A serious complication in hemophilia care is the development of factor VIII (FVIII) neutralizing antibodies (inhibitors). The authors used V gene phage display technology to define human anti-FVIII antibodies at the molecular level. The IgG4-specific, variable, heavy-chain gene repertoire of a patient with acquired hemophilia was combined with a nonimmune, variable, light-chain gene repertoire for display as single-chain variable domain antibody fragments (scFv) on filamentous phage. ScFv were selected by 4 rounds of panning on immobilized FVIII light chain. Sequence analysis revealed that isolated scFv were characterized by VH domains encoded by germline genes DP-10, DP-14, and DP-88, all belonging to the VH1 gene family. All clones displayed extensive hypermutation and were characterized by unusually long CDR3 sequences of 20 to 23 amino acids. Immunoprecipitation revealed that all scFv examined bound to the C2 domain of FVIII. Furthermore, isolated scFv competed with an inhibitory murine monoclonal antibody for binding to the C2 domain. Even though scFv bound FVIII with high affinity, they did not inhibit FVIII activity. Interestingly, the addition of scFv diminished the inhibitory potential of patient-derived antibodies with C2 domain specificity. These results suggest that the epitope of a significant portion of anti-C2 domain antibodies overlaps with that of the scFv isolated in this study.
Functional absence of blood coagulation factor VIII (FVIII) is associated with the X-linked bleeding disorder hemophilia A. The bleeding tendency in patients with hemophilia A can be corrected by the administration of plasma-derived or recombinant FVIII concentrates. After multiple transfusions, FVIII neutralizing antibodies (FVIII inhibitors) develop in approximately 25% of patients severely affected with hemophilia A.1 Spontaneous development of FVIII inhibitors in persons without hemophilia with normal FVIII levels occurs with a frequency of 1 case per million persons per year.2 FVIII inhibitors in both patient groups are associated with severe and sometimes life-threatening bleeding episodes.
Most of the inhibitors are directed toward epitopes located within the A2, A3, and C2 domains of the FVIII molecule.3 More detailed epitope mapping using a series of recombinant human/porcine FVIII hybrids revealed that residues Arg484-Ile508 contain a major determinant of the inhibitory epitope in the A2 domain of FVIII.4 Within the C2 domain, it has been proposed that residues Val2248through Ser2312 constitute a binding site for FVIII inhibitors.5 Recent evidence suggests that residues Glu2181-Val2243 contribute to the inhibitor epitope located in the C2 domain.6 A third inhibitor epitope has been localized to Gln1778 through Met1823 within the A3 domain.7,8 Studies on FVIII inhibitors are complicated because of the heterogeneity of anti-FVIII antibodies in patients' plasma.3 V gene phage display technology provides an opportunity to isolate human monoclonal antibodies from the total immunoglobulin repertoire.9Human immunoglobulin genes are assembled early in B-cell ontogeny by random rearrangement of variable (V), diversity (D), and joining (J) gene segments on the heavy (H) chain locus and V and J on either of the light (L) chain loci.10 Insertion and deletion of nucleotides at the junctions of the V, D, and J gene segments create additional diversity. On antigen stimulation, somatic hypermutation and receptor editing finally result in the formation of a repertoire of high-affinity antibodies.11 In the current study, we used phage display to isolate anti-FVIII light chain antibodies from a patient with acquired hemophilia. Our analysis indicates that antibodies with specificity for the C2 domain of FVIII have a large CDR3 and are encoded by gene segments of the VH1 family.
Materials and methods
Materials
Plasma-derived FVIII light chain was obtained from immunopurified FVIII concentrate.12 Anti-FVIII murine monoclonal antibodies (mAbs) CLB-CAg A, 9, 12 and 117 used in this study have been characterized previously12,13; mAbs ESH4 and ESH8 were purchased from American Diagnostica (Greenwich, CT). Recombinant FVIII fragments were expressed and metabolically labeled in insect cells using the Baculovirus system as described previously.7 14Taq DNA polymerase and restriction enzymes were purchased from Life Technologies (Breda, The Netherlands).
Patient's characteristics
After abdominal surgery, a previously healthy 44-year-old woman had severe hemorrhages. The level of FVIII appeared to be < 1%, and an inhibitor with a titer of 123 Bethesda units (BU)/mL was detected.15 Ten weeks later, the inhibitor titer reached a maximum value of approximately 1200 BU/mL. Plasma samples and peripheral blood mononuclear cells obtained at this time were used. Domain specificity and isotype of FVIII inhibitors were determined by immunoprecipitation.7,14 FVIII inhibitor neutralization was performed essentially as described previously.7
Phage display library construction
Peripheral blood lymphocytes obtained by Ficoll density centrifugation were used to isolate RNA, which was then used for cDNA synthesis with random hexamer primers. VH genes were amplified using each of the family-based back primers9 in combination with an IgG constant region primer 5′-CTTGTCCACCTTGGTGTTGCTGGG-3′. The repertoire was reamplified with an IgG4 subclass-specific oligonucleotide primer 5′-ACGTTGCAGGTGTAGGTCTTC-3′. Purified polymerase chain reaction products were subjected to a final round of amplification using a combination of family-based back primers, together with forward primers matching the different heavy chain joining (JH) germline genes; both primers were appended with NcoI orSalI restriction sites, respectively.16 The IgG4-specific VH gene repertoire was cloned in the vector pHEN-1-VLrep, which already contained a VL gene repertoire of nonimmune origin.17,18 The final repertoire was electroporated into Escherichia coli TG1 as described.9
Selection of phage library
Recombinant phages obtained by infection of the library with VSCM-13 helper phage (Stratagene, La Jolla, CA) were selected for binding to the FVIII light chain. A noninhibitory antibody specific for the light chain of FVIII, mAb CLB-CAg 12, was immobilized onto microtiter wells (Dynatech, Plockingen, Germany) at a concentration of 5 μg/mL in 50 mmol/L NaHCO3, pH 9.6. Wells were blocked with 3% human serum albumin (HSA) in Tris-buffered saline (TBS; 150 mmol/L NaCl, 50 mmol/L Tris, pH 7.4) for 2 hours at 37°C. Phages in TBS, 3% (wt/vol) HSA, and 0.5% (vol/vol) Tween-20 were preabsorbed to CLB-CAg 12-coated wells for 2 hours at room temperature. Subsequently, nonbound phages were transferred to microtiter wells containing FVIII light chain (100 ng/well) captured by mAb CLB-CAg 12 in 1 mol/L NaCl, 50 mmol/L Tris, pH 7.4, 2% (wt/vol) HSA. Alternatively, phages were selected against an FVIII light chain coated at a concentration of 2 μg/mL in 50 mmol/L NaHCO3, pH 9.6, overnight at 4°C in immunotubes (Nunc; Life Technologies, Breda, The Netherlands). After 20 washes with TBS/0.1% (vol/vol) Tween-20 and 20 washes with TBS, bound phages were eluted with 100 mmol/L triethylamine and used to infect E. coli TG1 cells.9 After each round of selection, phages from single-infected colonies were tested for binding to FVIII light chain immobilized by mAb CLB-CAg 12. Binding of phages was monitored by incubation with horseradish peroxidase-conjugated anti-M13 antibody as described.19 DNA sequences encoding the VH and VL domains of FVIII light-chain–specific clones were determined on an Applied Biosystems (Foster City, CA) 377XL automated DNA sequencer using primers LMB3, fdSEQ,9 and linkSEQ20 as described.21 Sequences were compared to germline V genes as compiled in the V-BASE sequence database.22
Characterization of scFv
To facilitate purification of scFv, V gene cassettes of FVIII light-chain–specific clones were subcloned in the expression vector pUC119-Sfi/Not-His6 as NcoI/NotI fragments.21 Expression and purification of scFv by immobilized metal chelate-affinity chromatography was performed essentially as described previously.23 Eluted fractions were dialyzed against TBS and analyzed by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE). Protein concentration was determined spectrophotometrically at A280.
Immunoprecipitation analysis was performed as follows: metabolically labeled FVIII fragments in immunoprecipitation buffer were pre-cleared by 2 successive incubations for 2 hours at room temperature with Ni-NTA agarose (Qiagen, Hilden, Germany) and 1 incubation with gelatin Sepharose 4B (Pharmacia-LKB, Woerden, The Netherlands). Immunoprecipitation buffer consisted of 50 mmol/L Tris, pH 7.6, 150 mmol/L NaCl, 20 mmol/L Imidazole, 1.2% (vol/vol) Triton X-100, 0.1% (vol/vol) Tween-20, 1% (wt/vol) bovine serum albumin, 10 μg/mL soybean trypsin inhibitor, 10 mmol/L benzamidine, and 5 mmol/L N-ethylmaleimide. Specific adsorption was performed by incubating pre-cleared medium with pre-formed scFv/Ni-NTA complexes overnight at 4°C. After extensive washing with immunoprecipitation buffer, SDS sample buffer was added and samples were analyzed under reducing conditions by 20% (wt/vol) SDS-PAGE.
Inhibitor neutralization by scFv
Patient's plasma or inhibitory mAb was diluted to a final concentration of 2 BU/mL in 50 mmol/L Tris, pH 7.3, and 0.2% (wt/vol) HSA. Serial dilutions of purified scFv were made in the same buffer. Diluted plasma or inhibitory mAb was incubated for 2 hours at 37°C with an equal volume of scFv and an equal volume of pooled normal plasma. Residual FVIII activity was measured relative to a control sample that was incubated in the absence of FVIII inhibitor in a one-stage clotting assay.
Results
Inhibitor characteristics and library construction
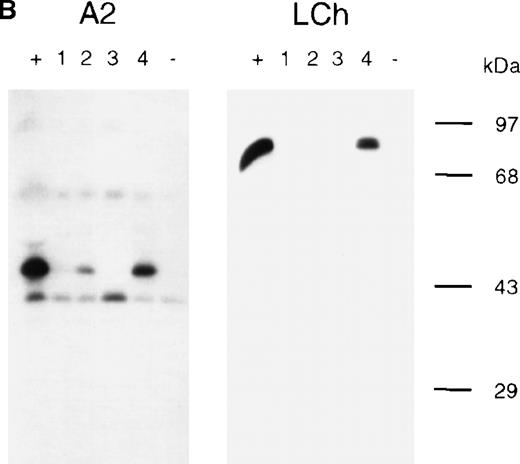
The domain specificity of anti-FVIII antibodies in the plasma of a patient with acquired hemophilia was evaluated by immunoprecipitation using metabolically labeled FVIII fragments. Patient's antibodies reacted with recombinant FVIII light-chain (A3-C1-C2), A2, and C2 domains (Figure 1A). The extent to which each epitope contributed to FVIII inhibition was determined by neutralization assays. Antibodies directed toward the A2 domain accounted for 50% of the FVIII inhibitory activity. Adding FVIII light chain resulted in 50% inhibitor neutralization, whereas only 20% neutralization was observed after the addition of the C2 domain (data not shown). These results indicated that the patient's antibodies interacted with the A2, C2, and A3-C1 domains of FVIII. Isotyping revealed a predominance of subclasses IgG2 and IgG4 for A2 domain-specific antibodies, whereas anti-FVIII light-chain antibodies consisted exclusively of subclass IgG4 (Figure 1B). The IgG4-specific VH gene repertoire was used to construct a phage display library consisting of 2.5 × 106 clones.
Characterization of anti-FVIII antibodies in the plasma of a patient.
Binding of antibodies to metabolically labeled FVIII fragments corresponding to the FVIII heavy chain (HCh), the A2 domain (A2), the FVIII light chain (LCh), and the C2 domain (C2) was evaluated by immunoprecipitation. (A) Reactivity of anti-FVIII antibodies in the patient's plasma. (lane 1, +) Positive control. mAb CLB-CAg 9 for HCh and A2, mAb CLB-CAg 117 for LCh and C2. (lane 2, −) Control plasma. (lane 3, P) Antibodies in the patient's plasma. (B) Subclass typing of anti-FVIII antibodies. (left panel, A2) Anti-A2 domain antibodies. (right panel, LCh) FVIII light-chain–specific antibodies. (lane 1, +) Total IgG. (lanes 2-5)1-4 IgG1, IgG2, IgG3, IgG4. (lane 6, −) Control plasma. In the patient's plasma no anti-FVIII antibodies of the IgM class could be detected (data not shown). Molecular weight markers are indicated at the right of the figures.
Characterization of anti-FVIII antibodies in the plasma of a patient.
Binding of antibodies to metabolically labeled FVIII fragments corresponding to the FVIII heavy chain (HCh), the A2 domain (A2), the FVIII light chain (LCh), and the C2 domain (C2) was evaluated by immunoprecipitation. (A) Reactivity of anti-FVIII antibodies in the patient's plasma. (lane 1, +) Positive control. mAb CLB-CAg 9 for HCh and A2, mAb CLB-CAg 117 for LCh and C2. (lane 2, −) Control plasma. (lane 3, P) Antibodies in the patient's plasma. (B) Subclass typing of anti-FVIII antibodies. (left panel, A2) Anti-A2 domain antibodies. (right panel, LCh) FVIII light-chain–specific antibodies. (lane 1, +) Total IgG. (lanes 2-5)1-4 IgG1, IgG2, IgG3, IgG4. (lane 6, −) Control plasma. In the patient's plasma no anti-FVIII antibodies of the IgM class could be detected (data not shown). Molecular weight markers are indicated at the right of the figures.
Isolation and sequence analysis of FVIII-specific clones
Recombinant phages expressing the patient's IgG4-specific VH gene repertoire were selected on immobilized FVIII light chain. After 4 rounds of panning, phages derived from 57 of 60 single-infected colonies displayed specificity for the FVIII light chain as determined by enzyme-linked immunosorbent assay (data not shown). The nucleotide sequences of the VH and VL genes of FVIII light-chain–specific clones were determined and aligned to the most homologous germline genes in the V-BASE sequence directory.22 In total, 5 unique VH domains were identified that were encoded by VH genes most likely derived from germline genes DP-10, DP-14, and DP-88, all from the VH1 gene family (Table1). Two VH domains (EL-16 and EL-25) were found in several clones in combination with different VL domains. The deduced amino acid sequences of the VH domains are compiled in Table2. The level of somatic mutation in FVIII light-chain–specific VH domains ranged from 11 to 16 amino acid substitutions (18 to 27 nucleotide substitutions) when compared with the most homologous germline genes. It should be noted that VH domains of clones EL-5, EL-16, and EL-25 are all derived from germline DP-14 (Table 2). All have a similar CDR3 sequence, and their patterns of somatic hypermutation suggest that the VHgenes of clones EL-5, EL-16, and EL-25 originate from a common B-cell precursor. The length of VH CDR3 (residues 95-102) of the FVIII light-chain–specific VH domains ranges from 20 to 23 amino acids (Table 2). In clones EL-5, EL-16, and EL-25, the rearranged JH segment, encoding the carboxy terminal part of the CDR3, was most homologous to gene segment JH6b.22Clones EL-9 and EL-14 have been assembled using gene segment JH3b. The 5 different VH domains identified paired with a variety of VL domains (Table 1). In total, we identified 13 unique VH-VL pairings, and in 7 of 13 the VL domain was encoded by Vκ1 family gene. Of the remaining 6, 4 were DPL16 (Vλ3 family gene) derived and the other 2 were Vκ4 and Vλ2 derived. Each unique VH-VL gene combination was subcloned and was expressed as scFv using the prokaryotic expression vector pUC119-Sfi/Not-His6.21
Most homologous germline genes used in FVIII light-chain–specific clones
| Clone . | VH Domain . | VL Domain . | ||
|---|---|---|---|---|
| Germline . | Family . | Germline . | Family . | |
| EL-5 | DP-14 | VH1 | L12a | VκI |
| EL-9 | DP-88 | VH1 | DPK8 | VκI |
| EL-14 | DP-10 | VH1 | DPK5 | VκI |
| EL-16 | DP-14 | VH1 | DPK5 | VκI |
| DPK8I,II | VκI | |||
| DPK24 | VκI | |||
| DPL11 | Vλ2 | |||
| DPL16I-III | Vλ3 | |||
| EL-25 | DP-14 | VH1 | DPK7 | VκI |
| DPL16 | Vλ3 | |||
| Clone . | VH Domain . | VL Domain . | ||
|---|---|---|---|---|
| Germline . | Family . | Germline . | Family . | |
| EL-5 | DP-14 | VH1 | L12a | VκI |
| EL-9 | DP-88 | VH1 | DPK8 | VκI |
| EL-14 | DP-10 | VH1 | DPK5 | VκI |
| EL-16 | DP-14 | VH1 | DPK5 | VκI |
| DPK8I,II | VκI | |||
| DPK24 | VκI | |||
| DPL11 | Vλ2 | |||
| DPL16I-III | Vλ3 | |||
| EL-25 | DP-14 | VH1 | DPK7 | VκI |
| DPL16 | Vλ3 | |||
Deduced protein sequences of isolated FVIII light-chain–specific scFv
| Heavy chains |
| FR1 CDR1 FR2 CDR2 FR3 CDR3 FR4 |
| ------------------------------ ----- -------------- ----------------- -------------------------------- ----------------------- ----------- |
| 111 |
| 12 3 4 56 789001 |
| 123456789012345678901234567890 12345 67890123456789 012a3456789012345 67890123456789012abc345678901234 567890abcdefghijklmno12 34567890123 |
| DP-10 QVQLVQSGAEVKKPGSSVKVSCKASGGTFS SYAIS WVRQAPGOGLEWMG GIIPIFGTANYAQKFQG RVTITADESTSTAYMELSSLRSEDTAVYYCAR |
| EL-14 ----------A---------------D--N -FP-- -------------- -------STK------- ---M---G---------N--------I----- QQNGGWYEGPLLE..PRPDALDI WGQGTMVTVSS |
| DP-14 QVQLVQSGAEVKKPGASVKVSCKASGYTFT SYGIS WVRQAPGQGLEWMG WISAYNGNTNYAQKLQG RVTMTTDTSTSTAYMELRSLRSDDTAVYYCAR |
| EL-5 ---- L--AT--------M----M----P-- --D-- ------------V- ---------H----F-- ---------RR--------------------- DGGGGAYEDVWSGEYPEYYAMDV WGQGTTVTVSS |
| EL-16 ---- L--AT--------M----M----P-- --D-- -------------- ---I-S---D----F-- ---------RR--------------------- ----------------------- ----------- |
| EL-25 ---- L--A---R---------------P-- --D-- -------------- ---I-S---D----F-- ---------RR--------------------- ----------------------- ----------- |
| DP-88 QVQLVQSGAEVKKPGSSVKVSCKASGGTFS SYAIS WVRQAPGQGLEWMG GIIPIFGTANYAQKFQG RVTITADKSTSTAYMELSSLRSEDTAVYYCAR |
| EL-9 -----------------------T----L- ----- ---------P--I- ------D-SKS--R--D ------NI----T-------------M-F-V- GASGIRYIDWP...PIPVDAFDI WGQGTMVTVSS |
| Light chains |
| FR1 CDR1 FR2 CDR2 FR3 CDR3 FR4 |
| ----------------------- ----------- --------------- ------- -------------------------------- --------- ----------- |
| 1 |
| 12 3 4 5 678 9 0 |
| 12345678901234567890123 45678901234 567890123456789 0123456 78901234567890123456789012345678 901234567 89012345678 |
| DPK5 DIQMTQSPSSVSASVGDRVTITC RASQGISSWLA WYQQKPGKAPKLLIY AASSLQS GVPSRFSGSGSGTDFTLTISSLQPEDFATYYC QQANSFP |
| EL-14 --V-------------------- ----------- --------------- ------- -------------------------------- -------LT FGGGTKVEIKR |
| L12a DIQMTQSPSTLSASVGDRVTITC RASQSISSWLA WYQQKPGKAPKLLIY KASSLES GVPSRFSGSGSGTEFTLTISSLQPDDFATYYC QQYNSYS |
| EL-5 E-VL----------I-------- ---EG-YH--- --------------- -----A- -A-----------D------------------ -HL---PLT FGGGTKVEIKR |
| DPK7 DIQMTQSPSSLSASVGDRVTITC RASQGISSWLA WYQQKP EKAPKSLIY AASSLQS GVPSRFSGSGSGTDFTLTISSLQPEDFATYYC QQYNSYP |
| EL-25 ---------F------------- ----H-N---- ------G----L--- ---R--- -------------E------------------ --L----LT FGGGTKLEIKR |
| DPK8 DIQLTQSPSFLSASVGDRVTITC RASQGISSYLA WYQQKPGKAPKLLIY AASTLQS GVPSRFSGSGSGTEFTLTISSLQPEDFATYYC QQLNSYP |
| EL-9 ETT------S------------- ---R-L-R--- --------T------ ------- ------------------V----A---G---- --YHTISRT FGPGTKLEIKR |
| EL-16 --VM---------F----I---- -------G--- --------------- ------- --------------------G-----V----- -KY--A-WT FGQGTKVEIKR |
| Heavy chains |
| FR1 CDR1 FR2 CDR2 FR3 CDR3 FR4 |
| ------------------------------ ----- -------------- ----------------- -------------------------------- ----------------------- ----------- |
| 111 |
| 12 3 4 56 789001 |
| 123456789012345678901234567890 12345 67890123456789 012a3456789012345 67890123456789012abc345678901234 567890abcdefghijklmno12 34567890123 |
| DP-10 QVQLVQSGAEVKKPGSSVKVSCKASGGTFS SYAIS WVRQAPGOGLEWMG GIIPIFGTANYAQKFQG RVTITADESTSTAYMELSSLRSEDTAVYYCAR |
| EL-14 ----------A---------------D--N -FP-- -------------- -------STK------- ---M---G---------N--------I----- QQNGGWYEGPLLE..PRPDALDI WGQGTMVTVSS |
| DP-14 QVQLVQSGAEVKKPGASVKVSCKASGYTFT SYGIS WVRQAPGQGLEWMG WISAYNGNTNYAQKLQG RVTMTTDTSTSTAYMELRSLRSDDTAVYYCAR |
| EL-5 ---- L--AT--------M----M----P-- --D-- ------------V- ---------H----F-- ---------RR--------------------- DGGGGAYEDVWSGEYPEYYAMDV WGQGTTVTVSS |
| EL-16 ---- L--AT--------M----M----P-- --D-- -------------- ---I-S---D----F-- ---------RR--------------------- ----------------------- ----------- |
| EL-25 ---- L--A---R---------------P-- --D-- -------------- ---I-S---D----F-- ---------RR--------------------- ----------------------- ----------- |
| DP-88 QVQLVQSGAEVKKPGSSVKVSCKASGGTFS SYAIS WVRQAPGQGLEWMG GIIPIFGTANYAQKFQG RVTITADKSTSTAYMELSSLRSEDTAVYYCAR |
| EL-9 -----------------------T----L- ----- ---------P--I- ------D-SKS--R--D ------NI----T-------------M-F-V- GASGIRYIDWP...PIPVDAFDI WGQGTMVTVSS |
| Light chains |
| FR1 CDR1 FR2 CDR2 FR3 CDR3 FR4 |
| ----------------------- ----------- --------------- ------- -------------------------------- --------- ----------- |
| 1 |
| 12 3 4 5 678 9 0 |
| 12345678901234567890123 45678901234 567890123456789 0123456 78901234567890123456789012345678 901234567 89012345678 |
| DPK5 DIQMTQSPSSVSASVGDRVTITC RASQGISSWLA WYQQKPGKAPKLLIY AASSLQS GVPSRFSGSGSGTDFTLTISSLQPEDFATYYC QQANSFP |
| EL-14 --V-------------------- ----------- --------------- ------- -------------------------------- -------LT FGGGTKVEIKR |
| L12a DIQMTQSPSTLSASVGDRVTITC RASQSISSWLA WYQQKPGKAPKLLIY KASSLES GVPSRFSGSGSGTEFTLTISSLQPDDFATYYC QQYNSYS |
| EL-5 E-VL----------I-------- ---EG-YH--- --------------- -----A- -A-----------D------------------ -HL---PLT FGGGTKVEIKR |
| DPK7 DIQMTQSPSSLSASVGDRVTITC RASQGISSWLA WYQQKP EKAPKSLIY AASSLQS GVPSRFSGSGSGTDFTLTISSLQPEDFATYYC QQYNSYP |
| EL-25 ---------F------------- ----H-N---- ------G----L--- ---R--- -------------E------------------ --L----LT FGGGTKLEIKR |
| DPK8 DIQLTQSPSFLSASVGDRVTITC RASQGISSYLA WYQQKPGKAPKLLIY AASTLQS GVPSRFSGSGSGTEFTLTISSLQPEDFATYYC QQLNSYP |
| EL-9 ETT------S------------- ---R-L-R--- --------T------ ------- ------------------V----A---G---- --YHTISRT FGPGTKLEIKR |
| EL-16 --VM---------F----I---- -------G--- --------------- ------- --------------------G-----V----- -KY--A-WT FGQGTKVEIKR |
FR, framework region; CDR, complementarity-determining region. Dashes indicate sequence identity to germline. Residues encoded by JH gene segments are underlined. Sequence numbering is according to Kabat.24 Sequences are available from GenBank under accession numbers AF146400 (VH EL-5); AF146401 (VH EL-9); AF146402 (VH EL-14); AF146403 (VH EL-16); AF146404 (VH EL-25); AF146405 (VL EL-5);AF146406 (VL EL-9); AF146407 (VL EL-14); AF146408 (VL EL-16); AF146409(VL EL-25).
FVIII specificity of isolated scFv
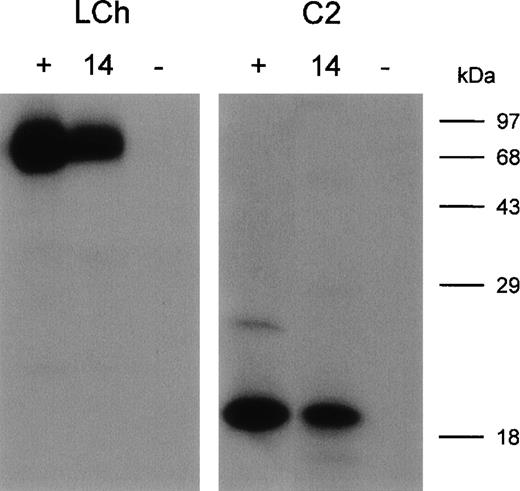
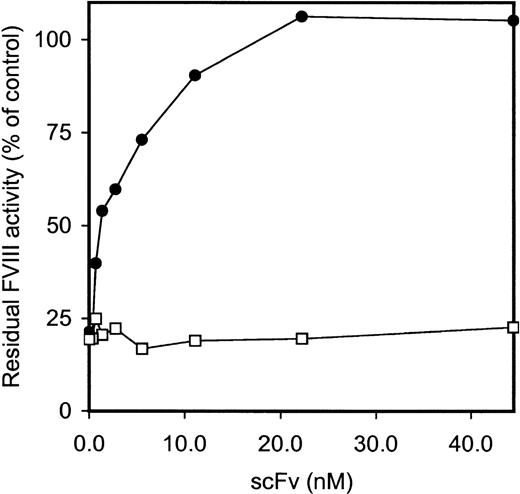
Five clones reacting with the FVIII light chain were selected for further analysis (Table 2). E. coli TG1-expressed scFv were purified as described in Materials and Methods. All 5 scFv showed specific binding to the FVIII light chain, whereas scFv derived from a randomly picked control clone (O4) did not react under our experimental conditions (data not shown). Within the FVIII light chain, 2 dominant B-cell epitopes for inhibitory antibodies are located within the A3 and C2 domains.5-8 To investigate the domain specificity of scFv, immunoprecipitations with metabolically labeled FVIII light chain and C2 domain were performed. ScFv EL-14 reacted with the radiolabeled FVIII light chain and the C2 domain (Figure2). Identical results were obtained for the other 4 scFv (data not shown). Preliminary experiments demonstrated that scFv were fully capable of competing for binding with the murine mAb CLB-CAg 117 to FVIII. This C2 domain-specific antibody has been described as efficiently interfering with FVIII activity.13The ability of scFv to inhibit FVIII procoagulant activity was compared to that of IgG purified from a patient's plasma. Surprisingly, no inhibition of FVIII procoagulant activity was observed for the scFv up to a concentration of 200 nmol/L (Figure3). In contrast, the patient's purified IgG inhibited FVIII activity with a specific activity of 160 BU/mg.
Immunoprecipitation of metabolically labeled FVIII light chain (LCh) and C2 domain (C2) by scFv.
(lane 1, +) Positive control; mAb CLB-CAg 117. (lane 2, 14) scFv EL-14. (lane 3, −) Negative control; scFv O4. Molecular weight markers are given at the right of the figure.
Immunoprecipitation of metabolically labeled FVIII light chain (LCh) and C2 domain (C2) by scFv.
(lane 1, +) Positive control; mAb CLB-CAg 117. (lane 2, 14) scFv EL-14. (lane 3, −) Negative control; scFv O4. Molecular weight markers are given at the right of the figure.
Functional characterization of isolated scFv.
Various concentrations of purified patient's IgG (○) and scFv EL-14 (•) were incubated with an equal volume of normal plasma for 2 hours at 37°C. FVIII activity, determined by a one-stage clotting assay, is depicted relative to a control incubation in the absence of IgG and scFv. Similar results were obtained for scFv EL-5, EL-9, EL-16, EL-25, and negative control scFv O4.
Functional characterization of isolated scFv.
Various concentrations of purified patient's IgG (○) and scFv EL-14 (•) were incubated with an equal volume of normal plasma for 2 hours at 37°C. FVIII activity, determined by a one-stage clotting assay, is depicted relative to a control incubation in the absence of IgG and scFv. Similar results were obtained for scFv EL-5, EL-9, EL-16, EL-25, and negative control scFv O4.
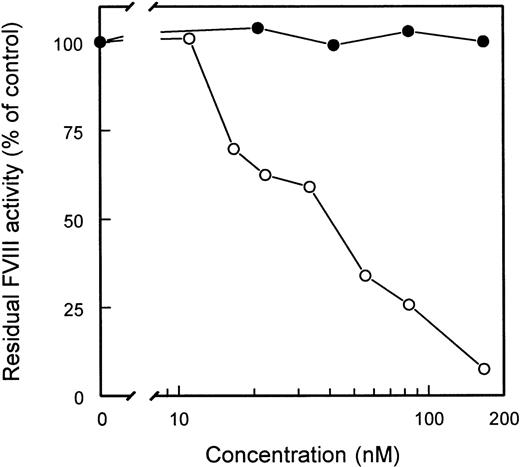
Inhibitor neutralizing capacity of scFv
The ability of scFv to interfere with FVIII inhibitory activity of CLB-CAg 117, a C2 domain-specific antibody, was tested. Adding increasing amounts of scFv EL-14 completely eliminated FVIII inhibition by CLB-CAg 117 (Figure 4). In contrast, scFv EL-14 did not affect the inhibitory activity of CLB-CAg A, a monoclonal antibody directed against residues Lys1804-Lys1818 in the A3 domain of FVIII.25 In addition, scFv EL-5, EL-9, EL-16, and EL-25 were capable of neutralizing the inhibition of FVIII by CLB-CAg 117. Complete neutralization of CLB-CAg 117 was reached at concentrations of 100 to 400 nmol/L for these scFv.
Inhibitor neutralization by isolated scFv.
CLB-CAg A and CLB-CAg 117 were diluted to a concentration that corresponded to approximately 2 BU/mL. Increasing concentrations of scFv EL-14 were added, and the mixture was incubated for 2 hours at 37°C. Residual FVIII activity was determined relative to a control sample that was incubated in the absence of mAb. CLB-CAg A (□); CLB-CAg 117 (•).
Inhibitor neutralization by isolated scFv.
CLB-CAg A and CLB-CAg 117 were diluted to a concentration that corresponded to approximately 2 BU/mL. Increasing concentrations of scFv EL-14 were added, and the mixture was incubated for 2 hours at 37°C. Residual FVIII activity was determined relative to a control sample that was incubated in the absence of mAb. CLB-CAg A (□); CLB-CAg 117 (•).
Similarly, we tested whether scFv could abrogate the inhibition of FVIII by the patient's purified IgG. First, the contribution of anti-C2 antibodies to the total FVIII inhibitory activity of the patient's IgG was assessed. A recombinant C2 domain could neutralize 23% ± 5% of FVIII inhibitory activity of the patient's IgG. The addition of scFv EL-14 resulted in similar levels of neutralization (23% ± 4%). The same results were obtained with the other 4 scFv (data not shown). Simultaneously adding all scFv did not result in higher levels of neutralization. These findings suggested that the isolated scFv were capable of competing with the patient's C2-specific IgG for binding to FVIII.
Discussion
Development of neutralizing antibodies to FVIII constitutes a major complication in hemophilia care. Despite considerable insights into epitope specificity and mode of action of FVIII inhibitors, limited information is available on the primary structure of human antibodies directed against FVIII. In this study, we used V gene phage display to explore the properties of scFv with specificity for the FVIII light chain. Thirteen scFv were isolated, all directed against the C2 domain of FVIII. It should be noted that FVIII inhibitors with A3-C1 specificity were detected in the patient's plasma. However, we were unable to isolate scFv directed against the A3-C1 domains. During the selection procedure, potential binding sites in the A3-C1 domains may be masked by the methods used for immobilization of the FVIII light chain.
Sequence analysis revealed that heavy chains of these scFv were encoded by VH genes, most homologous to the germline gene segments DP-10, DP-14, and DP-88, all belonging to the VH1 gene family. The extensive hypermutation observed suggests that these FVIII-specific VH genes originate from antigen-stimulated B cells.11 Germline gene sequences DP-10, DP-14, and DP-88 all encode an identical combination of loop conformations or canonical structures.26 Previously, using Epstein-Barr virus immortalization, a monoclonal IgG4κ antibody (BO2C11) was derived from the B-cell repertoire of a hemophilia A patient with an inhibitor.27 The heavy chain of this C2 domain-specific antibody was encoded by the gene segment DP-5, also belonging to the VH1 gene family.27 These data suggest that FVIII antibodies with C2 domain specificity preferentially use VH gene segments derived from the VH1 family. The scFv described in this study are derived from a single patient with acquired hemophilia. Further analysis of the VH gene use of additional C2-specific anti-FVIII antibodies is required to substantiate our findings. In healthy individuals, the random rearrangement of V, D, and J segments may generate autoreactive antibodies that will be deleted from the repertoire on encountering antigen.11 Therefore, high-affinity autoreactive antibodies are unlikely to be isolated from the repertoire of healthy persons.28 Consequently, we do not expect to find anti-C2 domain antibodies in the repertoire of a nonimmune donor similar to the ones described in this study.
In this study we used a nonimmune VL gene repertoire to assemble FVIII-specific scFv. Vκ1 and Vλ3 gene segments primarily encoded the variable light chains identified in this study. Preferential use of the latter light chain genes cannot be explained by the limited diversity of the used VL gene repertoire because various antibodies with different light chains have been isolated from this VL gene library.9,17 18
FVIII inhibitors with C2 specificity have been shown to inhibit FVIII binding to von Willebrand factor, phospholipids, or both.27 29-31 Surprisingly, the scFv described in this study did not inhibit FVIII activity (Figure 3). The size of scFv, smaller than that of complete IgG antibodies (30 vs. 150 kd), may explain the lack of FVIII inhibition. Alternatively, the isolated scFv may correspond to noninhibitory antibodies in a patient's plasma. We are currently constructing complete IgG4 molecules using the variable domains of scFv. Functional analysis of these complete IgG4 molecules will reveal whether the variable heavy-chain domains identified in this study are representative of either inhibitory or noninhibitory antibodies. Competition experiments revealed that scFv neutralized the inhibitory activity of mAb and human anti-FVIII antibodies with C2 specificity, suggesting that the binding sites for scFv are in proximity to the inhibitor epitope in the C2 domain.
Acknowledgments
The authors thank M-J. S. H. Donath for the purified FVIII light chain. They also thank W. G. van Aken, R. C. Aalberse, K. Mertens, P. J. Lenting, and J. A. van Mourik for critical evaluation of the manuscript.
Supported by a travel grant from the Haemophilia Foundation and by grant G9410995 from the Medical Research Council, United Kingdom.
Reprints:Jan Voorberg, Department of Blood Coagulation, CLB, Plesmanlaan 125, 1066 CX Amsterdam, The Netherlands; e-mail:j_voorberg@clb.nl.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal