Three of at least 8 Fanconi anemia (FA) genes have been cloned (FANCA, FANCC, FANCG), but their functions remain unknown. Using the yeast 2-hybrid system and full-length cDNA, the authors found a strong interaction between FANCA and FANCG proteins. They also obtained evidence for a weak interaction between FANCA and FANCC. Neither FANCA nor FANCC was found to interact with itself. These results support the notion of a functional association between the FA gene products.
An important way in which gene function can be elucidated is in the analysis of protein–protein interactions. Intracellular distribution and putative interactions remain controversial in Fanconi anemia (FA) gene products, as illustrated by recent articles1,2 and by subsequent correspondence in this journal.3,4 Immunoprecipitation data suggest that the FA gene products interact in a multimeric protein complex.1,5We applied the yeast 2-hybrid system6 to test independently the concept of FA protein interaction.
Materials and methods
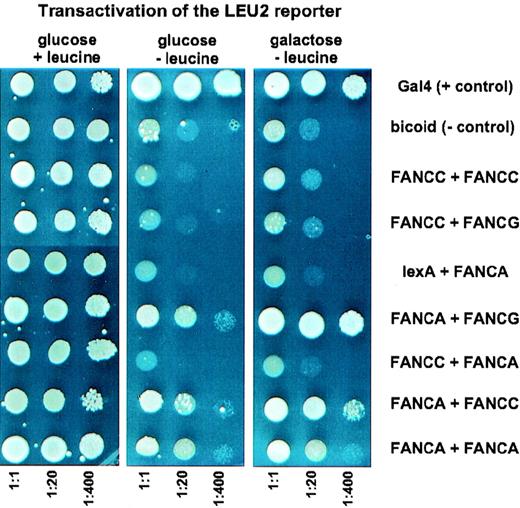
For constructing lexA fusions, FANCA, FANCC, and FANCG full-length cDNA was subcloned to the plasmid pEG202. To generate B42 fusions, the respective FA cDNA was subcloned to pJG4-5. These constructs were transformed to the yeast strain EGY48 and tested for protein expression by immunoblot analysis (data not shown). The control plasmid pRFMHI encodes the Gal4 transactivator and serves as a positive control for activation of the reporters. Similarly, the pSH17-4 plasmid encodes theDrosophila bicoid protein, which does not activate the reporters. Because the lexA-FANCG hybrid protein resulted in transactivation of the reporters, FANCG could only be tested in fusions with B42.
Yeast strain EGY48 was cotransfected in pairs with different combinations of vectors expressing the full-length FA lexA or B42 fusion protein. Positive interaction was documented by the ability of yeast cells to grow on galactose medium, assayed by the expression of the chromosomal leucine reporter and by episomal LacZ expression. Reduced growth on glucose minus leucine, or on glucose medium containing X-Gal (compared to the corresponding galactose plates) served as controls for an interaction because expression of the B42 fusion protein is repressed on glucose plates. The percentage of yeast recombinants that may generate false-positive results by activation of the reporter genes was less than 4%.
Results and discussion
The FANCA protein interacted strongly with FANCG, as evidenced by LEU2 and LacZ expression (Figure 1). Activation of the LEU2 reporter gene was also observed when lexA–FANCA was cotransfected with B42–FANCC. However, this interaction was much weaker and could not be detected by the less sensitive LacZ reporter assay (data not shown). There was no evidence for interaction using the inverse combination. There also was no detectable interaction between the FANCC and FANCG gene products, nor was there evidence that FANCA and FANCC interact with themselves. Marginal growth of the FANCA/FANCA cotransformants on leucine-deficient galactose plates did not result from interaction because such cotransformants spotted onto glucose medium and lacking leucine behaved like those spotted onto galactose plates. In contrast, there was a consistent difference between the growth performance of the FANCA/FANCC cotransformants, depending on whether they were plated onto leucine-free glucose or galactose medium (Figure 1). To determine whether a mutant FANCC protein could abolish the FANCA/FANCC interaction, we generated L554P–FANCC mutant fusion constructs. These mutant hybrid proteins did not enter the yeast nucleus reliably, so these interaction assays were not informative. In the liquid beta-galactosidase assay, activation of LacZ by the FANCA/FANCG interaction was nearly as efficient as activation by the natural yeast transcriptional activator Gal4, confirming a very strong interaction. Quantification of activation by all other cotransformants resulted in beta-galactosidase activities of less than 5% of the FANCA/FANCG interaction (data not shown). Given the permissiveness of the yeast 2-hybrid system, the relatively weak interaction between FANCA and FANCC could also be a consequence of transcriptional activation by spurious hydrophobic interactions. In addition, the detection of minor interactions may critically depend on the choice of fusion partners and on other technical features of the 2-hybrid system.7 However, the likely importance of FANCA as a core protein in a multimeric complex is strongly supported by the prominent FANCA/FANCG interaction.5
Yeast plates showing growth response under different dilution and testing conditions.
Yeast strain EGY48 was cotransfected in pairs, with the indicated combinations of vectors for the expression of full-length Fanconi anemia LexA or B42 fusion proteins. Three dilutions each were spotted onto complete minimal dropout media. The first protein of each of the combinations represents the LexA fusion, the second protein the B42 fusion. Each result was confirmed by 4 independent assays.
Yeast plates showing growth response under different dilution and testing conditions.
Yeast strain EGY48 was cotransfected in pairs, with the indicated combinations of vectors for the expression of full-length Fanconi anemia LexA or B42 fusion proteins. Three dilutions each were spotted onto complete minimal dropout media. The first protein of each of the combinations represents the LexA fusion, the second protein the B42 fusion. Each result was confirmed by 4 independent assays.
Acknowledgment
The authors thank Hans Joenje (Free University, Amsterdam, The Netherlands) for providing the FANCA and FANCC cDNA.
Supported by the Schroeder-Kurth Fonds, Germany.
Reprints:Tanja Reuter, Department of Biochemistry, University of Würzburg, Biozentrum, D-97074 Würzburg, Germany; email: reuter@biozentrum.uni-wuerzburg.de.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal