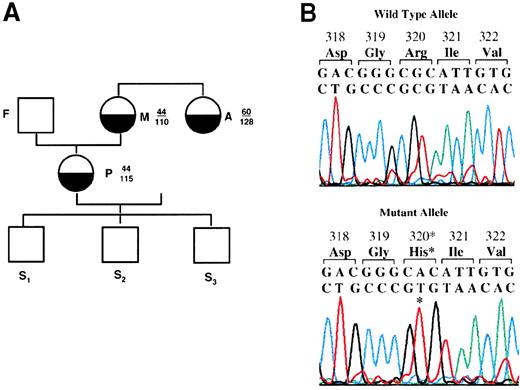
Three members of a San Antonio, Texas, family were identified with prothrombin activity levels half the normal level but to have normal levels of antigen. All exons of the prothrombin gene from the proband were sequenced. A G-to-A mutation at nucleotide 7543 was found that resulted in the substitution of His for Arg at residue 320. The Arg320-Ile321 bond is 1 of 2 sites in prothrombin cleaved by Factor Xa in the prothrombinase complex to form thrombin. Substitution of His for Arg at this site resulted in the blockage of Factor Xa cleavage, forming a dysfunctional molecule. The proband, her mother, and her maternal aunt were found to be heterozygous for this mutation. This is the first known observation of an amino acid substitution at this site that resulted in dysprothrombinemia.
Thrombin is the enzymatically active form of its zymogen, prothrombin, and it plays a key role in blood coagulation and anticoagulation.1 Human prothrombin is composed of 579 amino acid residues. It has 5 functional domains that include a pre-pro leader sequence required for secretion and γ-carboxylation, the γ-carboxyglutamic acid containing Gla domain, 2 kringle domains for interaction with cofactors, and the catalytically active serine protease domain. On the initiation of coagulation, Factor Xa—in the presence of Factor Va, calcium ions, and phospholipid membrane— cleaves prothrombin at Arg271-Thr272 and Arg320-Ile321 to release the catalytic domain from the carboxy-terminal of the protein. Human thrombin contains an A-chain of 36 amino acids and a B-chain of 259 amino acids. Thrombin has diverse functions that include the stimulation of platelets to form a platelet plug and the cleavage of fibrinogen to form the fibrin clot, both of which prevent excessive loss of blood from injured blood vessels. In addition, thrombin activates protein C and protein S to initiate the inhibition of the coagulation process.2
Defects in the prothrombin gene cause two types of congenital disorders, hypoprothrombinemia and dysprothrombinemia, that result in excessive bleeding. Prothrombin antigen levels in plasma decrease significantly in hypoprothrombinemia, whereas in dysprothrombinemia normal prothrombin antigen levels are detectable. These disorders are rare, and only approximately 30 families with dysprothrombinemia or hypoprothrombinemia have been reported.3 A small amount of thrombin activity has always been detectable in these patients, which is consistent with the finding that complete deficiency in prothrombin in mice is lethal.4 5 Genetic analysis shows that hypothrombinemia and dysprothrombinemia result from the substitution, deletion, or insertion of a single nucleotide in the prothrombin gene, which leads to the substitution of an amino acid in the protein. To date, mutations have been identified for 16 amino acid residues in prothrombin. In this article we report a new genetic mutation in the prothrombin gene that causes dysprothrombinemia.
Materials and methods
Prothrombin activity and antigen level determination
A 1-stage assay using prothrombin-deficient plasma (Precision Biologic, Nova Scotia, Canada) was used to measure prothrombin activity in plasma samples.6 Prothrombin antigen levels were determined using the Laurell-immunodiffusion rocket assay with rabbit antihuman antibody (Diagnostica Stago, Asnieres, France). Levels of other coagulation factors for the proband were within normal ranges.
Genetic analysis
Leukocytes of the proband and her family members were used to isolate genomic DNA.7 Polymerase chain reaction (PCR) amplified the 14 exons of the prothrombin gene with sets of primers that have proved successful in the past.8 These products were used for single-strand conformation polymorphism analysis (SSCP)8 and restriction enzyme digestion, and they were cloned into the vector pCR2.0 (Invitrogen, Carlsbad, CA). All exons and the intervening sequences C, H, and M, as well as the 5′ and 3′ ends of all other intervening sequences, were sequenced using vector-specific primers on an Applied Biosystems sequencer (PE Applied Biosystems, Foster, CA). Restriction enzyme analysis of the PCR products confirmed specific DNA substitutions.
Ecarin activation of plasma prothrombin and Western blot analysis
Plasma samples (8 μL) were incubated with Echis carinatusvenom (Ecarin, 0.1 U; Sigma, St. Louis, MO) and hirudin (1 μg or 0.94 U; Sigma) at 37°C for 4, 7, 15, 30, and 60 minutes. EDTA (final concentration 15 mmol/L) was added to stop the reaction, and samples were immediately frozen on dry ice and stored at –20°C until Western blot analysis was performed. The reaction mixture (2 μL) was analyzed by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) in the absence or presence of a reducing agent (10% β-mercaptoethanol). Western blot analysis of the activation products of prothrombin was performed as described previously.4 Rabbit polyclonal antibody raised against human prothrombin (Nordic Immunological, San Clemente, CA) was used with biotinylated goat antirabbit antibody and a horseradish peroxidase staining system (Vectastain ABC; Vector Laboratories, Burlingame, CA) to detect prothrombin after the transfer of plasma proteins to a nylon membrane. The enhanced chemiluminescence system (Amersham, Piscataway, NJ) was used to detect the membrane-bound biotin–peroxidase complex.
Results and discussion
A 55-year-old woman (proband) had protracted vaginal bleeding from a uterine sarcoma. She had a longstanding bleeding disorder, including postpartum bleeding, and required blood transfusions even after dental procedures. Her mother and maternal aunt had similar histories of bleeding after minor surgical procedures. After the diagnosis of dysprothrombinemia was made, the patient was treated with fresh-frozen plasma, and she underwent successful surgery to remove the uterine tumor. Thrombin activity measurements for the proband, her mother, and her maternal aunt were approximately 50% of normal (Figure1a). Normal prothrombin antigen levels were found for all tested family members, including the proband. During the course of the experiments described here, the proband died of metastatic carcinoma.
Characterization of prothrombin San Antonio.
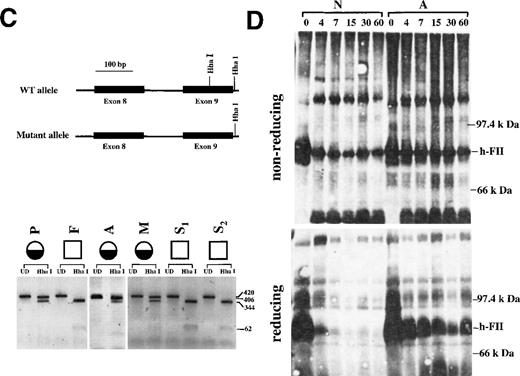
(A) The prothrombin San Antonio family. Half-filled circles: Family members identified to be heterozygous for the prothrombin San Antonio gene. Open squares: Family members identified to be homozygous for the normal allele. Prothrombin activity (numerator) and antigen (denominator) levels are indicated for several family members. P, proband; F, father; M, mother; A, Aunt; S1-S3, sons. (B) The cloned PCR product for exons 8 and 9 was analyzed by DNA sequence analysis. (asterisk) Mutation resulting in the substitution of His for Arg320. Sequences for the complementary strand of the wild-type allele (top) and the mutated prothrombin allele (bottom) were obtained directly from sequencing. Red, T; green, A; black, G; and blue, C. (C) Identification of the mutation at nucleotide 7543 in the prothrombin gene by Hha I-restriction enzyme digestion. (top) Schematic representation of the Hha I-restriction map of the PCR products for exons 8 and 9 from the wild-type (WT) and the mutant prothrombin allele. (bottom) Hha I digestion fragments of the PCR products from the proband and her family members (see part A). Fragment sizes in bp are indicated. UD, undigested. (D) Western blot analysis of the activation of prothrombin by Ecarin (in the presence of hirudin) from a normal control (N) and the maternal aunt of the proband (A). Plasma activated with Ecarin in the presence of hirudin was fractionated by 10% SDS-PAGE in the absence (top) or presence (bottom) of β-mercaptoethanol. The mobility of prothrombin is indicated (h-FII). The incubation time of plasma sample with Ecarin is indicated at the top.
Characterization of prothrombin San Antonio.
(A) The prothrombin San Antonio family. Half-filled circles: Family members identified to be heterozygous for the prothrombin San Antonio gene. Open squares: Family members identified to be homozygous for the normal allele. Prothrombin activity (numerator) and antigen (denominator) levels are indicated for several family members. P, proband; F, father; M, mother; A, Aunt; S1-S3, sons. (B) The cloned PCR product for exons 8 and 9 was analyzed by DNA sequence analysis. (asterisk) Mutation resulting in the substitution of His for Arg320. Sequences for the complementary strand of the wild-type allele (top) and the mutated prothrombin allele (bottom) were obtained directly from sequencing. Red, T; green, A; black, G; and blue, C. (C) Identification of the mutation at nucleotide 7543 in the prothrombin gene by Hha I-restriction enzyme digestion. (top) Schematic representation of the Hha I-restriction map of the PCR products for exons 8 and 9 from the wild-type (WT) and the mutant prothrombin allele. (bottom) Hha I digestion fragments of the PCR products from the proband and her family members (see part A). Fragment sizes in bp are indicated. UD, undigested. (D) Western blot analysis of the activation of prothrombin by Ecarin (in the presence of hirudin) from a normal control (N) and the maternal aunt of the proband (A). Plasma activated with Ecarin in the presence of hirudin was fractionated by 10% SDS-PAGE in the absence (top) or presence (bottom) of β-mercaptoethanol. The mobility of prothrombin is indicated (h-FII). The incubation time of plasma sample with Ecarin is indicated at the top.
Initially, SSCP analysis was used to localize nucleotide differences in the 14 exons of the prothrombin gene from the proband, her relatives, and a normal control. No differences were observed using this technique. Because SSCP analysis has limited sensitivity,9 the PCR products of all 14 exons and several intervening sequences of the proband were cloned and analyzed by DNA sequence analysis. The prothrombin gene sequence from the proband revealed 3 nucleotide substitutions within the exons and 5 nucleotide substitutions and 1 deletion within the intervening sequences of the gene.10 Substitution of a G for A554 in exon 2 and a C for A8908 in exon 10 did not result in amino acid substitutions. The differences within the intervening sequences were the deletion of a T at nucleotide 459 in intron A, the substitution of a C for T4048, a C for T4096, and a C for T4097 in intron E, the substitution of a G for T9393 in intron J, and the substitution of a G for A19 911 in intron M. All these differences, except that at nucleotide 9393, were identified as polymorphisms in the prothrombin gene.8 11-13 At nucleotide 7543 in exon 9, the substitution of an A for a G nucleotide did result in an amino acid substitution of Arg to His at residue 320 (Figure 1b). Six of 9 clones containing the PCR products of exons 8 to 9 (including the intervening sequence) had this substitution, indicating that the proband was heterozygous for the substitution. This is the first mutation identified to result in the substitution of Arg320 by another amino acid in prothrombin; Arg320-Ile321 is 1 of the 2 cleavage sites for Factor Xa required for the activation of prothrombin to thrombin.
The G7543A substitution found in the proband resulted in the loss of an Hha I restriction site in exon 9 of the prothrombin gene (Figure 1c). The 420-bp PCR product spanning exons 8 and 9 and the intervening sequence H of the prothrombin gene from the proband and her family members were digested with Hha I. With normal control DNA, Hha I digested the DNA at 2 sites, generating 3 fragments of 344, 62, and 14 bp (data not shown), whereas the digestion of DNA from the proband generated fragments of 406 and 14 bp (Figure 1c). The 62-bp band was faint. It was observable when 2 normal alleles were present, but it was not observable in heterozygotes. The 406- and 344-bp bands easily detectable in Figure 1c indicate that the proband was heterozygous for the mutation. Hha I restriction digestion of the same PCR product of exons 8-9 for 5 relatives of the proband showed that 2 additional relatives were heterozygous for this mutation (the mother and the maternal aunt of the proband), whereas her father and her 2 sons had 2 normal alleles. These results are consistent with the prothrombin activity assays that indicated that the proband, her mother, and her maternal aunt had approximately 50% of normal activity with normal levels of protein (Figure 1a).
To test whether the Arg-to-His substitution at residue 320 resulted in the inability of this mutant prothrombin to be activated to thrombin, plasma samples from a normal control and from the maternal aunt of the proband were used (Figure 1d). During the course of these experiments, the proband unexpectedly died from metastatic carcinoma; therefore, her plasma was unavailable for these studies. The snake venom Ecarin14 15 has been found to activate prothrombin at 1 of the 2 Factor Xa sites (Arg320-Ile321) through a meizothrombin intermediate. This reaction does not require Factor Va, phospholipid, and calcium. Ecarin and hirudin (an inhibitor of prothrombin activation that prevents autoactivation by meizothrombin or thrombin) were added to plasma samples for various lengths of time and were followed by Western blot analysis (Figure 1d). The activation of normal prothrombin results in 2 chains held together by a disulfide bond (meizothrombin). Under nonreducing conditions meizothrombin has the same mobility as native prothrombin, whereas under reducing conditions the 2 separated chains migrate off the gel. Activation of plasma from the aunt showed prothrombin that migrated to a similar position as native prothrombin under nonreducing conditions. Because the aunt was heterozygous for the mutation at residue 320, this prothrombin band was a combination of normal meizothrombin and mutant prothrombin. Under reducing conditions, mutant prothrombin maintained the same mobility as native unactivated prothrombin, whereas meizothrombin ran off the gel. These results indicated that the amino acid substitution at residue 320 resulted in a protein resistant to Ecarin cleavage.
One other dysfunctional prothrombin, prothrombin Clamart, has been proposed to have a mutation at Arg320 that results in a prothrombin that cannot be activated.16 Activation of prothrombin Clamart by Factor Xa results in the formation of prothrombin 2, indicating an impairment in cleavage at Arg320-Ile321. Thrombin activity was found to be half the normal level, and the proband was heterozygous for the mutation.16 It was suggested that a mutation may occur in the vicinity of the Arg320-Ile321 bond and may affect the interaction between Factor Xa and prothrombin. The mutation has not yet been identified at either the DNA or the protein level. Defective cleavage of the other bond in prothrombin cleaved by Factor Xa at Arg271-Thr-272 has been identified in several families.11 17-19
Identification of these naturally occurring mutations in prothrombin has given us insight into the cause of congenital disorders of prothrombin. These mutations, along with those generated by site-directed mutagenesis, have improved our understanding of the structure-function relationship of prothrombin and of other serine proteases.
Acknowledgment
The authors thank Ann Becker for performing some of the coagulation assays.
Supported in part by National Institutes of Health grant HL58103 (US Public Health Service) from the National Heart, Lung and Blood Institute; and by a postdoctoral fellowship from the American Heart Association, Ohio-West Virginia Affiliate.
Reprints:Sandra J. F. Degen, Division of Developmental Biology, Children's Hospital Research Foundation, 3333 Burnet Avenue, Cincinnati, OH 45229-3039; e-mail: sandra.degen@chmcc.org.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal