Center, 2002 Holcombe Boulevard, Houston, TX 77030; e-mail: mkroll@bcm.tmc.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


![Fig. 5. Forskolin-treatment of Chinese hamster ovary cells expressing wild-type GpIb/β/IX enhanced the interaction between 14-3-3ζ and the GpIb complex. / CHO β/IX cells expressing vector, wild-type GpIbα or GpIbα deleted from residues 570-590 were treated for 1 hour with 100 μmol/L forskolin, which stimulated cyclic adenosine monophosphate-dependent protein kinase. (A) Forskolin caused increased phosphorylation of recombinant GpIbβ in CHO cells radiolabeled with 32P] orthophosphate. (B) Forskolin increased the amount of recombinant GpIbβ that bound to the GST-14-3-3ζ fusion protein only in cells expressing wild-type GpIbα. (C) Forskolin increased the quantity of 14-3-3ζ that coimmunoprecipitated with recombinant GpIbα from CHO cells transduced with wild-type GpIbα, GpIbβ, and GpIX. (D) Quantitative analysis of the effect of forskolin-induced PKA activation on the amount of 14-3-3ζ that coimmunoprecipitated with GpIbα and the amount of GpIbβ and GpIbα that bound to the GST-14-3-3ζ fusion protein, in CHO cells stably expressing wild-type GpIbα, GpIbβ, and GpIX. The amount of protein is reported as arbitrary densitometry units. (n = 3; *P < .001, Student ttest).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/95/2/10.1182_blood.v95.2.551/6/m_bloo00227005aw.jpeg?Expires=1769136717&Signature=I~5FnPgcOQjjwEbeQ6roOZ-~pYONMxzmyXfLPrHa42-iZ5votHFRBPvmUUSQfpNNzVOqKfzhyk7OlCqsWWpeTYwMIcdtCTzpU2W68H3Wn9sT388svhknS-oAsY7znvUn70lit0SAGpEo8EJTRP3Vhao3RWtXA9EhID~5l0o4-nmHDRlcpPXoAO~NbWR2AM31RTl7STtWx46SLQP~yfVZS82IcFU4xSQSCmmtLZhSO359jV~FpVe3ZXpmjF61LPpHZnt3-YUX2i7Hx46DA4YW1RLT~Mj9ylTLCUvme2zO-Aq7feEOpbQmPINM91cHvBFF7ftZUIAb8prSxSaV~Mo9xw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 5. Forskolin-treatment of Chinese hamster ovary cells expressing wild-type GpIb/β/IX enhanced the interaction between 14-3-3ζ and the GpIb complex. / CHO β/IX cells expressing vector, wild-type GpIbα or GpIbα deleted from residues 570-590 were treated for 1 hour with 100 μmol/L forskolin, which stimulated cyclic adenosine monophosphate-dependent protein kinase. (A) Forskolin caused increased phosphorylation of recombinant GpIbβ in CHO cells radiolabeled with 32P] orthophosphate. (B) Forskolin increased the amount of recombinant GpIbβ that bound to the GST-14-3-3ζ fusion protein only in cells expressing wild-type GpIbα. (C) Forskolin increased the quantity of 14-3-3ζ that coimmunoprecipitated with recombinant GpIbα from CHO cells transduced with wild-type GpIbα, GpIbβ, and GpIX. (D) Quantitative analysis of the effect of forskolin-induced PKA activation on the amount of 14-3-3ζ that coimmunoprecipitated with GpIbα and the amount of GpIbβ and GpIbα that bound to the GST-14-3-3ζ fusion protein, in CHO cells stably expressing wild-type GpIbα, GpIbβ, and GpIX. The amount of protein is reported as arbitrary densitometry units. (n = 3; *P < .001, Student ttest).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/95/2/10.1182_blood.v95.2.551/6/m_bloo00227005dx.jpeg?Expires=1769136717&Signature=k5PFc7l4C9Ujfx~Wm4A0LVmedSHbRqReYwIGGO5eVHEVTelP30Hv1cGjHdDm8JYQnrIH04rwyP9JZsO1nspLAx5x1apvhWICkFRIAcr7VRezGuSsz7qWdDRJGEbEBh0I8DPwKn0I58XvjdZAQsmx6vCRHXLpbkZfvGaUArJEKJyuyq4~P8-2MWIblnZB9IiuXIhZbDj~ZNifDCHgcHlXgMel641B6Okdh4TeiepfQrOuF05gxWJqPIQqcM1TrIJjqqN-zi8elCR0vATpjSmLLnhFCIilBZh2MvdYqAVH~vMoWTotSRPvDlR-S7HOpu8IlMOWPwBGp3rYknlBKiFyqA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal