Long-term survivors of aplastic anemia (AA) have a high incidence of clonal disorders, in particular paroxysmal nocturnal hemoglobinuria (PNH), myelodysplastic syndromes (MDS), and acute nonlymphocytic leukemia. To investigate the potential involvement of N-RAS gene mutations in the predisposition to leukemic evolution, a subset of patients at potentially increased risk for clonal disease was selected based on evidence of existing clonal evolution. Nine patients showed a monoclonal pattern of X-chromosome inactivation, 18 demonstrated a PNH clone, and in 3 MDS developed during the course of this study. No mutations were detected during the aplastic phase of disease; 2 of 3 patients with MDS after AA also showed no mutations. However, in 1 patient in whom the disease transformed from AA/PNH to MDS, a mutation of GGT → GAT at N-RAS codon 13 became detectable, whereas the PNH mutation disappeared. The authors conclude that N-RAS mutations are not an early event preceding transformation of AA or AA/PNH to leukemia. In a subset of patients, RAS mutations may occur at the time of evolution to MDS, but preexisting RAS mutations do not explain the propensity of AA to leukemogenesis. Although PNH is also associated with leukemia, this may arise in the non-PNH cells, indicating that PIG-A gene mutation is not per se oncogenic.
Idiosyncratic aplastic anemia (AA) is a rare hematologic disease characterized by peripheral blood pancytopenia and bone marrow hypocellularity. Long-term survivors of AA after antithymocyte globulin (ATG) therapy have a high incidence of clonal disorders, including paroxysmal nocturnal hemoglobinuria (PNH), myelodysplastic syndromes (MDS), and acute nonlymphocytic leukemia (ANLL).1-6 PNH is itself an acquired clonal stem cell disorder, but the development of other clonal disorders, such as MDS and ANLL, has also been observed in patients with PNH as a late terminal event.7-13
An activating mutation of N-RAS in codons 12, 13, or 61 has been reported to occur in 20% to 30% of patients with ANLL and MDS.14-16 MDS with an N-RAS mutation has an increased likelihood of evolution to ANLL.17 If patients with AA or PNH had an increased incidences of N-RAS mutation, this might explain the evolution to malignant disease. Several groups,18-20including our own, have examined series of patients with AA or PNH and have not detected any activating mutations.
Although MDS or ANLL eventually develops in as many as 26% of patients with AA, this may occur over a period of 20 years, and previous studies have been subject to the criticism that individual patients may not have been at risk for clonal evolution at the time of the study. We therefore decided to extend our previous work by examining a selected subgroup of patients with evidence of clonal disease. Three were patients in whom frank MDS developed during the course of our study, 18 were patients with AA with a PNH clone detectable by flow cytometry, and 9 were female patients with AA with a clonal pattern of X-chromosome inactivation. None of the 30 patients had detectable mutations in codons 12, 13 or 61, of the N-RAS gene during the AA or PNH phase of disease. However, 1 patient with a PNH clone later evolved to MDS and to ANLL, and an acquired activating mutation gly-> asp at codon 13 on evolution to MDS, concurrent with the loss of the specific 19bp deletion mutation within his PIG-A gene.
Patients and methods
Patients
Thirty patients with AA were selected for study on the basis of evidence for evolution of clonal disease. Nine female patients with AA had a monoclonal pattern of X-chromosome inactivation in their peripheral blood cells, 3 patients (2 female and 1 male) developed MDS during the course of this study, and 18 patients (12 males and 6 females) showed evidence of a PNH clone by flow cytometry; of these, 5 females had shown a clonal pattern of X-chromosome inactivation in total peripheral blood cells, and 1 showed it only after enrichment of CD59-ve cells. No karyotype abnormalities were observed in the patients with AA or AA/PNH, while all 3 patients with MDS showed multiple abnormalities. Patient characteristics are listed in Table1.
Patient characteristics
| Patient No. . | Sex . | Age (y) . | Diagnosis . | Disease Duration (y) . | Hb g/dL . | WBC ×109/L . | Neut ×109/L . | Platelet ×109/L . |
|---|---|---|---|---|---|---|---|---|
| 1 | F | 36 | AA | 8 | 14.4 | 7.8 | 5.8 | 193 |
| 2 | F | 28 | NSAA | 5.9 | 13 | 3.2 | 7.3 | 49 |
| 3 | F | 71 | AA | 0.2 | NA | NA | NA | NA |
| 4 | F | 61 | NSAA | 2.3 | 8.3 | 7.8 | 5.9 | 279 |
| 5 | F | 26 | NSAA | <0.1 | 8.3 | 3.5 | 1.5 | 40 |
| 6 | F | 24 | AA | <0.1 | 6.3 | 2.3 | 0.9 | 19 |
| 7 | F | 33 | NSAA | 4 | 12.3 | 3 | 1.3 | 10 |
| 8 | F | 68 | NSAA | 0.5 | 11.3 | 5.9 | NA | 93 |
| 9 | F | 38 | NSAA | 3.6 | 11.4 | 6.5 | 3.7 | 107 |
| 10 | F | 64 | AA/MDS | 5.2 | 8.3 | 9.2 | 2.8 | 29 |
| 11 | F | 69 | AA/MDS | 7 | 8.4 | 1.1 | 0.1 | 6 |
| 12 | M | 71 | AA/MDS | 3.6 | 9.1 | 4.8 | 0.8 | 74 |
| 13 | F | 21 | AA/PNH | 2.5 | 10.6 | 5.6 | 3 | 127 |
| 14 | F | 29 | AA/PNH | 12 | 7.8 | 1.9 | 2.7 | 62 |
| 15 | F | 54 | AA/PNH | 13 | 10.3 | 6.9 | 3.9 | 22 |
| 16 | F | 35 | AA/PNH | 21 | 6.4 | 3.4 | — | 105 |
| 17 | F | 34 | AA/PNH | 12 | 10 | 4 | 2.9 | 137 |
| 18 | F | 60 | AA/PNH | 12 | 10.3 | 3.1 | 2.1 | 75 |
| 19 | M | 67 | AA/PNH | 1.9 | 7.9 | 1.0 | 0.1 | 47 |
| 20 | M | 43 | AA/PNH | 4.5 | 11.2 | 2.7 | 1.5 | 47 |
| 21 | M | 29 | AA/PNH | 6.5 | 13.5 | 4.5 | 2.7 | 106 |
| 22 | M | 31 | AA/PNH | 1.8 | 10.2 | 3.9 | 2.6 | 173 |
| 23 | M | 30 | AA/PNH | 10 | 11.9 | 3.1 | — | 64 |
| 24 | M | 30 | AA/PNH | 1.2 | 13.4 | 4.3 | — | 63 |
| 25 | M | 24 | AA/PNH | 1 | 13.5 | 3.4 | — | 159 |
| 26 | M | 51 | AA/PNH | 1.4 | 10.3 | 0.7 | 0.1 | 10 |
| 27 | M | 31 | AA/PNH | 0.1 | 10 | 4.9 | 2.8 | 62 |
| 28 | M | 55 | AA/PNH | 3 | 9 | 1.4 | 0.3 | 10 |
| 29 | M | 42 | AA/PNH | 24 | 9.9 | 3.2 | — | 36 |
| 30 | M | 41 | AA/PNH | 1.6 | 9 | 1.7 | 0.8 | 25 |
| Patient No. . | Sex . | Age (y) . | Diagnosis . | Disease Duration (y) . | Hb g/dL . | WBC ×109/L . | Neut ×109/L . | Platelet ×109/L . |
|---|---|---|---|---|---|---|---|---|
| 1 | F | 36 | AA | 8 | 14.4 | 7.8 | 5.8 | 193 |
| 2 | F | 28 | NSAA | 5.9 | 13 | 3.2 | 7.3 | 49 |
| 3 | F | 71 | AA | 0.2 | NA | NA | NA | NA |
| 4 | F | 61 | NSAA | 2.3 | 8.3 | 7.8 | 5.9 | 279 |
| 5 | F | 26 | NSAA | <0.1 | 8.3 | 3.5 | 1.5 | 40 |
| 6 | F | 24 | AA | <0.1 | 6.3 | 2.3 | 0.9 | 19 |
| 7 | F | 33 | NSAA | 4 | 12.3 | 3 | 1.3 | 10 |
| 8 | F | 68 | NSAA | 0.5 | 11.3 | 5.9 | NA | 93 |
| 9 | F | 38 | NSAA | 3.6 | 11.4 | 6.5 | 3.7 | 107 |
| 10 | F | 64 | AA/MDS | 5.2 | 8.3 | 9.2 | 2.8 | 29 |
| 11 | F | 69 | AA/MDS | 7 | 8.4 | 1.1 | 0.1 | 6 |
| 12 | M | 71 | AA/MDS | 3.6 | 9.1 | 4.8 | 0.8 | 74 |
| 13 | F | 21 | AA/PNH | 2.5 | 10.6 | 5.6 | 3 | 127 |
| 14 | F | 29 | AA/PNH | 12 | 7.8 | 1.9 | 2.7 | 62 |
| 15 | F | 54 | AA/PNH | 13 | 10.3 | 6.9 | 3.9 | 22 |
| 16 | F | 35 | AA/PNH | 21 | 6.4 | 3.4 | — | 105 |
| 17 | F | 34 | AA/PNH | 12 | 10 | 4 | 2.9 | 137 |
| 18 | F | 60 | AA/PNH | 12 | 10.3 | 3.1 | 2.1 | 75 |
| 19 | M | 67 | AA/PNH | 1.9 | 7.9 | 1.0 | 0.1 | 47 |
| 20 | M | 43 | AA/PNH | 4.5 | 11.2 | 2.7 | 1.5 | 47 |
| 21 | M | 29 | AA/PNH | 6.5 | 13.5 | 4.5 | 2.7 | 106 |
| 22 | M | 31 | AA/PNH | 1.8 | 10.2 | 3.9 | 2.6 | 173 |
| 23 | M | 30 | AA/PNH | 10 | 11.9 | 3.1 | — | 64 |
| 24 | M | 30 | AA/PNH | 1.2 | 13.4 | 4.3 | — | 63 |
| 25 | M | 24 | AA/PNH | 1 | 13.5 | 3.4 | — | 159 |
| 26 | M | 51 | AA/PNH | 1.4 | 10.3 | 0.7 | 0.1 | 10 |
| 27 | M | 31 | AA/PNH | 0.1 | 10 | 4.9 | 2.8 | 62 |
| 28 | M | 55 | AA/PNH | 3 | 9 | 1.4 | 0.3 | 10 |
| 29 | M | 42 | AA/PNH | 24 | 9.9 | 3.2 | — | 36 |
| 30 | M | 41 | AA/PNH | 1.6 | 9 | 1.7 | 0.8 | 25 |
F, female; Hb, hemoglobin; M, male; NA, not available; Neut, neutrophil; WBC, white blood count.
A case report on patient 12 has been published.12
Cell separation and DNA and RNA extraction
Polymorphonuclear (PMN) cells were separated from mononuclear cells (MNC) by gradient centrifugation, using Ficoll Paque (Amersham Pharmacia, St. Albans, UK). PMNs were separated from red cells by sedimentation in 5% Dextran. Cells were lysed in Tri-reagent (Sigma, Poole, UK), and DNA and RNA were extracted according to the manufacturer's protocol.
Clonality analysis by RT-PCR
Reverse transcription–polymerase chain reaction.
Patients heterozygous for the nt.1311 C/T polymorphism in the G6PD gene21 22 were analyzed by reverse transcription–polymerase chain reaction (RT-PCR). A maximum 2 μg total cellular RNA from granulocytes or lymphocytes was incubated at 80°C for 3 minutes, then made up to 20.2 μL final volume containing 50 μmol/L antisense primer D 5′-GGTGCAGCAGTGGGGTGAAA-3′), 1 × first-strand buffer, 500μmol/L each dNTP, 25 U RNasin (Promega, Southampton, UK), and 250 U MMLV-RT (Gibco-BRL, Paisley, UK) and incubated at 42°C for 1 hour. The reaction was made up to 100 μL in 1 × PCR buffer, 10μmol/L primer sense A 5′-GACCAAGAAGCCGGGCATGTT-3′), and 1 U Taq DNA polymerase. The amplification was carried out for 40 cycles of 1 minute at 94°C, 1 minute at 56°C, and 1 minute at 72°C. One microliter of the first-round PCR product was added to 49μL of a fresh reaction mix and reamplified using15 pmol each of sense F 5′-TGTTCTTCAACCCCGAGGAGT-3′) and antisense M 5′-AAGACGTCCAGGATGAGGTGATC-3′) primers. Primer M contained 2 mismatches to the gene sequence to create a Bcl I restriction site if the T allele was present.
Digestion of RT-PCR products.
A reaction mix was made up containing 1 μL of appropriate 10× buffer, 1 μL Bcl I (New England BioLabs, Hitchin, UK), and 8 μL DNA amplified with the mismatched primer and incubated at 50°C for 1 hour. Digested products were separated by electrophoresis on 4% NuSieve agarose gels (FMC Bioproducts, Rockland, ME) containing ethidium bromide and visualized using a UVP Imager. The image was saved on disk, and the bands were quantitated using ImagerSoft 1D2D software. Control experiments using artificial mixtures of C/C and T/T DNA demonstrated a linear relationship with band intensity (data not shown).
Detection of N-RAS mutations
Amplification of the N-RAS gene by PCR.
Regions of the N-RAS gene spanning codons 12 and 13 or 61 were amplified from cellular genomic DNA; 100 ng DNA was used in a 50 μL PCR reaction. Primers sense NA12, 5′-GACTGAGTACAAACTGGTGG-3 and antisense NB12 5′-CTCTATGGTGGGATCATATT-3′) amplified an exon 1 fragment of 109 bp including codons 12 and 13. Primers sense NA61 5′-GGTGAAACCTGTTTGTTGGA-3 and antisense NB61 5′-ATACACAGAGGAAGCCTTCG-3′) amplified an exon 2 fragment of 103 bp including codon 61.
Blotting.
Thirty-microliter PCR products were denatured with 300 μL of a solution containing 0.4 mol/L NaOH/21 mmol/L EDTA at 95°C for 5 minutes, cooled on ice, and neutralized with 300 μL of a solution containing 0.92 mol/L Tris, pH 7.4. Equal portions of each denatured product were transferred to 3 replicate Hybond NFP nylon filters (Amersham Pharmacia) wetted with 20 × SSC (3 mol/L NaCl, 0.3 mol/L sodium citrate, pH 7), using a slot blot apparatus (Minifold II, Schleicher & Schuell). Filters were air-dried, and DNA was cross-linked to the membrane by exposure to ultraviolet light.
Labeling of oligonucleotide probes.
Twenty-mer oligonucleotide probes complementary to each potential wild-type and mutant N-RAS sequence for codons 12, 13, and 61 were obtained from Clontech.
2 pmol of each oligonucleotide was incubated at 65°C for 5 minutes, then 5′ end-labeled in a 15-μL reaction containing 3 μL γ-32P] adenosine, 5′ triphosphate (370 MBq/mL; ICN), and 15 U T4 polynucleotide kinase (Amersham Pharmacia) in the manufacturer's buffer at 37°C for 45 minutes. Labeled oligomers were separated from unincorporated nucleotide by Sephadex G50 chromatography.
Allele-specific oligonucleotide hybridization.
Hybridizations were performed with a mixture of 3 oligonucleotide probes with a common melting temperature. Filters were prehybridized at 56°C for 1 to 2 hours in hybridization buffer (5 × SSPE, 1% SDS, 0.1 mg/mL herring sperm DNA, 5 × Denhardt solution). This was replaced with fresh buffer containing the probe and hybridized at 56°C for another 1 to 2 hours. The filters were washed in 2 × SSPE, 0.1% SDS, for 20 minutes at room temperature and then in 5 × SSPE, 0.1% SDS, at 56°C for 1 hour. The final stringent wash was carried out at 66°C for codons 12 and 13 probes, or at 62°C for codon 61 probes, in 5 × SSPE, 0.1% SDS, for 10 minutes with continuous agitation. Membranes were exposed to Kodak XAR film (Eastman Kodak, Rochester, NY) at room temperature for 3 to 4 hours or at 70°C for longer periods. Membranes showing nonspecific signals were rewashed, increasing the temperature by 1°C each time until such signals were no longer detectable.
Quantification of GPI-linked surface proteins
Analysis of GPI-anchored membrane proteins on neutrophils, monocytes, lymphocytes, and erythrocytes was performed by flow cytometry using monoclonal antibodies specific to CD55, CD59, CD14, CD16, and CD66c as previously described.12
PCR amplification of the PIG-A gene
The entire coding region of the PIG-A gene was amplified in 8 overlapping fragments, to include the complete exons and the 5′ and 3′ intron/exon boundaries. Genomic DNA (100 ng) was used in a reaction containing 1 × standard buffer with 1.5 mmol/L MgCl2, 100μmol/L each dNTP, 15 pmol each of sense and antisense primers, and 1 U Taq DNA polymerase (PE Applied Biosystems, Warrington, UK) in a total volume of 50 μL, for 35 cycles of 45 seconds at 94°C, 30 seconds at 57°C, and 1 minute at 72°C, followed by 5 minutes at 72°C for 1 cycle.
RT-PCR for the PIG-A gene
A maximum 1μg cellular RNA from PMN or MNC was used as a template to make cDNA in a 20-μL reaction containing 0.4 μg random primers (Promega), 1 × first-strand buffer, 200 μmol/L of each dNTP, and 200 U RNase H free reverse transcriptase (Superscript ΙΙ; Gibco-BRL) at 42°C for 60 minutes. Two microliters RT reaction was amplified by PCR in a total volume of 50μL, as above.
Gel electrophoresis and DNA purification
PCR or RT-PCR products were analyzed by electrophoresis on 4% NuSieve agarose (FMC Bioproducts) gel and visualized by ethidium bromide staining. Before reamplification, DNA fragments were excised from the gel and purified using a QIAGEN DNA purification kit.
Direct sequencing of genomic DNA
Dideoxy sequencing was performed using an ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction Kit (Perkin Elmer) and was analyzed using an automated sequencer ABI PRISM (model 377, version 2.1.1; PE Applied Biosystems).
Single-strand conformation polymorphism and heteroduplex analysis
For single-strand conformation polymorphism (SSCP), 5μL PCR product was denatured for 5 minutes at 96°C in 10 μL 95% formamide loading buffer and placed on ice slurry. Then 5 μL sample was loaded onto an MDE polyacrylamide gel (FMC BioProducts) and electrophoresed at room temperature for 16 to 24 hours at 250 V. For heteroduplex analysis, the sample was denatured at 96°C for 2 minutes and allowed to cool to room temperature for 1 hour for duplex formation. Immediately before loading the sample onto the gel, 30% glycerol loading buffer (2 μL) was added to each sample.
Results
Identification of clonal patients
Patterns of X-chromosome inactivation consistent with clonal hemopoiesis have been observed in variable proportions of female patients with AA.18,25-28 Most studies have used DNA methylation as a surrogate marker of X-inactivation, but methylation may not always correlate with activation status. We therefore studied expression at the RNA level of the silent polymorphism at nt.1311 of the G6PD gene29 using an RT-PCR method (Mortazavi et al, manuscript submitted). To avoid the inclusion of patients with extreme lyonization, we used a stringent criterion of greater than 95% expression of 1 allele to define a clonal pattern. Thirty-eight percent of informative patients showed a clonal pattern, which was significantly higher than among controls, and there was a trend toward longer duration of disease and older age in the clonal group (data not shown). Nine female patients with clonal patterns of X-chromosome inactivation were included in this study; 18 patients with a PNH clone detectable by flow cytometry and 3 additional patients in whom MDS developed during the course of the study were also included
Amplification of the N-RAS gene
DNA was extracted from granulocytes and lymphocytes, and 2 regions of the N-RAS gene spanning either codons 12 and 13 or codon 61 were separately amplified by PCR. Amplification of exon 1 containing codons 12 and 13 produced a 109-bp DNA fragment, and amplification of exon 2 containing codon 61 produced a 103-bp DNA fragment (data not shown).
Detection of point mutations by ASO hybridization
To assess the prevalence of activating mutations of the N-RAS proto-oncogene, PCR products were transferred to 3 replicate Hybond NFP nylon filters by slot blotting and were probed for wild-type and each possible mutant sequence at codons 12, 13, or 61 of the human N-RAS gene by ASO hybridization. In the first instance, combinations of 3 or more oligonucleotide probes specific for each possible activating mutation at codons 12, 13, and 61 were used.
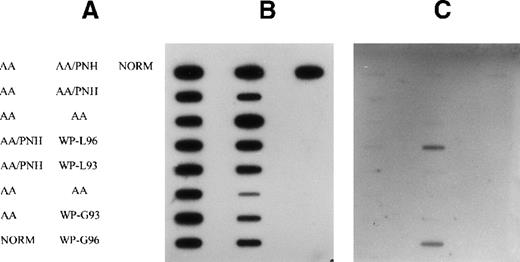
No mutation was detected either in AA or in AA/PNH samples (Figure1). However, a strong positive signal was detected in granulocytes and lymphocytes at codon 13 using a combination of probes (SER, CYS, and ASP) in 1 patient with MDS (patient 12) who had a history of AA/PNH (Figure 1). To find the exact mutation, the DNA sample was hybridized with individual mutant probes. An activating mutation of GGT→GAT at codon 13, representing a substitution of wild-type glycine by asparagine, was observed. No mutation was detectable in the DNA sample taken from the PNH stage (Figure 1), at which stage the GPI-anchored proteins CD59 and CD55 were markedly deficient in granulocytes and monocytes (data not shown). DNA from 2 other patients with MDS and histories of AA, who showed a clonal pattern of X-inactivation, was screened for activation of N-RAS in AA and MDS stages. No mutation was detectable in either stage (data not shown).
Detection of N-RAS mutation at codon 13 in peripheral blood cell DNA of patients with AA, AA/PNH, and MDS.
(A) Arrangement of samples on the filter (NORM, normal control). (B) Membrane probed with wild-type (Gly) probe. (C) Membrane probed with combinations of Ser, Cys, and Asp mutant-specific probes. Samples from patient 12 granulocytes (WP-G) and lymphocytes (WP-L) in 1993 (AA/PNH phase) and 1996 (MDS phase) are indicated
Detection of N-RAS mutation at codon 13 in peripheral blood cell DNA of patients with AA, AA/PNH, and MDS.
(A) Arrangement of samples on the filter (NORM, normal control). (B) Membrane probed with wild-type (Gly) probe. (C) Membrane probed with combinations of Ser, Cys, and Asp mutant-specific probes. Samples from patient 12 granulocytes (WP-G) and lymphocytes (WP-L) in 1993 (AA/PNH phase) and 1996 (MDS phase) are indicated
Detection of PIG-A gene mutation for patient 12
Flow cytometry results for patient 12 in October 1993 and October 1994 revealed that a substantial proportion of his neutrophils and monocytes were deficient in CD55 and CD59 GPI-anchored proteins. To identify the mutations within the PIG-A gene, the entire coding region of the PIG-A gene of this patient was amplified from polymorphonuclear genomic DNA in a series of overlapping fragments. PCR products of exon 6 showed a smaller abnormal band in addition to a normal-sized (361-bp) DNA band on a high-resolution agarose gel. The amplification was repeated, and the same abnormal band was consistently produced. RT-PCR on RNA also confirmed the presence of the abnormal band. This deletion was not present in PMN cells at the MDS stage. To localize the mutation further, the PCR product of exon 6 was subjected to Fok Ι restriction enzyme digestion. Normal control DNA for exon 6 produced 2 fragments of 229 and 132 bp, whereas PCR product from the patient produced 2 normal-sized bands of 229 bp and 132 bp, identical to those of normal control. In addition, another smaller band of 113 bp appeared, indicative of the presence of a deletion in this latter fragment (data not shown).
Heteroduplex analysis and SSCP was carried out for all DNA fragments for PIG-A coding regions. Abnormal bands were observed for exon 6. The heteroduplex analysis showed a normal homoduplex band plus a weak mutant homoduplex band that migrated faster than the normal control.
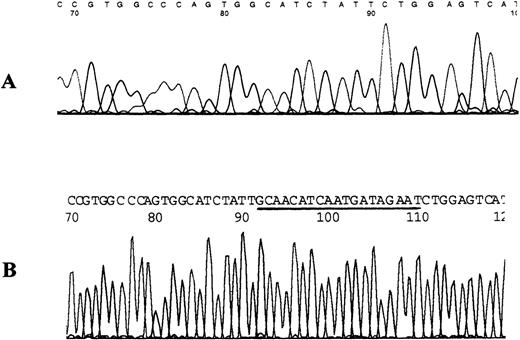
To identify the nature and exact position of the mutation, the gel-purified mutant fragment was reamplified and used for direct sequencing. Sequencing results revealed that 19 nt (from nt.1343 to nt.1361) were deleted from the abnormal fragment (Figure2). The mutation caused a frameshift and created a TAG stop codon immediately 1 codon after the deletion at nt.1364 to nt.1366. This frameshift would produce a truncated protein with 448 amino acids.
Nucleotide sequence for the mutant fragment of exon 6 for patient 12.
(A) DNA sequence from patient. Note that 19 bp was deleted from the sequence after the T numbered 90 in this sequencing run. (B) Normal sequence from control DNA. Note that the sequence deleted from the patient's DNA is underlined.
Nucleotide sequence for the mutant fragment of exon 6 for patient 12.
(A) DNA sequence from patient. Note that 19 bp was deleted from the sequence after the T numbered 90 in this sequencing run. (B) Normal sequence from control DNA. Note that the sequence deleted from the patient's DNA is underlined.
Discussion
Aplastic anemia and PNH have been considered preleukemic conditions because of their significant risk for transformation to clonal disorders.30,31 Reports from single and multicenter studies have shown a 10% to 26% occurrence of MDS and acute leukemia in patients 10 to 20 years after immunosuppressive therapy for AA.1,2,4-6 Cytogenetic analysis at diagnosis reveals that a few patients with AA have clonal abnormalities, but most are karyotypically normal.32,33 However, studies using X-chromosome inactivation methods show that some patients with AA may have monoclonal patterns in peripheral blood cells at presentation18 or later in the course of the disease.18,27,28 Therefore, it is possible that clonal evolution began during AA. There are alternative possible explanations for selective X-chromosome inactivation, including stem cell ablation, loss of stem cell heterogeneity,34 and selection for advantageous alleles on the active X-chromosome,35 but in the context of selecting patients at risk for clonal evolution, they all imply extreme hematopoietic stress.
Activating mutations of N-RAS in codons 12, 13, and 61 are among the most common abnormalities associated with hematologic malignancy. It has been shown that activation of the N-RAS gene is present in 20% to 30% of all patients with ANLL and MDS.14,15 Furthermore, patients with MDS who have the N-RAS mutation are at higher risk for ANLL,17 which might help explain the role of N-RAS in leukemogenesis. Because long-term survivors of AA after immunosuppressive therapy are at risk for clonal disorders, we have investigated the hypothesis that these patients may evolve to ANLL and MDS through activation of the N-RAS oncogene.
We have studied 30 patients with a diagnosis of AA, AA/PNH, and AA/MDS for possible mutations in the N-RAS gene. All 9 female patients with AA, 5 of 6 female patients with AA/PNH, and 2 of 2 female patients with AA/MDS had monoclonal patterns of X-chromosome inactivation either at the AA or the PNH stage. Despite the sensitivity of our technique, which can detect the mutation in as low as 2% of the cell population, no mutations were detected among patients with AA, AA/MDS, AA/PNH. This is in agreement with previous results reported for AA18,19and AA/PNH.20 The difference between our study and others is that all our patients were selected on the basis of evidence for existing clonal evolution. The risk for clonal disorders increases over time in AA or AA/PNH. Among our patients, 18 of 30 were observed for more than 2 years (between 2 and 24 years) from the initial diagnosis of AA, 12 of 30 were observed for more than 5 years, and 7 of 30 were observed for more than 10 years. Two of those latter patients were observed for 21 and 24 years, respectively. One patient with a diagnosis of AA/PNH (patient 12) and two patients with diagnoses of AA (patients 10 and 11), observed for 3.6, 5.2, and 7 years, respectively, then had MDS. The risk for MDS among patients with AA has been reported to increase with age4; these patients were 71, 64, and 69 years old, respectively.
Patients were analyzed at the MDS stage for possible mutations of N-RAS to determine whether activation of this gene could be implicated in the evolution from AA to MDS. No mutation was found in patients 10 and 11, but in patient 12 a mutation of GGT→GAT occurred at codon 13 that represented a substitution of wild-type glycine by asparagine. This mutation was shown to be present in PMN and MNC, indicating that the mutation occurred in a multipotent hematopoietic stem cell. Cytogenetic analysis revealed a karyotype of 45,XY, add(6)(q?1), −7, del(9)(q12q22) in 30 abnormal mitoses. A normal karyotype of 46,XY was observed in only 1 mitosis. We found no evidence of the PNH abnormality within the MDS cells as analyzed by flow cytometry and mutation detection techniques, indicating that MDS originated from a non-PNH clone, presumably from a distinct AA stem cell affecting myeloid and lymphoid lineages. Detection of the activation of a RAS oncogene in our patient provided evidence that RAS has a role in the induction of this transformation. RAS has been involved in approximately 20% to 30% of ANLL and MDS cases. It has been shown that ANLL is more likely to develop in patients with MDS who have a mutated RAS gene17; indeed, ANLL developed in our patient after a few months. Because there was a relatively strong mutant RAS signal for the MDS clone and because the PNH cells disappeared totally at the same time, RAS activation might have been the initiating event causing MDS.
In some patients with PNH, MDS or leukemia has been reported to develop within the PNH clone and to result in blast cells with a deficiency of GPI-anchored proteins.9,11,36,37 In others it has been reported to originate from a non-PNH stem cell and to result in the disappearance of the PNH clone.12 38 Because PNH usually arises in the context of AA, which already carries a predisposition to leukemia, we suggest that leukemia in these patients may be a manifestation of the AA stem cell as much as of PNH itself.
Both female patients in whom MDS developed showed clonal patterns of X-chromosome inactivation during the aplastic phase. None of 16 female patients with polyclonal hematopoiesis had malignant clonal disorders during the time of this study, with a mean follow-up of 2.7 years. Among patients with AA, it would be of interest to follow up those who have clonal hematopoiesis to determine whether they are at greater risk for MDS and ANLL than those who have polyclonal hematopoiesis.
Thus, in general, N-RAS mutations are not early events preceding the transformation of AA or AA/PNH to leukemia, and preexisting RAS mutations do not explain the propensity of AA to leukemogenesis. RAS mutations may, however, occur at the time of evolution from AA to MDS. Although PNH is also associated with leukemia, leukemia may arise in the non-PNH clone. Therefore, the PIG-A gene mutation is not per se oncogenic. In the absence of cytogenetic abnormalities and other clonal markers such as RAS, monoclonality in female patients with AA does not appear to be a sign of neoplastic transformation, since the bone marrow usually shows hypoplasia as the only abnormality.
Acknowledgments
The authors thank J. C. W. Marsh for helpful discussions, R. Chopra for assistance with DNA extraction, and all their normal controls for the generous donation of blood.
Supported by a grant from the United Kingdom Leukaemia Research Fund and by a research studentship from the Islamic Republic of Iran.
Reprints:T. R. Rutherford, Department of Haematology, St. George's Hospital Medical School, Cranmer Terrace, London SW17 0RE, United Kingdom; e-mail: trutherf@sghms.ac.uk.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal