Graft rejection after histocompatibility locus antigen (HLA)-identical stem cell transplantation results from the recognition of minor histocompatibility antigens on donor stem cells by immunocompetent T lymphocytes of recipient origin. T-lymphocyte clones that specifically recognize H-Y epitopes on male target cells have been generated during graft rejection after sex-mismatched transplantation. Previously, 2 human H-Y epitopes derived from the same SMCY gene have been identified that were involved in bone marrow graft rejection. We report the identification of a new male-specific transplantation antigen encoded by the Y-chromosome-specific gene DFFRY. The DFFRY-derived peptide was recognized by an HLA-A1 restricted CTL clone, generated during graft rejection from a female patient with acute myeloid leukemia who rejected HLA-phenotypically identical bone marrow from her father. The identification of this gene demonstrates that at least 2 genes present on the human Y-chromosome code for male-specific transplantation antigens.
Histocompatibility locus antigen (HLA)-identical bone marrow transplantation (BMT)1 is frequently complicated by graft-versus-host disease (GVHD) or graft rejection. Each complication is thought to be initiated by HLA-restricted T lymphocytes that recognize minor histocompatibility (mH) antigens.1-4 These polymorphic antigens, which differ between donor and recipient, are peptides derived from intracellular proteins. The male-specific H-Y antigens are the most extensively studied mH antigens. The involvement of H-Y antigens in transplant rejection was identified by the observation that, within an inbred mouse strain, females rapidly rejected primary syngeneic male skin grafts.5,6 In humans, female patients who undergo transplantation with male bone marrow have a higher risk for graft rejection than when they undergo transplantation with marrow of female origin.1,7-9 In mice and humans, H-Y-specific T-lymphocyte clones could be generated from the peripheral blood of patients during GVHD or graft rejection after sex-mismatched transplantation.1,10 11 These H-Y-specific T-lymphocyte clones can be used as tools to identify H-Y antigens and the corresponding Y-specific genes.
The first identified gene encoding for an H-Y antigen was SMCY. In mouse, the gene was identified by transfection of a series of cosmids of the short arm of the Y chromosome into cells expressing the appropriate restriction molecules. These cells were then tested for their ability to stimulate an H-Y epitope-specific T-lymphocyte clone. Transfection of 1 cosmid encoding for the SMCY gene resulted in stimulation of the H-Y-specific T-lymphocyte clone.10 In humans, 2 H-Y antigens were identified by analysis of eluted peptides from male target cells using a combination of microcapillary liquid chromatography–electrospray ionization mass spectrometry with T-cell epitope reconstitution assays. Both H-Y antigens, recognized by an H-Y-specific HLA-B7-restricted cytotoxic T-lymphocyte (CTL) clone and by an HLA-A2 restricted CTL clone, were encoded by different peptides from the SMCY protein.12 13
Although SMCY encodes for most H-Y antigens identified to date, genetic mapping of the mouse Y chromosome suggested that at least 2 and up to 5 distinct loci, including SMCY, encode H-Y antigens.14Recently, a systematic search of the nonrecombinant region of the human Y chromosome identified 3 known and 5 novel Y-specific genes with a ubiquitous tissue expression.15 Each gene had a homologue on the X chromosome encoding a very similar but nonidentical protein isoform. The amino acid sequence identity of the X-Y isoforms varied from 85% to 97%. Because all these ubiquitously expressed Y-specific genes have sufficient polymorphisms to generate male-specific peptides, they are all potential candidates to encode H-Y antigens. The identification of UTY as a mouse H-Y antigen-encoding gene confirmed the hypothesis that H-Y antigen is the product of more than 1 gene on the Y-chromosome.16 In humans, however, H-Y antigens published thus far are derived from the SMCY gene.12 13
In 1990, we reported a detailed analysis of a female patient with acute myeloid leukemia (AML) who rejected a bone marrow graft from her HLA-phenotypically identical father. Before transplantation and during graft rejection, a strong cellular recipient antidonor cytotoxic reactivity could be demonstrated. This reactivity was directed against mH antigens, including a male-specific antigen. Cloning of this recipient–antidonor response resulted in the generation of a CTL clone shown to be H-Y specific and HLA-A1 restricted.1
In this study, we report the identification of a novel male-specific transplantation antigen, recognized by the HLA-A1 restricted CTL clone. The H-Y antigen presented by HLA-A1 is the 9-residue peptide IVDCLTEMY derived from DFFRY, a gene coding for a new male-specific transplantation antigen. The identification of this H-Y antigen demonstrates that the human H-Y antigen involved in graft rejection may consist of the products of more than 1 gene on the human Y-chromosome.
Materials and methods
CTL and cell lines
The CD8+ CTL clone HLA-A1 HY was derived by limiting dilution from peripheral blood mononuclear cells (PBMC) of a female patient with AML, who had rejected the phenotypically HLA-identical male bone marrow derived from her father.1 The CTL clone was cultured by stimulation with irradiated allogeneic PBMC and donor-derived Epstein-Barr virus (EBV)-transformed B cells (EBV-LCL) in RPMI-1640 medium (Bio-Whittaker, Verviers, Belgium) containing 10% pooled human serum and IL-2 (Roussel Uclaf, Paris, France) at 300 IU/mL. As a control, an SMCY-specific, HLA-B7-restricted CTL clone was used.12 HeLa cells and WEHI-164 clone 13 cells were obtained from American Type Culture Collection (Rockville, MD) and maintained in complete medium consisting of RPMI-1640 medium with 10% fetal calf serum. The HLA-A1 gene was cloned to the vector PLXSN and stably transfected into HeLa (HeLa/HLA-A1) using retroviral transfection as described by Millar et al.17 After transfection, HeLa/HLA-A1 cells were maintained in complete culture medium supplemented with G418 (500 μg/mL).
Cloning of Y-specific genes
Total RNA was isolated from male EBV-LCL with Trizol (Gibco BRL, Gaithersburg, MD) according to the manufacturer's procedure. cDNA was prepared from RNA using M-MLV BRL reverse transcriptase (Gibco BRL) for 60 minutes. at 37°C as described.18 19 One fiftieth of each cDNA reaction was individually amplified using specific primers for each Y-gene (Table 1) identified by a systematic search of the nonrecombinant region of the human Y chromosome. Each ubiquitously expressed gene had a homologue on the X chromosome encoding a similar but nonidentical protein isoform. Underlined bases are mutations compared to the original sequence of the Y-specific gene to introduce artificial start or stop codons or to introduce restriction sites for cloning experiments. Because SMCY and DFFRY have large open reading frames, both genes are cloned in 3 overlapping cDNA constructs. With the exception of SMCY, all genes were amplified using the expanded long-template polymerase chain reaction (PCR) system (Boehringer Mannheim GmbH, Mannheim, Germany). The amplification was started with a denaturation step of 2 minutes at 92°C, followed by 30 cycles, with each cycle consisting of 20 seconds at 92°C, 1 minute at 60°C, and 1 minute at 68°C. SMCY was amplified using the Marathon cDNA amplification kit (Clontech Laboratories, Palo Alto, CA) according to the manufacturer's instructions. After amplification, each Y-specific cDNA was visualized on an ethidium bromide-stained low melt agarose gel (Sigma Chemical, St Louis, MO). cDNA was isolated from the low-melt agarose gel using agarase (Boehringer Mannheim GmbH) according to the manufacturer's procedure. Each individual cDNA was cloned in the expression vector PCR3.1 using the eukaryotic TA cloning kit (Invitrogen, Carlsbad, CA).
Y-specific primers
| Y-Specific Gene . | Sequence (5′-3′) . |
|---|---|
| DBY | Forward: CGCTTGAATTCAAGGATGAGTCATGTGGTG |
| Reverse: GGGTGATATCGCTGCAAAGCAGATTCAGTTG | |
| DFFRY 1 | Forward: AGATCTAAGCTTTGTCAAGCATGACAGCCATC |
| Reverse: TCCTAATTAATATCAGGATCCAAGTCTGGTTC | |
| DFFRY 2 | Forward: GCAAAACAAATAAGGATGTGCTTAGCAGAA |
| Reverse: AATTAATGAGGATATCCAAATACATCATCTCG | |
| DFFRY 3 | Forward: CGACATGTTCGGGGATGAGAAGCAGGACAG |
| Reverse: CATAGCGGCCGCAGTTAATTATTGCTTTTCAC | |
| EIF-1AY | Forward: TCACCGCCATGCCCAAGAATAAAGG |
| Reverse: CACTTGGTTCAATTTAGATATCATCAATG | |
| RPS4Y | Forward: CAGAATTCCGCCATGGCCCGGGGC |
| Reverse: AAAAAAAGATATCCTGCTACTGCAATTTAG | |
| SMCY 1 | Adaptor primer: CCATCCTAATACGACTCACTATAGGGC |
| Reverse: TCAATGTACAGGCATGTTGAAGTAGTCGGAC | |
| SMCY 2 | Forward: CACCATGGCTACCCAGGAGTACAGTTTGC |
| Reverse: TCAGGAGGCTATCTTGGCACCCATA | |
| SMCY 3 | Forward: CACCATGGCCCTGGCCCCTTCTGCTCAC |
| Adaptor primer: CCATCCTAATACGACTCACTATAGGGC | |
| TB4Y | Forward: CGCAGCCATGTCTGACAAACCTGG |
| Reverse: GATATTGGTGGCACATGCCTGTTTAA | |
| UTY | Forward: AATTTGTTGGTACCATGAAATCCTGCGC |
| Reverse: AATACTCGAGGAACTATATCAAGATGAGGA | |
| ZFY | Forward: GACCATGGATGAAGATGAATTTGAATTGCAG |
| Reverse: CAAGCTCGAGGACACACTGTTAGGGCAGA |
| Y-Specific Gene . | Sequence (5′-3′) . |
|---|---|
| DBY | Forward: CGCTTGAATTCAAGGATGAGTCATGTGGTG |
| Reverse: GGGTGATATCGCTGCAAAGCAGATTCAGTTG | |
| DFFRY 1 | Forward: AGATCTAAGCTTTGTCAAGCATGACAGCCATC |
| Reverse: TCCTAATTAATATCAGGATCCAAGTCTGGTTC | |
| DFFRY 2 | Forward: GCAAAACAAATAAGGATGTGCTTAGCAGAA |
| Reverse: AATTAATGAGGATATCCAAATACATCATCTCG | |
| DFFRY 3 | Forward: CGACATGTTCGGGGATGAGAAGCAGGACAG |
| Reverse: CATAGCGGCCGCAGTTAATTATTGCTTTTCAC | |
| EIF-1AY | Forward: TCACCGCCATGCCCAAGAATAAAGG |
| Reverse: CACTTGGTTCAATTTAGATATCATCAATG | |
| RPS4Y | Forward: CAGAATTCCGCCATGGCCCGGGGC |
| Reverse: AAAAAAAGATATCCTGCTACTGCAATTTAG | |
| SMCY 1 | Adaptor primer: CCATCCTAATACGACTCACTATAGGGC |
| Reverse: TCAATGTACAGGCATGTTGAAGTAGTCGGAC | |
| SMCY 2 | Forward: CACCATGGCTACCCAGGAGTACAGTTTGC |
| Reverse: TCAGGAGGCTATCTTGGCACCCATA | |
| SMCY 3 | Forward: CACCATGGCCCTGGCCCCTTCTGCTCAC |
| Adaptor primer: CCATCCTAATACGACTCACTATAGGGC | |
| TB4Y | Forward: CGCAGCCATGTCTGACAAACCTGG |
| Reverse: GATATTGGTGGCACATGCCTGTTTAA | |
| UTY | Forward: AATTTGTTGGTACCATGAAATCCTGCGC |
| Reverse: AATACTCGAGGAACTATATCAAGATGAGGA | |
| ZFY | Forward: GACCATGGATGAAGATGAATTTGAATTGCAG |
| Reverse: CAAGCTCGAGGACACACTGTTAGGGCAGA |
Cloning of subgenic fragments of the DFFRY2 gene
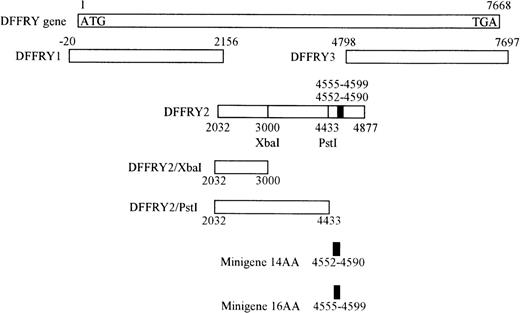
Restriction analysis on the DFFRY2 gene revealed an XbaI site on position 3000 and a PstI site on position 4433 of the DFFRY cDNA. Each restriction endonuclease also has a recognition site in the multiple cloning site of the PCR3.1 plasmid directly after the 3′ end of the DFFRY2 cDNA. Deletion mutants DFFRY2/XbaI and DFFRY2/PstI were generated by incubating the plasmid PCR3.1 containing the DFFRY2 gene with the restriction enzyme XbaI and PstI, deleting a 1877-bp and a 444-bp long fragment at the 3′ site of the DFFRY2 gene, respectively. After digestion, the linear truncated constructs were visualized on an ethidium bromide-stained, low-melt agarose gel. After isolating the constructs with agarase, the linear constructs were ligated with the rapid DNA ligation kit (Boehringer Mannheim GmbH) to form circular plasmid DNA consisting of vector PCR3.1 with the truncated DFFRY2/XbaI and DFFRY2/PstI cDNA.
Cloning of DFFRY2 minigenes
DFFRY2 minigenes were generated by hybridization of 2 oligo nucleotides, followed by incorporation into the BamHI and HindIII site of PCR3.1. For the generation of minigene 14AA, we used the following oligo nucleotides 5′3′: AGCTTCACCATGCTCAAACAGATAGTAGACTGTTTGACTGAAATGTATTACTAG and GATCCTAGTAATACATTTCAGTCAAACAGTCTACTATCTGTTTGAGCATGGTGA.
For minigene 16AA we used these oligo nucleotides: AGCTTCACCATGAAACAGATAGTAGACTGTTTGACTGAAATGTATTACATGGGCACATAG and GATCCTATGTGCCCATGTAATACATTTCAGTCAAACAGTCTACTATCTGTTTCATGGTGA.
Transfection of HeLa cells and screening of transfectants
Y-specific cDNA was transfected into HeLa cells by the DEAE–dextran-chloroquine method as described.20 The day before transfection, HeLa cells were seeded in 96-well, flat-bottom microtiter plates at 15 000 cells/well in 100 μL complete culture medium. Before transfection, medium was discarded and replaced by 45 μL DEAE–dextran/DNA mixture. This mixture was prepared for duplicate transfections in 96-well V-bottom microtiter plates by sequentially adding 98 μL RPMI-1640 supplemented with 10% decomplemented NuSerum IV (Collaborative Biomedical Products, Bedford, MA) containing 0.3 mg/mL DEAE-dextran (Sigma Chemical) and 100 μmol/L chloroquine, 1 μL TE containing 200 ng Y-specific cDNA in plasmid PCR3.1 and 1 μL (TE)10 mmol/L Tris, 1 mmol/L EDTA, pH 7.4, containing 200 ng plasmid pcDNAI/AMP-A1 (plasmid pcDNAI/AMP containing the HLA-A1 gene) or no HLA-A1 construct as a control. If the stably HLA-A1-transduced HeLa/HLA-A1 cells were used, no plasmid pcDNAI/AMP-A1 was added to the DEAE-dextran/DNA mixtures. The HeLa cells were incubated for 4 hours at 37°C, after which the DEAE-dextran/DNA was discarded and replaced by 80 μL phosphate-buffered saline containing 10% dimethyl sulfoxide. After 2 minutes at room temperature, phosphate-buffered saline–dimethyl sulfoxide was replaced by 200 μL complete culture medium. Transfected HeLa cells were incubated for 48 hours at 37°C. After removing the medium, 4000 HLA-A1 HY was added in 200 μL RPMI-1640 containing 10% pooled human serum and 300 IU IL-2 per milliliter. After 24 hours, the tumor necrosis factor (TNF) content in 50 μL supernatant was determined by adding it to 50 μL complete culture medium containing 50 000 WEHI-164 clone 13 cells, on which TNF has a cytolytic effect.21 After 18 hours, 10 μL cell proliferation reagent WST I (Boehringer Mannheim GmbH) was added, and the TNF concentration was calculated in comparison with recombinant TNF standards measured in the same assay.
Protein synthesis
The Y-specific genes were transcribed and translated using wheat germ extract and 3H-labeled leucine according to the manufacturer's procedure as is described in the TNT T7-coupled transcription/translation system (Promega, Madison, WI). The3H-labeled proteins were separated by 12% sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) for 2 hours. After fixation in a 10% methanol/10% glacial acetic acid solution for 30 minutes, amplification of radiation by the 3H-labeled proteins was achieved by using amplified fluorographic reagent (Amersham). Then the gel was dried for 2 hours at 65°C, and the proteins were visualized by autoradiography. The molecular weights of the proteins were determined by using kaleidoscope prestained protein standards (Bio-Rad Laboratories, Hercules, CA).
Peptide synthesis and peptide recognition assays
Peptides were synthesized by solid-phase fluorenylmethoxy-carbonyl (FMOC) chemistry and Wang resins on an AMS 422 multiple peptide synthesizer (Gilson Medical Electronics, Middletown, WI) and characterized by mass spectrometry. Standard 51Cr-release assays were used to determine lysis of target cells. EBV-LCL target cells were labeled with 100 μCi Na51CrO4. After 1 hour of incubation at 37°C, the cells were washed 3 times with RPMI supplemented with 2% fetal calf serum. Then 2000 Na51CrO4 labeled EBV-LCL cells/well were plated in a 96-well, V-bottom microtiter plate in 100 μL RPMI-1640 + 10% pooled human serum containing various concentrations of peptide. After 1 hour of incubation at 37°C, 20 000 CTLs were added in 100 μL RPMI-1640 + 10% pooled human serum. 51Cr-release was measured after incubation at 37°C for 4 hours.
Results
Cloning of Y-specific genes
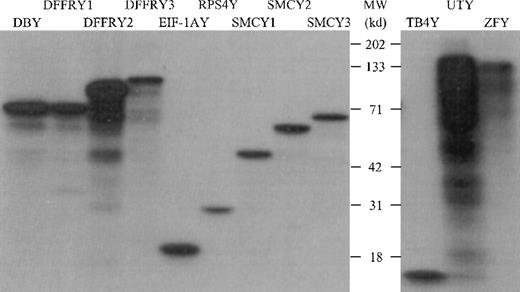
The PCR primers used, as shown in Table 1, were designed to amplify only the Y-specific gene and not the X-homologue. Restriction and sequence analysis of amplified Y-specific cDNA confirmed the Y specificity (data not shown). To determine whether the cloned Y cDNA was transcribed and translated appropriately, we performed an in vitro transcription/translation assay in which the Y-specific proteins were labeled with 3H and visualized on an autoradiogram after separation by SDS-PAGE, as shown in Figure1. All proteins showed the molecular weight (MWt) as deduced from their primary sequence, with the exception of ZFY. This protein had a MWt of 125 kd, whereas the sequence deduced that the MWt was 90 kd. However, DNA sequence analysis of both the 3′ and 5′ sites of ZFY revealed the correct ZFY DNA sequence, suggesting that this protein had undergone posttranslational modification.
In vitro transcription/translation of Y-specific cDNA.
The Y-specific genes were transcribed and translated using wheat germ extract and 3H-labeled leucine. The 3H-labeled proteins were separated on a 12% SDS-PAGE gel and were visualized by autoradiography.
In vitro transcription/translation of Y-specific cDNA.
The Y-specific genes were transcribed and translated using wheat germ extract and 3H-labeled leucine. The 3H-labeled proteins were separated on a 12% SDS-PAGE gel and were visualized by autoradiography.
Identification of the gene encoding for the HLA-A1-restricted H-Y epitope
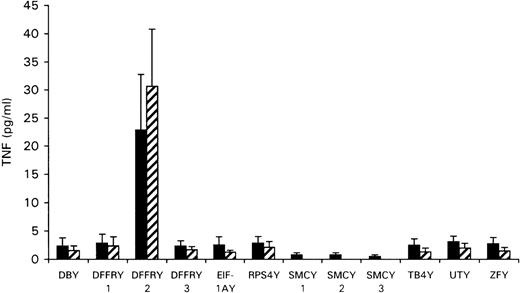
To determine whether any of the Y-specific genes encoded the HLA-A1-restricted H-Y epitope, the cloned Y-cDNA was transfected to the HeLa cells stably transfected with HLA-A1 (HeLa/HLA-A1) or cotransfected with HLA-A1 cDNA to HeLa cells. As shown in Figure2, transfection of DFFRY2 cDNA resulted in a significant amount of TNF production by HLA-A1 HY, whereas transfection of all other Y-genes induced no TNF release. To investigate whether this TNF release by HLA-A1 HY was HLA-A1 restricted, the DFFRY2 cDNA was transfected with and without HLA-A1 into HeLa cells. No significant TNF release by HLA-A1 HY took place when the DFFRY2 gene was transfected in the absence of the HLA-A1 molecule (TNF production 3.9 ± 1.5 pg/mL).
TNF production by HLA-A1 HY after stimulation with HeLa transfected with Y-specific cDNA.
HeLa cells were cotransfected with each Y-specific cDNA and HLA-A1 cDNA (black bars), or HLA-A1 stably transfected HeLa cells were transfected with each Y-specific cDNA only (hatched bars). Forty-eight hours after transfection, HLA-A1 HY was added. The culture supernatants were harvested 1 day later and were tested for TNF production.
TNF production by HLA-A1 HY after stimulation with HeLa transfected with Y-specific cDNA.
HeLa cells were cotransfected with each Y-specific cDNA and HLA-A1 cDNA (black bars), or HLA-A1 stably transfected HeLa cells were transfected with each Y-specific cDNA only (hatched bars). Forty-eight hours after transfection, HLA-A1 HY was added. The culture supernatants were harvested 1 day later and were tested for TNF production.
Localization of the HLA-A1 associated H-Y epitope in the DFFRY gene
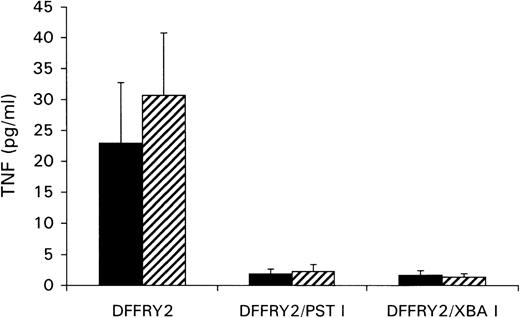
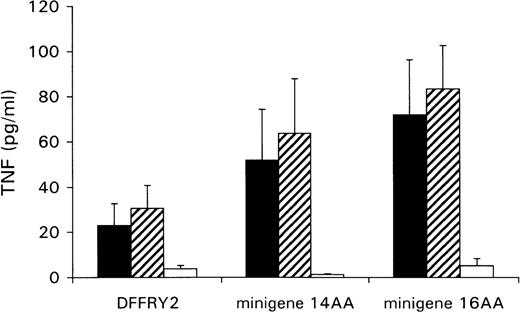
A genetic approach was used to localize the DFFRY sequence coding for the H-Y epitope. First, deletion mutants were obtained by digestion with restriction enzymes PstI and XbaI, resulting in a deletion at the 3′ site of the DFFRY2 gene of 444 bp (DFFRY2/PstI) and 1877 bp (DFFRY2/XbaI), respectively (Figure 3). As is demonstrated in Figure 4, TNF production by HLA-A1 HY was abolished when the CTL clone was stimulated with HeLa transfected with these DFFRY2 deletion mutants and HLA-A1. We concluded that the HLA-A1-associated H-Y antigenic peptide was encoded by the sequence of the DFFRY gene located between positions 4433 and 4828, 30 bp after the starting base of DFFRY3. To identify the H-Y epitope, the 4433 to 4828 region of the DFFRY gene was screened for peptides that would bind to HLA-A1 and at least would differ for 1 amino acid compared to the X-homologue. The DFFRY region 4561 to 4587 or 4590 codes for a HLA-A1-binding nonapetide and decapeptide, respectively. To determine whether this region indeed coded for the H-Y epitope, 2 pairs of oligo nucleotides were synthesized. Each pair of oligo nucleotides consisted of 2 complementary single-stranded DNA, which formed double-stranded DNA minigenes with a BamHI site and HindIII site after hybridization. These minigenes were cloned into the BamHI/HindIII-digested PCR3.1 expression vector. Minigene 14AA codes for the peptide MLKQIVDCLTEMYY (DFFRY position 4552-4590) and minigene 16AA for MKQIVDCLTEMYYMGT (DFFRY position 4555-4599) (Figure 3). Both minigenes were transfected into HeLa/A1 or cotransfected with HLA-A1 into HeLa. As is demonstrated in Figure 5, both minigenes induced TNF production by HLA-A1 HY, indicating that they encoded the H-Y epitope. The TNF production was abolished when no cotransfection of the HLA-A1 molecule into HeLa cells was performed.
Location of DFFRY deletion mutants and HLA-A1 HY epitope-encoding minigenes.
No recognition of DFFRY2 deletion mutants by HLA-A1 HY.
HeLa cells were cotransfected with DFFRY2 cDNA and HLA-A1 cDNA (black bars), or HLA-A1 stably transfected HeLa cells were transfected with DFFRY2 cDNA only (hatched bars). Forty-eight hours after transfection, HLA-A1 HY was added. The culture supernatants were harvested 1 day later and were tested for TNF production.
No recognition of DFFRY2 deletion mutants by HLA-A1 HY.
HeLa cells were cotransfected with DFFRY2 cDNA and HLA-A1 cDNA (black bars), or HLA-A1 stably transfected HeLa cells were transfected with DFFRY2 cDNA only (hatched bars). Forty-eight hours after transfection, HLA-A1 HY was added. The culture supernatants were harvested 1 day later and were tested for TNF production.
Minigenes encode epitope recognized by HLA-A1 HY.
HeLa cells were cotransfected with DFFRY2 or minigene cDNA and HLA-A1 cDNA (black bars), or HLA-A1 stably transfected HeLa cells were transfected with DFFRY2 cDNA only (hatched bars). As a control, HeLa cells were transfected with DFFRY2 or minigene cDNA only (white bars). Forty-eight hours after transfection, HLA-A1 HY was added. The culture supernatants were harvested 1 day later and were tested for TNF production.
Minigenes encode epitope recognized by HLA-A1 HY.
HeLa cells were cotransfected with DFFRY2 or minigene cDNA and HLA-A1 cDNA (black bars), or HLA-A1 stably transfected HeLa cells were transfected with DFFRY2 cDNA only (hatched bars). As a control, HeLa cells were transfected with DFFRY2 or minigene cDNA only (white bars). Forty-eight hours after transfection, HLA-A1 HY was added. The culture supernatants were harvested 1 day later and were tested for TNF production.
Identification of the HLA-A1 associated H-Y epitope
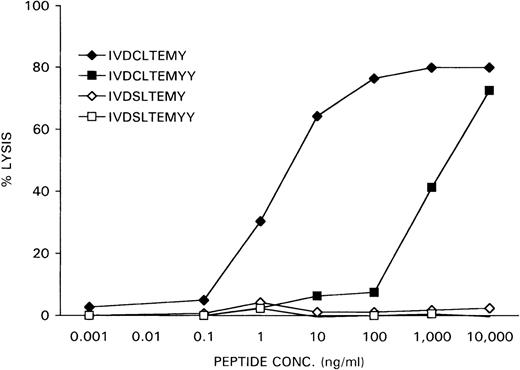
The nonapeptide IVDCLTEMY and the decapeptide IVDCLTEMYY, encoded by the minigenes, were synthesized and loaded at various concentrations on female HLA-A1-positive EBV-LCL cells, derived from the original patient who rejected her male bone marrow graft. The X-homologues IVDSLTEMY and IVDSLTEMYY were used as controls. Recognition of the peptide-loaded EBV-LCL cells by the CTL clone HLA-A1 HY was determined in a51Cr-release assay. As is shown in Figure6, the IVDCLTEMY-loaded EBV-LCL cells were efficiently killed by HLA-A1 HY with half-maximal target cell lysis at a peptide concentration of 1 ng/mL, whereas the IVDCLTEMYY-loaded EBV-LCL cells were only killed at high peptide concentrations. Neither X-homologue peptide was recognized by HLA-A1 HY.
Specific lysis of peptide-loaded female EBV-LCL target cells by HLA-A1 HY.
Female recipient HLA-A1–positive EBV-LCL cells were51Cr-labeled for 1 hour. After they were washed, the cells were incubated for 1 hour with DFFRY- and DFFRX-derived peptides at various concentrations. HLA-A1 HY was added at an effector-to-target ratio of 10, and 51Cr release was measured after 4 hours.
Specific lysis of peptide-loaded female EBV-LCL target cells by HLA-A1 HY.
Female recipient HLA-A1–positive EBV-LCL cells were51Cr-labeled for 1 hour. After they were washed, the cells were incubated for 1 hour with DFFRY- and DFFRX-derived peptides at various concentrations. HLA-A1 HY was added at an effector-to-target ratio of 10, and 51Cr release was measured after 4 hours.
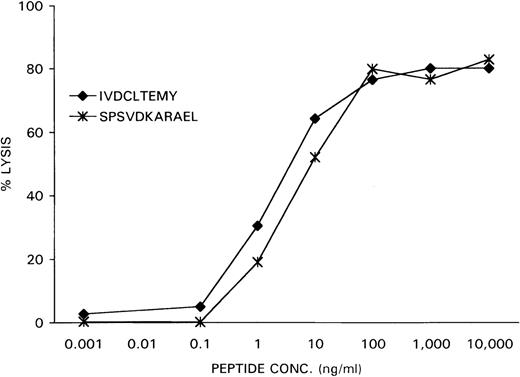
After 51Cr-labeling, the female recipient HLA-A1–positive EBV-LCL cells were pulsed with the DFFRY-derived peptide IVDCLTEMY and female HLA-B7–positive EBV-LCL cells with the SMCY-derived peptide SPSVDKARAEL at various concentrations.
HLA-A1 HY and HLA-B7 HY were added at an effector-to-target ratio of 10, and 51Cr release was measured after 4 hours.
After 51Cr-labeling, the female recipient HLA-A1–positive EBV-LCL cells were pulsed with the DFFRY-derived peptide IVDCLTEMY and female HLA-B7–positive EBV-LCL cells with the SMCY-derived peptide SPSVDKARAEL at various concentrations.
HLA-A1 HY and HLA-B7 HY were added at an effector-to-target ratio of 10, and 51Cr release was measured after 4 hours.
As a control, the previously identified epitope SPSVDKARAEL, derived from the SMCY protein and recognized by an HLA-B7 restricted H-Y specific CTL clone, was loaded at various concentrations on female HLA-B7-positive EBV-LCL cells. Recognition of the peptide-loaded EBV-LCL cells by the CTL clone HLA-B7 HY was determined in a51Cr-release assay. As is demonstrated in Figure 7, the CTL clone HLA-A1 HY recognized the HLA-A1-associated H-Y epitope IVDCLTEMY with the same dose-dependent efficiency as the CTL clone HLA-B7 HY recognized its specific target SPSVDKARAEL in the context of HLA-B7.
Discussion
Graft rejection and GVHD after HLA-identical stem cell transplantation are thought to result from the recognition of mH antigens by immunocompetent T lymphocytes from recipient or donor origin, respectively.1,4 The involvement of male-specific mH antigens in graft rejection was identified by the observation that female patients who underwent transplantation with HLA-phenotypically identical male bone marrow had a higher risk for graft rejection than those who underwent transplantation of bone marrow with HLA-phenotypically identical female origin.9 T-lymphocyte clones specifically recognizing male target cells could be generated during graft-rejection after sex-mismatched transplantation, and can be used as tools to identify H-Y antigens and the corresponding Y-specific genes.11 22
Previously, 2 human H-Y epitopes were identified that are involved in bone marrow graft rejection. The HLA-A2- and the HLA-B7-associated H-Y epitope recognized by an HLA-A2 and an HLA-B7 restricted CTL clone, respectively,12,13 were both encoded by the ubiquitously expressed SMCY gene, located in the same Yq deletion interval as the human H-Y antigen-controlling locus HY.23 24 In this article, we report the identification of a Y-chromosomal gene coding for a new male-specific transplantation antigen in the context of HLA-A1: DFFRY. The DFFRY-derived peptide was recognized by an HLA-A1-restricted CTL clone, which was also generated during graft rejection from a female patient with AML who rejected her HLA-phenotypically identical bone marrow derived from her father. The identification of this gene demonstrates that human H-Y antigen consists of at least 2 genes present on the human Y-chromosome.
DFFRY maps to proximal Yq11.2 within the H-Y antigen-controlling locus HY.15,25 The complete coding region of DFFRY shows 91% identity at the amino acid level to the X-homologue DFFRX.25 Both Y- and X-linked genes are similar to theDrosophila fat facets (faf) gene.25 Recently, it has been shown that faf is a member of a family of deubiquitinating genes, which play an important regulatory role at the level of protein turnover by preventing degradation of proteins by the proteasome through the removal of conjugated ubiquitin.26 The DFFRY and DFFRX gene contains the conserved Cys and His domains that are characteristic of ubiquitin-specific hydrolases. The high degree of conservation of these domains found in the genes argues strongly that they have a homologous biochemical function. Ubiquitous expression of DFFRX and DFFRY appears to support this view. However, it remains possible that the differences in amino acid sequence between DFFRY and DFFRX may lead to subtle differentiation of biologic function.
As was examined by RT-PCR with DFFRY specific primers, the human DFFRY mRNA is expressed in a wide range of tissues, whereas the mouse DFFRY gene is expressed specifically in the testis.25Consequently, the mouse DFFRY gene cannot encode male-specific transplantation antigens, whereas the human DFFRY gene may also code for male rejection antigens after transplantation of nonhematopoietic cells or organs. Previously, it was demonstrated that transplantation of a human male donor heart into a female recipient significantly increases the number of graft rejections as compared to sex-matched transplantation.27 Similarly, transplantation of a liver from a male donor to a female recipient increases the occurrence of chronic graft rejection compared to liver transplantation of a female donor.28 These reports indicate that H-Y antigens may be involved in organ allograft rejections as well.
The DFFRY gene codes for a very large protein (> 2500 residues) that differs from the female homologue by more than 230 residues, relatively uniformly distributed throughout its length. There is sufficient polymorphism in the DFFRY sequence to allow the generation of different peptides that are male specific and are capable of binding to different MHC molecules. Hence, DFFRY has at least the same potential to generate a large number of distinct DFFRY-specific H-Y epitopes as SMCY, which differs from the X-homologue by more than 200 residues. Identification of other H-Y epitopes may elicit the role of DFFRY as H-Y epitope encoding gene.
The 9 amino acid HLA-A1-associated H-Y epitope derived from the DFFRY gene contains 1 polymorphism compared to the X-homologue peptide—a cysteine residue at position 4 instead of a serine residue. Recently, an HLA-A2-associated H-Y antigen derived from the SMCY gene was identified that contained a cysteine residue that had undergone a posttranslational modification that significantly affected T-cell recognition.13 This described modification involves attachment of a second cysteine residue to the cysteine in the primary sequence through a disulfide bond. Half-maximal target cell lysis of T2 cells by the specific CTL clone was achieved by using a 100 pmol/L concentration of the cysteinylated peptide, whereas a concentration of 180 nmol/L of the unmodified peptide was required to achieve the same result. Half-maximal target cell lysis of EBV-LCL cells by the DFFRY-specific CTL clone was achieved by using a DFFRY peptide concentration of approximately 1 nmol/L. Although this peptide concentration was comparable to the concentration required for reconstitution of the HLA-B7-associated H-Y epitope derived from the SMCY gene, we cannot exclude that cysteinylation of the DFFRY epitope can take place during the 51Cr-release assay. We used normal RPMI-1640 medium containing 0.21 mmol/L cystine; in this peptide, cysteinylation by disulfide exchange with cysteine can occur rapidly.
Characterization of H-Y antigens involved in graft rejection may be useful in selection of donor-recipient combinations. As was shown previously, a strong recipient antidonor cellular cytotoxic reactivity directed against the DFFRY-derived H-Y antigen in the female patient with AML could be demonstrated before transplantation.1 The recipient rejected her bone marrow 60 days after BMT, even after highly immunosuppressive conditioning regimens. The frequency of H-Y-specific CD8+ T cells in the female recipient can be determined using HLA-peptide tetrameric complexes.29 30 Determination of the frequency of H-Y-specific CD8+ T cells in female recipients by HLA/H-Y peptide tetramerics may improve donor selection and allow the determination of bone marrow transplantation recipients at high risk for H-Y-induced graft rejection.
Supported by the Dutch Cancer Society (grant RUL 94-803) and the J. A. Cohen Institute for Radiopathology and Radiation Protection.
Reprints:J. H. Frederik Falkenburg, Leiden University Medical Center, Department of Hematology, C2-R-140, P.O. Box 9600, 2300 RC Leiden, The Netherlands; e-mail: falkenburg@hematology.azl.nl.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal