In rare cases of B-cell chronic lymphocytic leukemia (B-CLL), large cells morphologically similar to or indistinguishable from Hodgkin/Reed-Sternberg (HRS) cells of Hodgkin's disease (HD) can be found in a background of otherwise typical B-CLL. To test these HRS-like cells for a potential clonal relationship to the B-CLL cells, single cells were micromanipulated from immunostained tissue sections, and rearranged immunoglobulin genes were amplified from HRS-like cells and B-CLL cells and sequenced. The same variable (V) gene rearrangements with shared and distinct somatic mutations were found in HRS-like and B-CLL cells from 1 patient, which indicates derivation of these cells from 2 distinct members of a germinal-center B-cell clone. Separate clonal Vgene rearrangements were amplified from HRS-like and B-CLL cells from 2 other patients, showing concomitant presence of 2 distinct expanded B-cell clones. Epstein-Barr virus (EBV) was detected in the HRS-like cells of these 2 latter cases, indicating clonal expansion of an EBV-harboring B cell in the setting of B-CLL. There is evidence that HRS-like cells in B-CLL, like HRS cells in HD, derive from germinal-center B cells. In all cases, somatic mutations have been detected in the rearranged V genes of the HRS-like cells, and in 1 of the EBV-positive HRS-like cell clones, somatic mutations rendered an originally functional V gene rearrangement nonfunctional. We speculate that the HRS-like cells in B-CLL represent potential precursors for HRS cells causing HD.
Hodgkin and Reed/Sternberg (HRS) cells represent a histological hallmark and, if surrounded by a typical mixed cellular infiltrate, are indicative of the diagnosis of Hodgkin's disease (HD). The occurrence of cells with the morphology and often the immunophenotype of HRS cells (eg, positivity for the surface antigens CD30 and CD15) has been described in rare cases of B-cell chronic lymphocytic leukemia (B-CLL).1,2 In some instances, the HRS-like cells reside scattered in a background of CLL in the absence of the rich cellular infiltrate of HD.2,3Often Epstein-Barr virus (EBV) is present in the HRS-like cells, suggesting that EBV infection may play a role in the generation of these cells.3,4 Since it is now known that the HRS cells in HD are usually derived from germinal-center (GC) B cells,5it is an intriguing question whether also the HRS-like cells in diseases other than HD are B-lineage derived and related to GC B cells. In approximately half of the cases,3 a B-lineage origin of the HRS-like cells in B-CLL is indicated by the expression of B-cell–specific antigens. Furthermore, it has long been unclear whether the HRS-like cells in B-CLL are clonally related to the underlying CLL and perhaps represent phenotypic variants of the original lymphoma. A recent study by Ohno et al6 described rearranged immunoglobulin V region genes (IgV) in HRS-like and B-CLL cells in Richter's syndrome, which represents a blastic transformation of a B-CLL. Three cases studied indeed showed that the HRS-like cells can represent B cells clonally related to the concurrent B-CLL cells because identical V gene rearrangements were amplified from those cells in 2 of the cases.
We applied polymerase chain reaction (PCR) analysis of single micromanipulated cells for IgV gene rearrangements7to further study the clonal relationship of HRS-like cells and concurrent B-CLL cells and to investigate the differentiation stage of the HRS progenitors, which was not addressed by Ohno et al.6
Materials and methods
Patients and tissues
Patient 1 was an 85-year-old female with a previous history of CLL; the white blood cell count was 19 × 109/L at diagnosis of CLL in November 1988. Adenocarcinoma was diagnosed and BII gastrectomy performed. No further therapy, such as radiotherapy or chemotherapy, was given. Biopsy of a supraclavicular lymph node in January 1995 showed an infiltration of B-CLL with Hodgkin-like cells and a small number of Reed-Sternberg–like cells interspersed in the CLL tumor B cells. The white blood cell count in January 1995 was 32 × 109/L. No further biopsies were performed, the CLL continued, and the patient died in April 1995.
Patient 2 was a 61-year-old female who presented in May 1981 with an axillary lymph node infiltrated by CLL and a high proliferation activity of the tumor. In December 1986, the patient presented with enlarged lymph nodes at different locations. The white blood cell count was 42 × 109/L. Chemotherapy was performed according to the COP regimen (cyclophosphamide, vincristine [Oncovin], and prednisone). The histology of a lymph node biopsy taken in January 1987 revealed B-CLL with intermingled HRS-like cells. HRS-like and B-CLL cells were isolated from this biopsy specimen. The white blood cell count in February 1987 was 17 × 109/L. The patient received 6 cycles of chemotherapy according to the COP-ABV-IMEP regimen, which comprises COP–doxorubicin (Adriamycin), bleomycin, and vinblastine–ifosfamide, methotrexate, etoposide, and prednisone. Lymph node histology in November 1987 revealed infiltration by CLL with no indication of HRS-like cells or HD. The white blood cell count was 42 × 109/L. In April 1988 the patient presented again with enlarged lymph nodes; the lymphoma had progressed, and the white blood cell count was 84 × 109/L. The patient received 1 cycle of chemotherapy according to the CHOP-regimen (cyclophosphamide, doxorubicin, vincristine [Oncovin], prednisone). We did not follow this patient further because she did not return to follow-up.
Patient 3 was admitted to the hospital in September 1997 at the age of 47 because of left-sided axillary lymphadenopathy. The patient's clinical history did not reveal preceding neoplastic or autoimmune disorders; however, the diagnosis of infectious mononucleosis was made at an outside hospital in 1995. An axillary lymph node biopsy was taken, resulting in a diagnosis of B-CLL with scattered HRS-like cells. The white blood cell count was 8.0 × 109/L with 51% lymphocytes and no eosinophilia or monocytosis. Flow cytometry immunophenotyping of peripheral blood and bone marrow revealed the presence of an abnormal CD19+CD5+CD23+ lymphoid cell population, and bone marrow trephine showed 60% diffuse infiltration with small lymphocytes consistent with B-CLL. Atypical large cells with or without features of HRS cells were not present. The patient was treated with local radiation therapy (total 36 Gy) to the left axilla involving the adjacent supraclavicular and cervical regions in addition to 6 cycles of combination chemotherapy according to the CHOP regimen. Complete clinical and hematological remission was achieved in March 1998. Restaging in August 1998 and abdominal computed tomography (CT) scans revealed a solitary lesion localized between the aorta and inferior vena cava; it was not biopsied. In September 1998 the patient presented with paraaortal lymphadenopathy, and an autologous peripheral blood stem cell transplantation was performed in January 1999. Staging at day 100 by thoracic and abdominal CT scans was negative; however, bone marrow phenotyping revealed 41% CD5+CD19+mononuclear cells, which indicated persistent marrow involvement by B-CLL.
Immunostaining and in-situ hybridization
Immunological studies were performed on frozen and/or paraffin-embedded tissues. Sections were stained with antibodies against CD30, CD20, and LMP1 (BerH2, L26, and CS1-4, respectively; Dako, Hamburg, Germany); CD15 and CD5 (LeuM1 and DK23, respectively; Becton Dickinson, Mountain View, CA); and CD3 (OKT3; Ortho Diagnostic Systems, Raritan, NJ) using the avidin-biotin-complex technique (Dako) as described.8 Fast red or new fuchsin was used as substrate for the alkaline phosphatase (AP).
In-situ hybridization (ISH) for detection of EBV-encoded small nuclear RNAs (EBER1 and 2) was performed using in vitro transcribed digoxygenin-labeled sense and antisense EBER probes.9 In brief, dewaxed and rehydrated tissue sections were treated with hydrochloride (0.1 N) and Pronase (1.25 μg/mL) (Boehringer Mannheim, Mannheim, Germany). Sections were refixed in 4% paraformaldehyde, dehydrated through graded ethanols, and hybridized for 18 hours at 37°C. The excess on the probe was removed by washing it with 2 × SSC (standard saline citrate)/50% formamid and 0.1 × SSC/50% formamid, followed by digestion with 20 μg/mL ribonuclease for 20 minutes at 37°C. Slides were washed with 2 × SSC and 0.1 × SSC. Bound digoxygenin-labeled probes were detected by digoxygenin-AP–coupled Fab fragment, and bound AP was visualized by fast red substrate.
Isolation of DNA from tissue sections
DNA was extracted from frozen or paraffin-embedded tissue sections by standard methods10 or by using a tissue kit (QiaAmp Tissue kit; Qiagen, Hilden, Germany).
Micromanipulation of cells
Immunostained frozen sections (6-μm to 8-μm thick) were overlaid with tris(hydroxymethyl) aminomethane–buffered (Tris-buffered) saline, and single cells were mobilized with the help of hydraulic micromanipulators (Narishige, Tokyo, Japan) on an inverted microscope (Olympus, Hamburg, Germany).8Cells were transferred into 20 μL PCR buffer supplemented with 1 ng/μL 5S ribosomal RNA (rRNA) (Boehringer Mannheim) and stored at −80°C. Single CD30-positive HRS-like cells and single CD3-positive T cells as well as CD20-positive B cells of the CLL were obtained from adjacent sections. In some micromanipulation experiments, the B cells of the CLL were identified as small CD3-negative or CD30-negative cells. Samples of the buffer covering the sections were aspirated, and they served as negative controls for the PCR amplification. In the repeat experiments, separate sections were analyzed following the procedure described above.
Preamplification of genomic sequences
In some experiments, the genomic DNA of single cells was amplified following the primer-extension preamplification (PEP) approach of Zhang et al,11 using proteinase K incubation (2 hours at 50°C with 0.3 mg/mL proteinase K [PCR grade, Boehringer Mannheim]) instead of alkali denaturation to isolate DNA. From these reactions, 4 μL aliquots were analyzed for rearranged IgV genes, as described below.
PCR analysis
Rearranged VH,Vκ, and Vλ genes were amplified in a seminested PCR using family-specific V gene primers together with 2 sets of the respective joining (J) gene segment primers as described.8 12-14 The PCR conditions are summarized in Table 1. The sequences of theVλ3 gene family-specific leader primers are: Vλ3L1 5′ CAC CAT GGC CTG GAC CCC TCT CTG 3′, Vλ3L2 5′ CAC CAT GGC CTG GAY CCC TCT MCT 3′, Vλ3L3 3′ ATG GCA TGG ATC CCT CTC TTC CTC G 3′, Vλ3L4 5′ GCC ATG GCC TGG ACC GYT CTC CT 3′.
PCR conditions
| First Round of PCR | |||||
| Primer | FRI VH | FRI VH | FRI Vλ | Leader VH | Leader VH3,4 |
| Combination | FRI Vκ | FRI Vλ | 3′Jλ | FRI Vκ | Leader Vλ3 |
| 3′JH, 3′Jκ | 3′JH, 3′Jλ | 3′JH, 3′Jκ | 3′JH, 3′Jλ | ||
| Primer (nmol/L each) | 6.9 | 14.3 | 22.0 | 40.0 | 19.0 |
| dNTP (μmol/L each) | 100 | 200 | 100 | 200 | 100 |
| MgCl2 (mmol/L) | 2.5 | 2.0 | 1.5 | 2.0 | 2.5 |
| Annealing temp. | 59°C | 60°C | 59°C | 61°C | 59°C |
| Second Round of PCR | |||||
| Primer (nmol/L each) | 125 | 125 | 125 | 125 | 125 |
| dNTP (μmol/L each) | 100 | 100 | 100 | 200 | 100 |
| MgCl2 (mmol/L) | VH1-6: 1.5 Vκ1-4: 2.5 | 1.5 | 1.5 | VHL1,5: 1.5 VHL2,3: 2.0 VHL4: 3.0 Vκ1-4: 2.5 | VHL4: 3.0 VλL3: 1.5 |
| Annealing temperature | 61°C: VH1,2,5/6 Vκ1-4 65°C: VH3,4 | 61°C: VH1,2,5/6 65°C: VH3,4 63°C: Vλ1-4,6-9 | 63°C | 63°C: VHL1-5 61°C: Vκ1-4 | 63°C |
| Cells/Samples Analyzed | |||||
| Patient 1 | |||||
| HRS-like | 22 | 8 | 7 | 13 | |
| B-CLL | 22 | 6 | 5 | 29 | |
| Patient 2 | |||||
| HRS-like | 5 | 28 | |||
| B-CLL | 4 | 24 | 8 | ||
| Patient 3 | |||||
| HRS-like | 16 | ||||
| B-CLL | 16 |
| First Round of PCR | |||||
| Primer | FRI VH | FRI VH | FRI Vλ | Leader VH | Leader VH3,4 |
| Combination | FRI Vκ | FRI Vλ | 3′Jλ | FRI Vκ | Leader Vλ3 |
| 3′JH, 3′Jκ | 3′JH, 3′Jλ | 3′JH, 3′Jκ | 3′JH, 3′Jλ | ||
| Primer (nmol/L each) | 6.9 | 14.3 | 22.0 | 40.0 | 19.0 |
| dNTP (μmol/L each) | 100 | 200 | 100 | 200 | 100 |
| MgCl2 (mmol/L) | 2.5 | 2.0 | 1.5 | 2.0 | 2.5 |
| Annealing temp. | 59°C | 60°C | 59°C | 61°C | 59°C |
| Second Round of PCR | |||||
| Primer (nmol/L each) | 125 | 125 | 125 | 125 | 125 |
| dNTP (μmol/L each) | 100 | 100 | 100 | 200 | 100 |
| MgCl2 (mmol/L) | VH1-6: 1.5 Vκ1-4: 2.5 | 1.5 | 1.5 | VHL1,5: 1.5 VHL2,3: 2.0 VHL4: 3.0 Vκ1-4: 2.5 | VHL4: 3.0 VλL3: 1.5 |
| Annealing temperature | 61°C: VH1,2,5/6 Vκ1-4 65°C: VH3,4 | 61°C: VH1,2,5/6 65°C: VH3,4 63°C: Vλ1-4,6-9 | 63°C | 63°C: VHL1-5 61°C: Vκ1-4 | 63°C |
| Cells/Samples Analyzed | |||||
| Patient 1 | |||||
| HRS-like | 22 | 8 | 7 | 13 | |
| B-CLL | 22 | 6 | 5 | 29 | |
| Patient 2 | |||||
| HRS-like | 5 | 28 | |||
| B-CLL | 4 | 24 | 8 | ||
| Patient 3 | |||||
| HRS-like | 16 | ||||
| B-CLL | 16 |
HRS indicates HRS.
Combinations of primers: FRI family-specific primers forVH,Vκ, andVλ genes; VH leader (VHL) region family-specific primers; and VHL3, VHL4, and VλL3 primers specific for the leader regions of VH3, VH4, andVλL3 genes, respectively.
The basic PCR program in the first round of PCR was: 1 cycle for 2 minutes at 95°C, 80°C hold (addition of enzyme); 30 seconds at 59°C, 60°C, or 61°C; and 1 minute at 72°C. This was followed by 34 cycles for 1 minute at 95°C; 30 seconds at 59°C, 60°C, or 61°C; and 1 minute at 72°C, followed by 5 minutes at 72°C. The basic PCR program in the second round of PCR was: 1 cycle for 2 minutes at 95°C, 80°C hold (addition of enzyme); 30 seconds at 61°C, 63°C, or 65°C; and 1 minute at 72°C. This was followed by 44 cycles of 1 minute at 95°C; 30 seconds at 61°C, 63°C, or 65°C; and 1 minute at 72°C, followed by 5 minutes at 72°C.
In the second round of PCR, a 1.5-μL aliquot from the first round was further amplified with the same V gene primers in separate reactions and internal primer sets for the J gene segments.8 12-14 During this second round, each primer was used at a concentration of 125 nmol/L, with the exception of the JH primers, which were used at a concentration of 31.3 nmol/L. In the first round of amplification, 1.5 units of enzyme mix (Expand, Boehringer Mannheim) was added; in the second round, 1.25 units of Taq DNA polymerase (Gibco BRL, Karlsruhe, Germany) was added.
Rearranged IgV genes from DNA extracted from fresh tissue sections were amplified as described for the second round of PCR using 35 cycles of amplification. PCR products were gel-purified and directly sequenced. For sequencing of PCR products obtained from B-CLL cells of patient 3, primers binding to the complimentarity determining region II (CDRII) of the in-frame VH3 gene rearrangement (IF 5′ ATT AGT GGT AGT GGT GGT AG 3′) or to the CDRII of the out-of-frame VH3 gene rearrangement (OOF 5′ GCT ATT GGT ACT GCT GGT GA 3′) were used. This was done because the 2 VH3 gene rearrangements were repeatedly coamplified from the same samples, which resulted in mixed sequences when the VH3 FRI primer was used for sequencing. Sequences were analyzed using software (DNASIS; Pharmacia Biotech, Uppsala, Sweden); a genetic library (GenBank library, http://www.ncbi.nlm.nih.gov/BLAST); and a database (IMGT;http://www.genetik.uni-koeln.de/dnaplot/).
Results
Histology and immunohistology of B-CLL–affected lymph nodes
Lymph node biopsies from 3 patients with B-CLL were investigated, and histological sections of all cases showed features consistent with B-CLL. The typical polymorphic lympho-histiocytic cellular infiltrate of HD was absent in all cases.
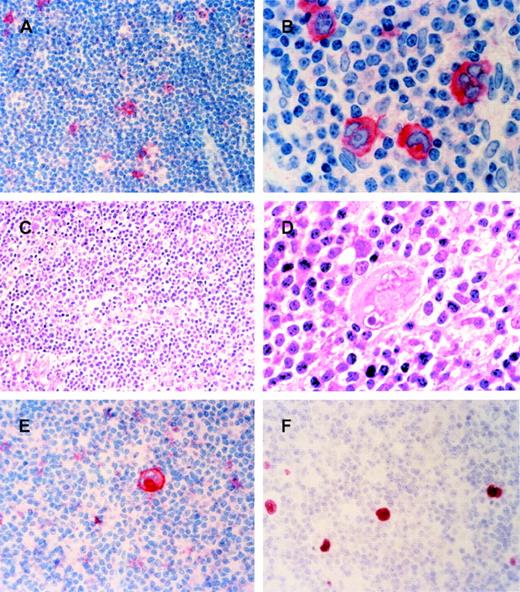
In case 1, the supraclavicular lymph node was infiltrated with small lymphocytes (Figure 1A). Some histiocytes with broad cytoplasm were intermingled. Medium- to large-size cells showing features of Hodgkin cells were randomly admixed, and binucleated cells resembling Reed-Sternberg cells were low in number. In case 2, the lymph node showed an infiltrate consisting of lymphocytes with round to slightly irregularly shaped nuclei and histiocytes (some with epitheloid differentiation). Hodgkin-like and numerous Reed-Sternberg–like cells were solitarily intermingled (Figure 1B-D). Occasionally small- to medium-size proliferation centers could be identified. In case 3, the lymph node structure was dominated by small lymphocytes showing some centers of proliferation. In addition to HRS-like cells, Hodgkin, classical Reed-Sternberg, and “mummified” cells were seen occasionally (Figure 1E). Many blood vessels were found, and histiocytes occasionally showed an epitheloid differentiation. In all 3 cases a slight to moderate positive immunostaining for the λ light chain and IgM was found in small- to medium-size lymphocytes (not shown). In the proliferation centers of all cases, some of the blast cells showed a perinuclear positivity for immunoglobulin. HRS-like cells were either negative for Ig or could show both light chains (likely reflecting unspecific adsorption of serum Ig).
Three cases of B-CLL with intermingled HRS-like cells.
(A, B, E) Anti-CD30 antibody staining of cases 1-3, hematoxylin counterstain. Single CD30-positive HRS-like cells are interspersed in the B-CLL infiltrated lymph node. (A) Case 1, 200 × magnification. (B) Case 2, 1000 × magnification. (E) Case 3, 400 × magnification. (C) Hematoxylin-eosin staining of case 2. The infiltrate mainly comprises small lymphocytes, a few blast cells, and histiocytes. A Reed-Sternberg–like cell is seen in the middle of the picture, 200 × magnification. (D) Hematoxylin-eosin staining of case 2. A Reed-Sternberg cell surrounded by small lymphocytes and histiocytes is seen in the middle of the picture, 1000 × magnification. (F) In situ hybridization of frozen section from CLL-infiltrated lymph node of case 3 for EBER-RNA. The HRS-like cells carry EBER-RNA, 400 × magnification. (A-F) Sections of formalin-fixed, paraffin-embedded, or frozen tissue.
Three cases of B-CLL with intermingled HRS-like cells.
(A, B, E) Anti-CD30 antibody staining of cases 1-3, hematoxylin counterstain. Single CD30-positive HRS-like cells are interspersed in the B-CLL infiltrated lymph node. (A) Case 1, 200 × magnification. (B) Case 2, 1000 × magnification. (E) Case 3, 400 × magnification. (C) Hematoxylin-eosin staining of case 2. The infiltrate mainly comprises small lymphocytes, a few blast cells, and histiocytes. A Reed-Sternberg–like cell is seen in the middle of the picture, 200 × magnification. (D) Hematoxylin-eosin staining of case 2. A Reed-Sternberg cell surrounded by small lymphocytes and histiocytes is seen in the middle of the picture, 1000 × magnification. (F) In situ hybridization of frozen section from CLL-infiltrated lymph node of case 3 for EBER-RNA. The HRS-like cells carry EBER-RNA, 400 × magnification. (A-F) Sections of formalin-fixed, paraffin-embedded, or frozen tissue.
Immunohistological studies showed that in all cases, the HRS-like cells expressed the CD30 antigen, and in cases 1 and 3, they also expressed CD15 (Table 2; Figure 1A, B, and E). The small lymphocytes of the CLL in all 3 patients were negative for CD30 and CD15; they were positive for the B-cell–associated CD20 antigen and for CD5 (Table 2). EBV was detected in the HRS-like cells of patients 2 and 3, but not in patient 1 (Table 2; Figure 1F). The small CLL tumor cells were EBV-negative in all cases.
Immunophenotype of HRS-like cells and B cells of the CLL
| Patient . | Cell Type . | CD30 . | CD15 . | CD20 . | CD3 . | CD5 . | Igλ* . | EBV† . |
|---|---|---|---|---|---|---|---|---|
| 1 | HRS-like | + | + | − | − | − | − | − |
| B-CLL | − | − | + | − | + | + | − | |
| 2 | HRS-like | + | − | − | − | −/+ | − | + |
| B-CLL | − | − | + | − | + | + | − | |
| 3 | HRS-like | + | + | − | − | − | − | + |
| B-CLL | − | − | + | − | + | + | − |
| Patient . | Cell Type . | CD30 . | CD15 . | CD20 . | CD3 . | CD5 . | Igλ* . | EBV† . |
|---|---|---|---|---|---|---|---|---|
| 1 | HRS-like | + | + | − | − | − | − | − |
| B-CLL | − | − | + | − | + | + | − | |
| 2 | HRS-like | + | − | − | − | −/+ | − | + |
| B-CLL | − | − | + | − | + | + | − | |
| 3 | HRS-like | + | + | − | − | − | − | + |
| B-CLL | − | − | + | − | + | + | − |
HRS-like cells were either negative or stained for both κ and λ, indicating unspecific adsorption of serum Ig. B-CLL cells were negative for κ light chain expression.
Indicates that the presence (+) or absence (−) of EBV was determined by staining with an antibody against LMP1 and EBER in situ hybridization (giving concordant results).
In patient 2, −/+ indicates that a small fraction of the cells are CD5-positive.
Very few solitary small cells were EBV-positive in the patients.
Micromanipulation and amplification of rearranged Vregion genes
Single large HRS-like cells were isolated by micromanipulation from CD30-stained frozen tissue sections of lymph node biopsies infiltrated by B-CLL. B cells of the CLL were identified and isolated as either CD20-positive, CD30-negative, or CD3-negative small cells. For patient 1, 3 micromanipulation experiments were performed, and 1 repeat experiment was performed for patient 2 (Table3). We used the following negative controls: (1) single T cells obtained from CD3-stained adjacent sections taken from the same tissue specimens on the same day as the HRS-like and B-CLL cells and (2) aliquots of the buffer covering the sections obtained during micromanipulation experiments. Genomic DNA of some micromanipulated cells and controls was amplified following the PEP protocol by Zhang et al.11 From aliquots of these reactions, rearranged Ig genes were amplified. The preamplification of genomic DNA was performed to investigate multiple aliquots of the amplified DNA of a cell with different combinations of primers and to preserve DNA for future analysis.
Summary of single cell analysis of HRS-like and B-CLL cells
| Pt. . | Cell Type . | V Gene . | No. of Positive Samples/Total No. . | PCR Products . | Repeated Rearrangements . | |
|---|---|---|---|---|---|---|
| Total . | Sequenced . | |||||
| 1 | HRS-like | VH | 13/43 | 13 | 13 | 13 VH4 (DP63)3-150 |
| Vκ | 0/22 | |||||
| Vλ | 7/28 | 7 | 7 | 2 Vλ3 (3r) | ||
| 5 Vλ3 (3a) | ||||||
| B-CLL | VH | 24/57 | 24 | 15 | 15 VH4 (DP63) | |
| Vκ | 0/22 | |||||
| Vλ | 19/40 | 19 | 10 | 7 Vλ3 (3r) | ||
| 3 Vλ3 (3a) | ||||||
| T cells | 1/25 | 1 | 1 | 1 VH4 (DP63)3-151 | ||
| Buffer | 0/13 | |||||
| 2 | HRS-like | VH | 10/33 | 10 | 10 | 10 VH3 (V3-30) |
| Vκ | 0/5 | |||||
| Vλ | 3/28 | 3 | 3 | 3 Vλ3 (GLL150) | ||
| B-CLL | VH | 7/36 | 7 | 7 | 7 VH3 (hv3005) | |
| Vκ | 2/12 | 4 | 3 | 1 Vκ2 (O1/O11) | ||
| 2 Vκ3 (L2) | ||||||
| Vλ | 2/24 | 2 | 2 | 2 Vλ3 (lv318) | ||
| T cells | 0/18 | |||||
| Buffer | 0/13 | |||||
| 3 | HRS-like | VH | 4/16 | 4 | 43-152 | 3 VH4 (VH4.16) |
| Vλ | 4/16 | 4 | 4 | 4 Vλ1 (1c-1) | ||
| B-CLL | VH | 5/16 | 10 | 9 | 4 VH3 (V3-23) | |
| 5 VH3 (DP48) | ||||||
| Vλ | 4/16 | 4 | 4 | 4 Vλ1 (1c-2) | ||
| T cells | 1/5 | 3 | 3 | 1 VH3 (V3-23) | ||
| 1 VH3 (DP48) | ||||||
| 1 Vλ1 (1c-2) | ||||||
| Buffer | 2/10 | 2 | 2 | 2 VH3 (V3-23) | ||
| Pt. . | Cell Type . | V Gene . | No. of Positive Samples/Total No. . | PCR Products . | Repeated Rearrangements . | |
|---|---|---|---|---|---|---|
| Total . | Sequenced . | |||||
| 1 | HRS-like | VH | 13/43 | 13 | 13 | 13 VH4 (DP63)3-150 |
| Vκ | 0/22 | |||||
| Vλ | 7/28 | 7 | 7 | 2 Vλ3 (3r) | ||
| 5 Vλ3 (3a) | ||||||
| B-CLL | VH | 24/57 | 24 | 15 | 15 VH4 (DP63) | |
| Vκ | 0/22 | |||||
| Vλ | 19/40 | 19 | 10 | 7 Vλ3 (3r) | ||
| 3 Vλ3 (3a) | ||||||
| T cells | 1/25 | 1 | 1 | 1 VH4 (DP63)3-151 | ||
| Buffer | 0/13 | |||||
| 2 | HRS-like | VH | 10/33 | 10 | 10 | 10 VH3 (V3-30) |
| Vκ | 0/5 | |||||
| Vλ | 3/28 | 3 | 3 | 3 Vλ3 (GLL150) | ||
| B-CLL | VH | 7/36 | 7 | 7 | 7 VH3 (hv3005) | |
| Vκ | 2/12 | 4 | 3 | 1 Vκ2 (O1/O11) | ||
| 2 Vκ3 (L2) | ||||||
| Vλ | 2/24 | 2 | 2 | 2 Vλ3 (lv318) | ||
| T cells | 0/18 | |||||
| Buffer | 0/13 | |||||
| 3 | HRS-like | VH | 4/16 | 4 | 43-152 | 3 VH4 (VH4.16) |
| Vλ | 4/16 | 4 | 4 | 4 Vλ1 (1c-1) | ||
| B-CLL | VH | 5/16 | 10 | 9 | 4 VH3 (V3-23) | |
| 5 VH3 (DP48) | ||||||
| Vλ | 4/16 | 4 | 4 | 4 Vλ1 (1c-2) | ||
| T cells | 1/5 | 3 | 3 | 1 VH3 (V3-23) | ||
| 1 VH3 (DP48) | ||||||
| 1 Vλ1 (1c-2) | ||||||
| Buffer | 2/10 | 2 | 2 | 2 VH3 (V3-23) | ||
HRS-like and B-CLL cells were analyzed together with T cells, and the controls were aspirated samples of the buffer covering the tissue sections during the micromanipulation. The data on patient 1 represent a summary of three independent micromanipulation experiments, and one repeat experiment was performed for patient 2. The clonalVH and Vγ genes from HRS-like cells were coamplified in one cell from patient 1 (VH4 and Vλ3gene 3a), in three cells from patient 2, and in one cell from patient 3. Coamplification of the VH andVL gene rearrangements from single cells was also seen for most B-CLL V region genes. In addition, amplification of the V region genes obtained from single B-CLL cells (and no additional other ones) and also from whole tissue DNA confirmed assignment of these genes to the B-CLL clones. Cells were analyzed with several different primer combinations, as outlined in Table 1.
The efficiency of the amplification of a V region gene from a single cell is usually below 50%,8 and therefore, sometimes two B cells of the CLL were micromanipulated and transferred into a PCR tube to minimize PCR work. We did not sequence 9VH4 and 9 Vλ3gene rearrangements from the B-CLL cells of patient 1, 1Vκ2 gene rearrangement from the B-CLL cells of patient 2, and 1 VH3 gene rearrangement from B-CLL cells of patient 3. The V gene segments used in the rearrangements are given in brackets. Identical V gene designations indicate clonally related V gene rearrangements. In patient 3,1c-1 and 1c-2 represent clonally unrelated rearrangements of the Vλ1 gene1c, amplified from HRS-like and B-CLL cells, respectively (but in both cases repeatedly obtained from the HRS-like and B-CLL cells).
Indicates that one sequence is identical to theVH4 gene rearrangement repeatedly amplified from B-CLL cells.
Indicates that this rearrangement is identical to theVH4 gene rearrangement of the B-CLL cells.
Indicates that one product (VH5-51, in-frame, 19% mutation) was obtained only once. IgD-positive B-cells were analyzed to control the efficiency of the single-cell PCR (data not shown).
From each of the 3 cases analyzed, between 16 and 50 individual HRS-like cells and at least 16 B-CLL cells were analyzed (Table 3). The amplification efficiences for clonal Ig gene rearrangements ranged from 7% to 30% for the HRS-like cells (Table 3). The corresponding efficiencies for the B-CLL cells could not be determined because often 2 B-CLL cells were put into 1 reaction tube. These values were in the same range as efficiencies obtained in previous micromanipulation studies.12,14 15 We believe that a given rearrangement is not amplified from all cells largely due to technical matters such as degradation or inaccessibility of the DNA. Moreover, part of the nucleus is missing for many micromanipulated cells, and some rearrangements may not be efficiently amplified because of mutations at primer binding sites (see below).
In patient 1, HRS-like cells and B cells of the CLL harbor clonally related V gene rearrangements with shared as well as distinct somatic mutations
From patient 1, a total of 50 HRS-like cells were analyzed forIgV gene rearrangements with 4 different combinations of primers (Tables 1 and 3). The same potentially functionalVH4 gene rearrangement was amplified from 13 of 43 HRS-like cells analyzed for rearranged VHgenes. From 2 of the 28 cells analyzed for Vλgene rearrangements, an identical potentially functionalVλ3 gene rearrangement was amplified, and a nonfunctional Vλ3 gene rearrangement was obtained from 5 cells. Sequencing of 25 out of 43 PCR products amplified from B-CLL cells revealed that the B-CLL cells carried the same VH and Vλ gene rearrangements (Tables 3 and 4). Among the 25 sequenced PCR products, the VH4 gene was obtained 15 times; the potentially functional Vλ3 gene 7 times; and the nonfunctional Vλ3 gene 3 times.
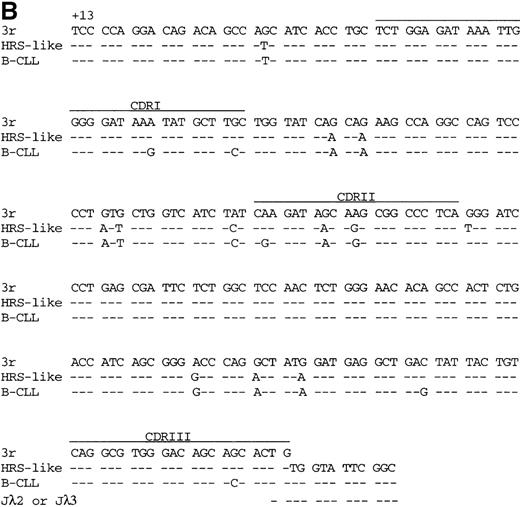
The VH and Vλ gene rearrangements amplified from HRS-like and B-CLL cells were somatically mutated (Figure 2 and Table4). No intraclonal sequence diversity was observed among the clonally related V gene rearrangements amplified repeatedly from HRS-like or B-CLL cells. The frequency of mutations in the VH4 gene segment of the HRS-like cells was 6.6%. In addition to the somatic mutations in theVH4 gene of the HRS-like cells, 2 further point mutations were present in the B cells of the CLL (mutation frequency, 7.3%). The potentially functionalVλ3 gene rearrangement carried 11 identical somatic point mutations in the HRS-like and B-CLL cells (Figure 2B) compared to the germline. One additional point mutation was found in the HRS-like cells (mutation frequency, 5.3%), and 5 further mutations were present in the respective gene rearrangement in the B-CLL cells (mutation frequency, 6.7%). The nonproductively rearrangedVλ3 gene of the HRS-like and B-CLL cells shared 25 mutations (sequences not shown). In the HRS-like cells, 1 additional point mutation was present, the gene rearrangement of the B-CLL cells carried 6 further mutations, and an 8–base pair (bp) duplication was found in framework region III (FRIII). The presence of a high load of somatic mutations in the V gene rearrangement amplified from the HRS-like and B-CLL cells may explain the low efficiency of the amplification of the 2Vλ3 gene rearrangements from the cells. Somatic mutations presumably also present at the primer binding sites could severely reduce the amplification efficiency.
V gene sequences from patient 1.
Sequences of (A) the VH4 gene rearrangement and (B) the potentially functional Vλ3 gene rearrangement of the HRS-like and B cells of CLL are compared to the most homologousV germline genes and J segments.35-38Dashes indicate sequence identity. Codons are numbered according to Kabat et al.34 CDRI-CDRIII are indicated.
V gene sequences from patient 1.
Sequences of (A) the VH4 gene rearrangement and (B) the potentially functional Vλ3 gene rearrangement of the HRS-like and B cells of CLL are compared to the most homologousV germline genes and J segments.35-38Dashes indicate sequence identity. Codons are numbered according to Kabat et al.34 CDRI-CDRIII are indicated.
Sequence analysis of clonal Ig gene rearrangements obtained from three cases of B-CLL with HRS-like cells
| Pt. . | Cell Type . | VH . | VL . | HRS-Like and CLL Cells: Clonally Related? . | ||||
|---|---|---|---|---|---|---|---|---|
| VH Gene . | Mut.(%) . | Potentially Functional . | VL Gene . | Mut.(%) . | Potentially Functional . | |||
| 1 | HRS-like | DP63 (4) | 6.6 | + | 3r (Vλ3) | 5.3 | + | Yes |
| 3a (Vλ3) | 13.3 | − | ||||||
| B-CLL | DP63 (4) | 7.3 | + | 3r (Vλ3) | 6.7 | + | ||
| 3a (Vλ3) | 15.3 | − | ||||||
| 2 | HRS-like | V3-30 (3) | 8.2 | +4-150 | GLL150 (Vλ3) | 9.5 | − | No |
| B-CLL | hv3005 (3) | 0 | + | lv318 (Vλ3) | 0 | + | ||
| O1/O11 (Vκ2) | 0 | − | ||||||
| L2 (Vκ3) | 0 | − | ||||||
| 3 | HRS-like | VH4.16 (4) | 9.1 | + | 1c-1 (Vλ1) | 5.0 | − | No |
| B-CLL | V3-23 (3) | 0 | + | 1c-2 (Vλ1) | 0 | + | ||
| DP-48 (3) | 0.5 | − | ||||||
| Pt. . | Cell Type . | VH . | VL . | HRS-Like and CLL Cells: Clonally Related? . | ||||
|---|---|---|---|---|---|---|---|---|
| VH Gene . | Mut.(%) . | Potentially Functional . | VL Gene . | Mut.(%) . | Potentially Functional . | |||
| 1 | HRS-like | DP63 (4) | 6.6 | + | 3r (Vλ3) | 5.3 | + | Yes |
| 3a (Vλ3) | 13.3 | − | ||||||
| B-CLL | DP63 (4) | 7.3 | + | 3r (Vλ3) | 6.7 | + | ||
| 3a (Vλ3) | 15.3 | − | ||||||
| 2 | HRS-like | V3-30 (3) | 8.2 | +4-150 | GLL150 (Vλ3) | 9.5 | − | No |
| B-CLL | hv3005 (3) | 0 | + | lv318 (Vλ3) | 0 | + | ||
| O1/O11 (Vκ2) | 0 | − | ||||||
| L2 (Vκ3) | 0 | − | ||||||
| 3 | HRS-like | VH4.16 (4) | 9.1 | + | 1c-1 (Vλ1) | 5.0 | − | No |
| B-CLL | V3-23 (3) | 0 | + | 1c-2 (Vλ1) | 0 | + | ||
| DP-48 (3) | 0.5 | − | ||||||
Clonal V gene sequences from the HRS-like and B-CLL cells are compared to the most homologous germline genes.35,36,39 40 The V gene families are given in parentheses after the gene name. Sequences are available from the EMBL/GenBank databases under accession number AJ242555-AJ242572.
The − sign indicates a nonfunctional rearrangement, and the + indicates a potentially functional rearrangement. Clonally unrelated rearrangements of the Vλ1 gene1c are represented by 1c-1 and 1c-2, which were amplified from HRS-like and B-CLL cells, respectively.
Indicates potentially functional rearrangement rendered nonfunctional due to a stop codon and a 1-bp deletion introduced by somatic mutation (Figure 3).
Further experiments were performed to amplify the region spanning from the promoter to FRI of the potentially functionalVH4 gene rearrangement in overlap with the sequences already generated. The purpose of this amplification was to compare the HRS-like cells with the B-CLL cells and to investigate whether somatic mutations may be present that would render this potentially functional rearrangement nonfunctional. Such “crippling” somatic mutations have been observed in HRS cells in some cases of classical HD.12 15 However, the sequences of the promoter and leader peptide region obtained were identical in HRS-like and B-CLL cells, and mutations that would lead to a loss of function of this gene rearrangement were not identified (sequences not shown).
From 1 of the 13 HRS-like cells and 1 of 25 T cells used as negative controls, a VH4 gene rearrangement was obtained whose sequences were identical to the corresponding sequence of the B-CLL cells (Table 3). Most likely these sequences result from cellular contamination by fragments of B cells of the CLL. Since almost all cells in the tissue represent members of the B-CLL tumor clone, these cells represent a major risk of contamination. No PCR product was amplified from a total of 13 buffer controls (Table 3).
The potentially functional VH4 andVλ3 gene rearrangements obtained from single B-CLL cells could also be amplified from genomic DNA extracted from tissue sections of the investigated lymph node. This result shows that the CLL tumor clone had been identified in the single-cell analysis.
The sequence of a second, slightly largerVλ3 PCR product could not be determined unambiguously, probably due to contamination of this amplificate with the functional Vλ3 gene rearrangement during isolation of the product from an agarose gel. This PCR product most likely represents the second Vλ3gene rearrangement of the B-CLL tumor clone.
HRS-like cells in patients 2 and 3 harbor somatically mutated Ig gene rearrangements that are clonal and not related to the V gene rearrangements of the CLL clone
From patient 2, we investigated 33 HRS-like cells. An identicalVH3 gene rearrangement was amplified from 10 cells (Table 3). This rearrangement is somatically mutated, with a mutation frequency of 8.2% (Figure3; Table 4). This in-frameVHDHJH gene rearrangement had lost its functionality due to a stop codon in CDRII and a 1-bp deletion in FRIII (Figure 3). A clonal nonfunctionalVλ3 gene was amplified from 3 of 28 HRS-like cells investigated for Vλ gene rearrangements. This rearrangement has a mutation frequency of 9.5% and shows a 13-bp deletion in FRI.
“Crippling” somatic mutations in theVH3 gene rearrangement from the HRS-like cells from patient 2.
Comparison of the VH3 gene rearrangement from HRS-like cells with the VH V3-30 germline gene39 and the JH4segment.38 ***Indicates a stop codon. A 1-bp deletion in FRIII is indicated by Δ. CDRI -CDRIII are indicated.
“Crippling” somatic mutations in theVH3 gene rearrangement from the HRS-like cells from patient 2.
Comparison of the VH3 gene rearrangement from HRS-like cells with the VH V3-30 germline gene39 and the JH4segment.38 ***Indicates a stop codon. A 1-bp deletion in FRIII is indicated by Δ. CDRI -CDRIII are indicated.
Of 36 samples containing 1 or 2 B cells of the CLL, 7 samples gave rise to a total of 13 PCR products. The V gene rearrangements of the B-CLL were not related to the rearrangements obtained from the HRS-like cells (Table 3). A potentially functionalVH3 gene rearrangement was amplified 7 times from independent samples. Two samples gave rise to the same potentially functional Vλ3 gene rearrangement. From 2 samples an identical nonfunctionalVκ3 gene rearrangement was obtained and a nonfunctional Vκ2 gene rearrangement was amplified once. The clonal VH3 and theVκ3 gene rearrangements and theVκ2 region gene were also obtained from genomic DNA extracted from the lymph node biopsy. (Vλ gene rearrangements were not investigated.) The clonal VH and VLgene rearrangements of the B-CLL were unmutated (Table 4).
From 16 HRS-like cells of patient 3, an identical potentially functional VH4 gene rearrangement was amplified from 3 cells, and an identical nonfunctionalVλ1 gene rearrangement was amplified from 4 cells (Table 3). The VH4 and theVλ1 gene rearrangements were somatically mutated with mutation frequencies of 9.1% and 5.0%, respectively. A 1-bp deletion was present in FRII of the Vλ1gene.
V gene rearrangements amplified from the B-CLL cells were not related to those of the HRS-like cells. From 16 samples containing 1 or 2 B-CLL cells, an identical potentially functionalVH3 gene rearrangement was obtained from 4 samples; a nonfunctional VH3 gene rearrangement was obtained from 5 samples; and an identical potentially functional Vλ1 gene rearrangement was obtained from 4 samples. One of the 5 control T cells and 2 of the 10 buffer controls gave rise to the clonal V gene rearrangements of the CLL, which was most likely due to cellular contamination. ClonalVH and VL gene rearrangements of the B cells of the CLL were also obtained from genomic DNA extracted from the lymph node biopsy. Except for a 1-bp substitution in the nonfunctional VH3 gene rearrangement, the V gene rearrangements of the B-CLL were unmutated (Table 4).
Discussion
Clonality and GC B-cell derivation of HRS-like cells in B-CLL
Clonal VH and VL gene rearrangements were amplified from HRS-like cells of each of the 3 B-CLL cases. This shows that the HRS-like cells represent clonal populations of mature B-lineage cells. These findings are in agreement with a previous report by Ohno et al6 on expanded populations of HRS-like cells with clonally relatedVH gene rearrangements. In 2 of 3 cases of Richter's syndrome with HD features, Ohno et al6 detected identical VH gene rearrangements in single HRS-like and B-CLL cells. In the third case, however, rearrangedVH genes were obtained from CLL cells but not from HRS-like cells, and in that case, the relationship of the 2 cell populations remained unexplained. Since only the CDRIII of IgHgene rearrangements were amplified and sequenced, the presence of somatic mutations remained uninvestigated, thereby preventing further insight into the stage of differentiation of the HRS cell progenitor.
In the present study, sequence analysis of the rearranged Vgenes of HRS-like cells revealed the presence of somatic mutations in all rearrangements. The frequency of somatic mutation ranged from 5% to 13.3% (Table 4). The detection of deletions or duplications in 4 of the somatically mutated V region genes (3 V genes from HRS-like cells and 1 V gene from B-CLL cells) is not unusual because such events have been repeatedly observed in normal and malignant human B cells.16,17 Thus, in the 3 cases of B-CLL with HRS-like cells analyzed in the present work, the precursors of the HRS-like cells are likely to be GC B cells or descendants of these cells since the process of somatic hypermutation is thought to be restricted to and specific for human B cells undergoing proliferation and antigen selection in GC.7
Studies on rearranged V genes in HRS cells in classical HD have established that these cells in most cases represent clonal populations of transformed GC B cells.5 In some of the cases of classical HD, crippling somatic mutations were detected that rendered originally productive V genes nonfunctional, thereby providing evidence that HRS cells (or their precursors) are not selected for antibody expression and have been rescued from apoptosis inside the GC by some transforming event.5,12,15 18
In the present study, crippling somatic mutations were detected in the originally potentially functional VH3 gene of the HRS-like cells of patient 2 (Figure 3). However, in this case (as opposed to several of the above-mentioned cases of HD with crippled in-frame rearrangements) we cannot rule out the possibility that another functional VH gene rearrangement was present on the second IgH allele.14 19Nevertheless, B cells carrying 2 productive VHregion genes are rare, and we did not detect a second heavy chain gene rearrangement. We consider it more likely that the HRS-like cells of patient 2 carry a DJ gene rearrangement or germline configuration in the second IgH allele and that the HRS-like cells had lost the capacity to express an antigen receptor by a crippling mutation. B cells acquiring crippling mutations during a GC reaction are usually efficiently eliminated within the GC. It is thus likely that the HRS-like cell clone in patient 2 is derived from a GC B cell that was rescued from apoptosis by some transforming event such as infection by EBV, which was detected in these cells. Thus, it appears that also in diseases other than classical HD, cells phenotypically resembling HRS cells can occur, which, like HRS cells of HD, derive from crippled GC B cells.
B-CLL and HRS-like cells derived from a shared GC B-cell precursor
In patient 1, the B-CLL cells carried somatically mutated clonalVH and Vλ gene rearrangements (Tables 3 and 4). Approximately 50% of B-CLL cases carry somatically mutated V genes.20 These cases are likely derived from precursors that had passed through a GC reaction. Interestingly, the vast majority of normal CD5-positive B cells carry unmutatedV region genes,21 which may indicate that CD5 B cells driven into a GC reaction have an increased risk to undergo malignant transformation.21
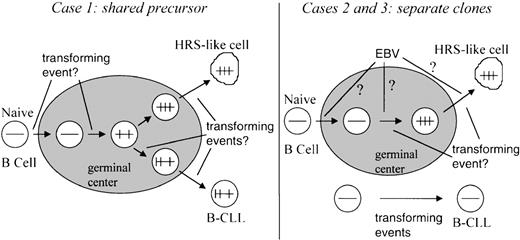
The HRS-like and B-CLL cells in patient 1 carried the same clonalV gene rearrangements (Tables 3 and 4). Thus, the 2 cell populations turned out to be derived from a common precursor B cell. Whereas most of the somatic mutations were shared between the HRS-like and B-CLL cells, some mutations were present only in one or the other of the cell types (Figure 2). This pattern of somatic mutation indicates that a GC B cell which had already acquired somatic mutations gave rise to 2 descendants, 1 of which developed to CLL, the other to the HRS-like cell population (Figure 4). Thus in this case, the pattern of somatic mutations also suggests a GC B-cell derivation of the HRS-like cells. In addition, the mutation pattern clearly argues against a disease scenario in which single mature cells of the B-CLL clone occasionally and accidentally acquire the phenotype of HRS-like cells. The derivation of the HRS-like and B-CLL cells from a common precursor cell may indicate that distinct molecular events caused the appearance of these 2 populations of clonally expanded cells (Figure 4). In search for a transforming event that could be involved in the generation of the 2 cell types, the cells were analyzed for the presence of EBV. However, both cell types were found to be EBV negative (Table 2).
Scenarios for the generation of B-CLL and associated HRS-like cells.
Horizontal bars schematically depict rearranged V region genes, and vertical bars depict somatic mutations. The shaded area represents a germinal center.
Scenarios for the generation of B-CLL and associated HRS-like cells.
Horizontal bars schematically depict rearranged V region genes, and vertical bars depict somatic mutations. The shaded area represents a germinal center.
The relationship of the B-CLL and the HRS-like cells in patient 1 is reminiscent of 2 cases, representing combinations of HD and B cell-non-Hodgkin's lymphoma (NHL) in the same patient, that we recently analyzed.22 In both cases (one composite follicular and Hodgkin's lymphoma, and a case of T-cell–rich B-cell lymphoma in the skin followed by HD in a lymph node 3 years later), the HRS and B-NHL cells turned out to be clonally related and to represent distinct descendents of a shared GC B cell precursor.22This was evident from the finding of shared as well as unique somatic mutations in the clonally related V genes of the 2 types of tumor cells. Assuming that the HRS-like cells in patient 1 of the present study represent malignant cells (a mere speculation at this point), the HRS-like and B-CLL tumor cells would have likely undergone shared as well as distinct transforming events, which would allow the appearance of 2 distinct populations of clonally related cells.
HRS-like cells in B-CLL: tumor precursors for HRS cells in HD?
Since the clonally related HRS-like and B-CLL cells in patient 1 likely share some transforming event (see above), one may speculate that these HRS-like cells have an increased risk to develop to an HRS-like cell clone of HD, perhaps upon acquiring additional transforming events. In light of the increased risk of B-CLL patients to develop HD as a secondary malignancy (see below), HRS-like cells clonally related to the B-CLL, such as in patient 1, may represent precursors for HRS tumor cells in HD following B-CLL.
In patients 2 and 3 a different scenario would have to be considered, as in these cases the HRS-like cells were not related to the CLL tumor clone (Tables 3 and 4, Figure 4). In patients 2 and 3, the HRS-like cell clones were found to harbour EBV. As seen in EBV-positive classical HD, the HRS-like cells analyzed here and observed previously in cases of B-CLL3,4 express the EBV-encoded latent membrane protein–1 (LMP-1), which is known for its B-cell transforming capability.23 To explain the expansion of an EBV-harboring B-cell clone in a setting of B-CLL, the following scenario can be envisioned: In healthy EBV carriers, very rare B cells (1-50 per 106 cells) harbor EBV.24 These cells are usually resting and express LMP-2 as the only EBV-encoded latent antigen.25 However, the virus may sometimes change its latency gene expression profile, thereby inducing proliferation of the B cell.26 In healthy individuals, such proliferating EBV-positive B cells are rapidly eliminated by an effective CD8 T-cell response.27 However, B-CLL patients are often immunosuppressed and have an impairment of cytotoxic T-cell functions.28 Thus, in the setting of a B-CLL tumor, reactivated EBV-positive B cells can perhaps clonally expand without being eliminated by cytotoxic T cells. If this reactivation by switching of the EBV gene expression profile is a rare event, the proliferating EBV-positive B cells appearing in the patient at a given point in time may often derive from a single precursor cell.
Alternatively, in the case of patient 3, the occurrence of HRS-like cells may relate to the observation that this patient suffered from infectious mononucleosis (the acute primary EBV infection) 2 years before B-CLL was diagnosed. Since HRS-like cells infected by EBV regularly develop during infectious mononucleosis,29 30 the HRS-like cell clone in patient 3 might originate from an HRS-like cell that was generated during the primary EBV infection and persisted in the patient.
Interestingly, patients with B-CLL have approximately an 8-fold increased risk to develop HD,31 with HD representing 1 of the most frequent secondary neoplasms (besides transformation of the B-CLL into a large cell lymphoma, ie, Richter syndrome). In 4 patients described in the literature, the EBV status of HRS-like cells of B-CLL and the HRS cells of the concurrent or subsequent HD was studied. In each instance, EBV was found in both types of HRS cells.3,32 Thus, the HRS-like EBV-positive cell clones seen in patients 2 and 3 may represent cells that sometimes, upon acquisition of additional transforming events (besides infection by EBV), can develop to HD. That other events in addition to EBV infection are needed to transform a B-CLL–associated HRS-like cell to an HRS tumor clone causing HD is supported by the finding that the lympho-histiocytic cellular infiltrate typical for HD is missing in B-CLL with HRS-like cells. This cellular infiltrate is likely caused by the HRS cells and is a major distinguishing feature of HD.33
The present study provides evidence that HRS-like cells in B-CLL, like HRS cells in HD, represent clonal populations of B lymphocytes derived from GC B cells. These cells can either derive from the precursor that also gave rise to the B-CLL or represent independent clones of EBV-transformed B cells that expanded in the setting of a B-CLL. In either case, HRS-like cells may potentially represent precursors for HRS cells in HD.
Acknowledgments
We are grateful to A. Klöckner, A. Fassbender, and J. Jesdinsky for excellent technical assistance and to T. Spieker for help with the EBER in-situ hybridization. We thank A. Bräuninger for critical reading of the manuscript and R. Dalla-Favera for helpful discussion.
Supported by grants from Deutsche Forschungsgemeinschaft (SFB502) and the Deutsche Krebshilfe, Dr Mildred Scheel Stiftung.
Reprints:Ralf Küppers, Department of Internal Medicine and Institute for Genetics, LFI E4 R706, University of Cologne, Joseph-Stelzmannstr 9, 50931 Cologne, Germany.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal