Two anhaptoglobinemic patients showing anaphylactic transfusion reactions by antihaptoglobin antibody were found. Southern blot analysis indicated that 2 patients were homozygous for the deleted allele of the haptoglobin gene (Hpdel) as reported previously. We have identified the junction region of the deletion from genomic DNA of 1 patient using cassette-mediated polymerase chain reaction (PCR). Then, the deleted region from the 5′ breakpoint to the promoter region of the Hpwas amplified from genomic DNA of a control individual using PCR. DNA sequence analysis of these regions indicated that the 5′ breakpoint of the Hpdel allele was located 5.2 kilobase (kb) upstream of exon 1 of the Hp and the 3′ breakpoint was positioned between 52 and 53 base pair (bp) upstream of exon 5 of the haptoglobin-related gene. There was no significant homology between the DNA sequences flanking the 5′ and 3′ breakpoints, except for a 2-bp (TG) identity. To examine the gene frequency, we have developed a simple PCR method to detect the gene deletion. We found 8, 16, and 17 Hpdelalleles in 157 Koreans, 523 Japanese, and in 284 Chinese, respectively, but did not find the Hpdel in 101 Africans or in 100 European-Africans. The incidence of individuals homozygous for the Hpdel allele was therefore expected to be 1/4000 in Japanese, 1/1500 in Koreans, and 1/1000 in Chinese. This incidence is higher than that of IgA deficiency in Japanese. More attention should be paid on haptoglobin deficiency and antihaptoglobin antibody as the cause of transfusion-related anaphylactic reactions in Asian populations.
Haptoglobin (Hp) is a hemoglobin-binding polymorphic plasma glycoprotein present in all vertebrates.1 Heritable normal variations in humans were first reported by Smithies and Walker,2 and there are 3 common genetic haptoglobin phenotypes, Hp1, Hp2, and Hp2-1, which are determined by a pair of codominant alleles—Hp1 andHp.2 Although the physiologic function of the protein is not completely understood, it may limit iron loss during normal erythrocyte turnover and during hemolysis. While certain pathologic states such as severe hemolysis and liver dysfunction are known to lead to secondary anhaptoglobinemia,3 previous reports have suggested that some individuals with hypohaptoglobinemia or anhaptoglobinemia (Hp0 phenotype) have a genetic origin.4-7 A silent allele Hp0at the haptoglobin locus (Hp) has been postulated by studies on the anomalous inheritance of haptoglobin phenotypes, and the homozygosity of Hp0 has been suggested as a cause of anhaptoglobinemia.8,9 However, the genetic mechanisms of Hp0 allele(s) have not been understood for a long time. Recently, we have identified 1 ofHp0 allele(s) that was an allelic deletion in the haptoglobin gene cluster (Hpdel).10 The gene deletion was larger than 20 kilobase (kb) and extended from upstream of the promoter region of the Hp to the 3′ region of intron 4 of the downstream haptoglobin-related gene (Hpr), which shares a high degree of nucleotide sequence homology with theHp1.11-13
We have encountered 2 anhaptoglobinemic patients with anti-Hp antibody, who suffered from severe anaphylactic shock after infusion of blood products.14,15 Briefly, the first was a 33-year-old woman (NK) in the 31st week of her first pregnancy. She was hospitalized for threatened premature delivery due to chronic polyhydroamnios. When she developed edema in her extremities with increased amniotic fluid and decreased serum albumin, 25% albumin solution was infused. After receiving several drops, she exhibited severe anaphylactic reactions, and immediate intravenous administration of corticosteroids improved her symptoms. The second case was a 94-year-old woman (SN) with myelodysplastic syndrome. She was transfused red blood cells 3 times and platelet-concentrate once during a 7-month period without any symptoms. However, when she was transfused platelet-concentrate 1 month later, she suffered from anaphylactic reactions, which responded to corticosteroids. She had borne 2 children without any trouble when she was young. Both these patients showed anti-Hp antibody and no detectable Hp in their sera.14 15
In the current study, we have determined that these 2 patients were homozygous for the Hpdel allele by Southern blot analysis. The junction region of theHpdel allele was amplified by cassette-mediated polymerase chain reaction (PCR) and sequenced. We then developed a simple method to detect this deletion by PCR amplification for screening anhaptoglobinemic individuals to examine the gene frequency in various populations.
Materials and methods
DNA preparations from patients and healthy volunteers
B lymphocytes from 2 patients were transformed by Epstein-Barr (EB) virus, and genomic DNA was extracted from EB virus-transformed lymphocytes using an organic solvent method. In addition, genomic DNA from peripheral leukocytes of family members of the patients, and randomly selected Japanese, Koreans, Chinese, Africans (Xhosa), and European-Africans of South Africa was isolated as described previously.16-20
Southern blot analysis
The Hpdel allele was detected by Southern blot analysis as described previously.10 Briefly, genomic DNA (7 μg) was separated in a 0.8% agarose gel after digestion by an appropriate restriction endonuclease, transferred onto a nylon membrane and hybridized with the digoxigenin-labeled cDNA probe coding for the α chain or β chain of haptoglobin (a kind gift from Dr R. Lippen, Université Libre de Bruxelles, Service de Génétique Appliquée).21 The digoxigenin-labeled DNA probe was prepared using a DIG DNA labeling kit (Boehringer Mannheim, Mannheim, Germany).
PCR amplification of the junction region of Hpdelallele
A Marathon cDNA amplification kit (Clontech Japan, Tokyo, Japan) was used for isolation of the junction region of theHpdel. Genomic DNA (5 μg) from EB virus-transformed lymphocytes of the patient (NK) homozygous for the Hpdel allele was digested with 1 of several endonucleases (50 U) (DraI, EcoRV,PvuII, RsaI, and HaeIII) and ligated with a Marathon cDNA adaptor (Clontech). Then, the junction region of the deletion was amplified using cassette-mediated PCR. Because there are AP1 and AP2 sequences within the adaptor, we used the gene-specific primers (Hpr-first and Hpr-nest) as downstream primers and the AP1 and AP2 primers (Clontech) as upstream primers. The Hpr-first primer 5′-GGG CTT CCC ACA TAC TGT CAA GGA G-3′, within intron 4 and exon 5 of the Hpr, and the AP1 primer were used for the first PCR, and then nested PCR was performed using the Hpr-nest primer 5′-CCA CAT ACT GTC AAG GAG AGC AAG A-3′, and the AP2 primer. The temperature profile of PCR for amplification of the junction region of theHpdel was at 94°C for 1 minute, followed by 25 cycles of denaturing at 98°C for 10 seconds, and annealing and extension at 68°C for 4 minutes. PCR amplifications were performed using 2.5 units of LA Taq polymerase (Takara, Shiga, Japan) in a 25 μL LA Taq buffer containing 5 pmol of each primer, 2.5 mmol/L MgCl2 and 400 μmol/L dNTP.
PCR amplification of the deleted region
The region between an upstream region of the 5′ breakpoint of the deletion of the Hpdel and the promoter region of the haptoglobin gene was amplified from genomic DNA of a control individual using primers Hp-del-U (5′-CTT TAT GGC ACT GGG GAA CAA GCA TTT TG-3′) (Figure 2) and Hp-promoter-L (5′-CCC ATC AAC AGG AGG AAG GGT CAA TAC TGC-3′, within a promoter region of the Hp).13 The terms “U” and “L” indicate upper and lower primers, respectively. The temperature profile was at 94°C for 1 minute, followed by 30 cycles of denaturing at 98°C for 10 seconds, annealing at 60°C for 30 seconds and extension at 72°C for 5 minutes. PCR amplification was performed using 2.5 units of LA Taq polymerase (Takara) in a 25 μL LATaq buffer containing 5 pmol of each primer, 2.5 mmol/L MgCl2 and 400 μmol/L dNTP.
PCR amplification of the junction region of the Hpdelallele and exon 1 of theHp
To amplify the Hpdel allele, PCR was performed in 25 μL Ex Taq buffer containing 5 pmol of primers Hp-del-U and H-del-L (5′-CAG GAA GAG ATT TTT AGC CGT GGT CAG CAG-3′, within exon 5 of the Hpr), 1 unit of ExTaq DNA polymerase and 200 μmol/L dNTP. The temperature profile was at 94°C for 1 minute, followed by 35 cycles of denaturing at 98°C for 10 seconds, annealing at 60°C for 30 seconds and extension at 72°C for 1 minute. As an amplification control, the exon 1 of the haptoglobin gene was coamplified with 5 pmol of primers Hp-Ex1-U (683 ∼ 707 bp from the ATG initiation codon of the Hp) and Hp-Ex1-L (1135 ∼ 1159 bp) as described previously.10
DNA sequencing
PCR products purified by a suprec-02 centrifugation tube (Takara) were directly sequenced in both directions using each PCR primer or several internal sequences (not shown) as a sequence primer using a Bigdye Terminator Cycle Sequencing Reaction Kit and analyzed using an ABI PRISM 310 genetic analyzer (Perkin Elmer Japan ABI).
Results
Characterization of the haptoglobin gene cluster of patients with anti-Hp antibody
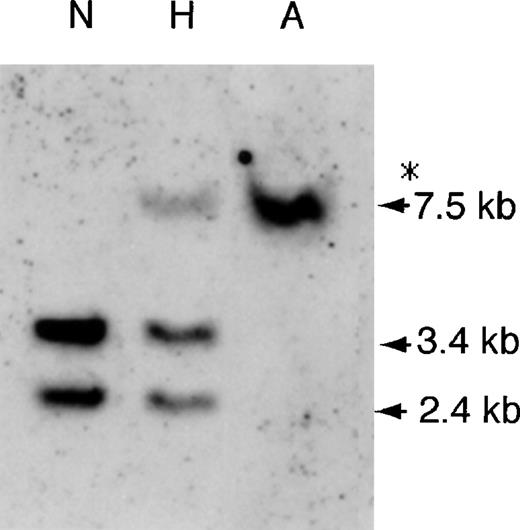
The haptoglobin gene cluster of 2 patients with anti-Hp antibody was analyzed by Southern blot analysis with cDNA probes coding for α or β chain of Hp. Figure 1shows typical hybridization results. When genomic DNA was digested bySacI, a 3.4-kb DNA fragment corresponding to the Hp and a 2.4-kb signal corresponding to the Hpr were identified by anHp β cDNA probe in a control individual (lane N). In addition to these 2 bands, an unexpected band of 7.5 kb was found in a hypohaptoglobinemic individual who was previously shown to be heterozygous for the Hpdel allele (lane H). Only the 7.5-kb band was found in 2 anhaptoglobinemic patients who had anti-Hp antibody (lane A). When genomic DNAs of the 2 patients were digested by XbaI, and by HindIII, only unexpected band of 6.5 kb and of 4.4 kb, corresponding to theHpdel, was detected by the Hp β cDNA probe, respectively, and no signal was detected by the Hp αcDNA probe (not shown). In addition, we failed to amplify exons 1 ∼ 4 of the Hp using PCR from genomic DNA of the patients. These results indicated that the 2 patients with anti-Hp antibody were homozygous for the Hpdel allele, which is the sole null allele of the Hp reported to date.10
Southern blot analysis of genomic DNA using theHp β cDNA probe.
Genomic DNAs (7 μg) from a patient (NK) with anhaptoglobinemia (lane A), from an individual with hypohaptoglobinemia (lane H) and from a control individual (lane N) were digested by SacI. The band indicated by an asterisk was unexpected. The sizes (kb) of the hybridized fragments are indicated.
Southern blot analysis of genomic DNA using theHp β cDNA probe.
Genomic DNAs (7 μg) from a patient (NK) with anhaptoglobinemia (lane A), from an individual with hypohaptoglobinemia (lane H) and from a control individual (lane N) were digested by SacI. The band indicated by an asterisk was unexpected. The sizes (kb) of the hybridized fragments are indicated.
Identification of the junction region of the Hpdel
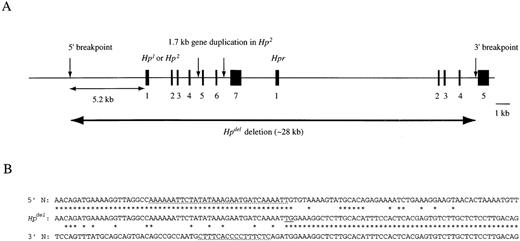
Previous mapping results suggested that the 3′ end of the gene deletion occurred within a 284-bp stretch between the AatI site within intron 4 and the BamHI site in exon 5 of theHpr. To identify the junction region of theHpdel allele, cassette-mediated PCR was performed using several endonuclease-digested libraries constructed from genomic DNA of the EB virus-transformed lymphocytes of 1 patient (NK). The longest PCR product (2.5 kb) was obtained from DraI library. DNA sequence analysis of the 2.5-kb PCR product indicated that the sequence of 2380 bp of 5′ region of the product has not been identified previously, and that 60 bp of 3′ region of the product was identical to the intron 4 and exon 5 of theHpr.13 The DNA sequence between 781-bp upstream of the 5′ breakpoint and 228-bp downstream of the 3′ breakpoint of the Hpdel allele is shown in Figure 2. BLAST searches (http://www.ncbi.nlm.nih.gov/BLAST/) indicated that the 5′ flanking region of the 5′ breakpoint contained sequences similar (more than 90% homologous) to the 5′ flanking sequence of the New World monkey Hp gene.
DNA sequence of the junction region of theHpdel allele.
The positions of PCR primers are underlined. An arrow indicates the junction. The LTR-like sequences are italicized.
DNA sequence of the junction region of theHpdel allele.
The positions of PCR primers are underlined. An arrow indicates the junction. The LTR-like sequences are italicized.
PCR amplification and sequencing of the nucleotides between the 5′ breakpoint of the Hpdeland the promoter region of the Hp
Because the DNA sequence of an upstream region of the 5′ breakpoint of the Hpdel was highly homologous with the 5′ flanking region of the Hp gene of black-handed spider monkey (Ateles geoffroyi), we tried to amplify a region between the 5′ breakpoint of the deletion and the promoter region of the Hp from genomic DNA of a control individual. A 4.5-kb fragment was amplified by the Hp-del-U and Hp-promoter-L primers, and DNA sequence analysis indicated that this fragment was the 5′ flanking region of the Hp gene. The 5′ breakpoint of the deletion located at about 5170-bp upstream of the 5′ end of exon 1 of Hp, and the 3′ breakpoint was positioned between 52- and 53-bp upstream of exon 5 of the Hpr (Figure 3A). From these results, the size of the deletion of theHpdel allele is estimated to be 28 kb. The DNA sequences flanking the 5′ and 3′ breakpoints showed no significant DNA sequence homology in the junction region of the deletion, except for 2 bases (TG) (Figure 3B). Computer analysis of the 6540-bp sequence obtained in this study by the Repeat Masker program through the Washington University Human Genome Center (http://ftp.genome.washington.edu/cig-bin/) identified that about 70% of this sequence were repetitive.
Deletion breakpoint junction.
(A), The physical map of the breakpoints of Hpdel. The exons of Hp and Hpr are indicated by black boxes. The gene deletion in the Hpdel allele is indicated by a double-headed horizontal arrow. (B), The alignment of the junction region (Hpdel) with a region containing the 5′ breakpoint determined in this study (5′ N) and a region containing the 3′ breakpoint (3′ N) (DDBJ/EMBL/GenBank accession number M69197). Identical sequence is indicated by *. A polypyrimidine tract and an A-T rich sequence are underlined.
Deletion breakpoint junction.
(A), The physical map of the breakpoints of Hpdel. The exons of Hp and Hpr are indicated by black boxes. The gene deletion in the Hpdel allele is indicated by a double-headed horizontal arrow. (B), The alignment of the junction region (Hpdel) with a region containing the 5′ breakpoint determined in this study (5′ N) and a region containing the 3′ breakpoint (3′ N) (DDBJ/EMBL/GenBank accession number M69197). Identical sequence is indicated by *. A polypyrimidine tract and an A-T rich sequence are underlined.
Screening and identification of the Hpdelallele using PCR
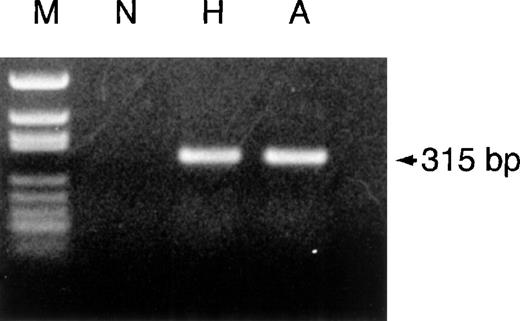
For screening for the Hpdel, we amplified the Hpdel allele using conventional PCR with primers (Hp-del-U and Hp-del-L), encompassing the junction of the Hpdel (Figure 2). As shown in Figure 4, a 315-bp DNA fragment was amplified from genomic DNA of individuals homozygous or heterozygous for the Hpdel, whereas no DNA fragment was amplified from genomic DNA of a control individual. Direct DNA sequencing of this 315-bp PCR product from the 2 patients indicated the specific amplification of the junction region of the breakpoints. Homozygosity for the Hpdel allele was confirmed by the absence of amplified product of exon 1 of theHp. These PCR results were consistent with those from Southern blot analysis in 3 individuals homozygous and 10 individuals heterozygous for the Hpdel allele.
PCR amplification of the junction region of theHpdel from a control individual (N), hypohaptoglobinemia (H) and anhaptoglobinemia (A).
PCR products underwent electrophoresis in a 1.2% agarose gel and were stained by ethidium bromide. The size of the PCR product is indicated.MspI-digested pBluescript (M) was used as a molecular size marker.
PCR amplification of the junction region of theHpdel from a control individual (N), hypohaptoglobinemia (H) and anhaptoglobinemia (A).
PCR products underwent electrophoresis in a 1.2% agarose gel and were stained by ethidium bromide. The size of the PCR product is indicated.MspI-digested pBluescript (M) was used as a molecular size marker.
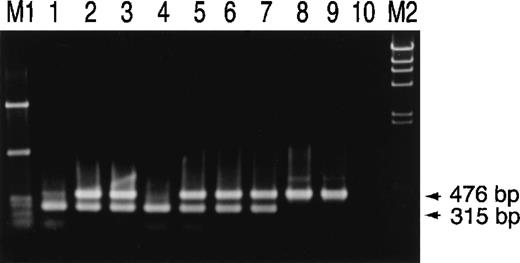
To amplify the Hpdel and the Hp(exon 1) in the same reaction tube using Hp-del-U and Hp-del-L primers, and Hp-Ex1-U and Hp-Ex1-L primers, PCR was performed from individuals with anhaptoglobinemia and hypohaptoglobinemia and from a control individual. A 315-bp band was amplified from an individual homozygous for the Hpdel, whereas only a 476-bp band was amplified from a control individual. Both 315-bp and 476-bp bands were amplified from individuals heterozygous for theHpdel allele. We then examined theHpdel in the members of the 2 families of the patients (SN and NK). Consistent with the results from Southern blot analysis, the 2 patients were homozygous for theHpdel allele (Figure5, lanes 1 and 4). Two children of SN were heterozygous for the Hpdel. The parents and a brother of NK were heterozygous for theHpdel.
Family study of the Hpdel allele in 2 families of anhaptoglobinemic patients with anti-haptoglobin antibody.
Simultaneous PCR amplification of the junction region of theHpdel allele and of exon 1 of theHp was carried out. PCR products underwent electrophoresis in a 1.2% agarose gel and were stained by ethidium bromide. Lanes 1, 2, and 3 are SN and 2 children of SN. Lanes 4, 5, 6, and 7 are NK, father, mother, and brother of NK. Lanes 8 and 9 are control individuals. Lane 10 is negative control. The size of the PCR product is indicated.TaqI-digested φ × 174 (M1) andHindIII-digested λ phage (M2) were used as molecular size markers.
Family study of the Hpdel allele in 2 families of anhaptoglobinemic patients with anti-haptoglobin antibody.
Simultaneous PCR amplification of the junction region of theHpdel allele and of exon 1 of theHp was carried out. PCR products underwent electrophoresis in a 1.2% agarose gel and were stained by ethidium bromide. Lanes 1, 2, and 3 are SN and 2 children of SN. Lanes 4, 5, 6, and 7 are NK, father, mother, and brother of NK. Lanes 8 and 9 are control individuals. Lane 10 is negative control. The size of the PCR product is indicated.TaqI-digested φ × 174 (M1) andHindIII-digested λ phage (M2) were used as molecular size markers.
To examine the gene frequency of the Hpdelallele, PCR amplification was performed from genomic DNAs of 523 randomly selected Japanese in 4 different areas of Japan (Akita, northeast part of Japan; Tokyo, central part of Japan; Fukuoka, west part of Japan; Okinawa, southwest island of Japan), 157 Koreans (Seoul), 284 Chinese (Shenyang, northeast part of China; Guangzhou, south part of China), 101 Africans (Xhosa) (Cape Town, South Africa), and 100 European-Africans (Cape Town). Sixteen individuals in 523 Japanese, 8 in Koreans, and 17 individuals in 284 Chinese were found to be heterozygous for the Hpdel allele (Table1). Because there was no significant difference between gene frequencies of theHpdel in 4 subpopulations of Japan and between those in 2 subpopulations of China, the allele frequency of theHpdel in Japanese, in Koreans, and in Chinese, was calculated to be 0.015, 0.025, and 0.030, respectively. On the other hand, we did not detect the Hpdelallele in any of the 101 randomly selected Africans (Xhosa) or in any of the 100 European-Africans. However, these findings cannot exclude the absence of the Hpdel in Africans and in European-Africans, because of the small number of samples examined.
Allele frequencies of theHpdel in various populations
| Populations . | Number of Individuals . | Number of Allele . | Allele Frequency . |
|---|---|---|---|
| Japanese (total) | 523 | 16 | 0.015 |
| Akita | 120 | 5 | 0.021 |
| Tokyo | 126 | 4 | 0.016 |
| Fukuoka | 140 | 3 | 0.011 |
| Okinawa | 137 | 4 | 0.015 |
| Korean | 157 | 8 | 0.025 |
| Chinese (total) | 284 | 17 | 0.030 |
| Shenyang | 132 | 10 | 0.038 |
| Guangzhou | 152 | 7 | 0.023 |
| African | 101 | 0 | 0 |
| European-African | 100 | 0 | 0 |
| Populations . | Number of Individuals . | Number of Allele . | Allele Frequency . |
|---|---|---|---|
| Japanese (total) | 523 | 16 | 0.015 |
| Akita | 120 | 5 | 0.021 |
| Tokyo | 126 | 4 | 0.016 |
| Fukuoka | 140 | 3 | 0.011 |
| Okinawa | 137 | 4 | 0.015 |
| Korean | 157 | 8 | 0.025 |
| Chinese (total) | 284 | 17 | 0.030 |
| Shenyang | 132 | 10 | 0.038 |
| Guangzhou | 152 | 7 | 0.023 |
| African | 101 | 0 | 0 |
| European-African | 100 | 0 | 0 |
Discussion
Southern blot analysis in this study indicated that the 2 patients with anti-Hp antibody were homozygous for theHpdel allele previously identified.10 The results indicated that the large haptoglobin gene deletion was the causal mutation for the anhaptoglobinemia, resulting in the production of antibody after blood transfusions. We have determined that the 5′ breakpoint of the gene deletion was near an LTR-like sequence that was located about 5.2-kb upstream of exon 1 of the Hp. From these results, the size of deletion was calculated to be 28 kb. In this study, we have determined the nucleotide sequence of the 5′ flanking region of the Hp (from −7551 to −1011 bp). About 70% of this sequence consisted of repeat sequences that contained 5 Alu, 2 L1, and 2 LTR-like sequences (data not shown). The remaining sequences were homologous to the 5′ flanking region of the Hp gene of the New World monkey. The alignment between the DNA sequences containing the 5′ and the 3′ breakpoints revealed short direct repeats of 2 bp (TG) (Figure 3B), indicating that the deletion occurred by nonhomologous recombination. Nonhomologous recombination has been characterized by the presence of short repeats (2 ∼ 6 bp),22 whereas homologous recombination needs larger homologous sequence (more than 14 bp).23 The sequence features such as A-T rich elements, alternating purine/pyrimidine tracts, polypurine-pyrimidine tracts, palindolomic sequences, and topoisomerase I and II cleavage sites are often found at or near the junction surrounding nonhomologous recombination breakpoints.24 Among these motifs, an A-T rich element was found in 5′ flanking region of the the 5′ breakpoint, and a polypyrimidine tract was detected in the 5′ flanking sequence of the 3′ breakpoint (Figure 3B). Thus, these sequences might play some role in this large haptoglobin gene deletion.
In this study, we have developed a method to identify theHpdel allele using PCR. This method can detect the Hpdel in all 13 individuals who were determined to be heterozygous or homozygous for theHpdel using Southern blot analysis. We then examined the gene frequency of the deletion in Japanese, Koreans, Chinese, Africans, and European-Africans. We estimated that the gene frequency of the Hpdel was 0.015 in Japanese, 0.025 in Koreans, and 0.030 in Chinese. Accordingly, we estimated that individuals with true anhaptoglobinemia, who are homozygous for the Hpdel, are 1 in every 4000 Japanese, 1 in every 1500 Koreans, and 1 in every 1000 Chinese. Recently, Yoshioka et al25 found 1 individual with true anhaptoglobinemia out of 9711 unrelated Japanese. Therefore, it is likely that the Hpdel allele is the most frequent Hp0 allele in Japanese. Our results also suggest that about 0.025% Japanese, 0.067% Koreans, and 0.1% Chinese are at risk to produce an antibody against haptoglobin after repeated blood transfusions. The first description of congenital absence of haptoglobin reported over 30 years ago in individuals of African decent.4 However, we did not find theHpdel in any of the 101 Africans or the 100 European-Africans. These results were consistent with previous reports that no detectable change was found in the haptoglobin gene cluster of African individuals with anhaptoglobinemia.26,27 Although we cannot rule out the possibility that the frequency of theHpdel allele in these populations was much lower than that in Asian populations, a recent study reported that the frequency of true congenital anhaptoglobinemia was at approximately 1 in every 1000 Europeans, and that anhaptoglobinemia was more frequent in Africans.7 Therefore, the genetic mechanisms for anhaptoglobinemia might be different between East Asian populations and other ethnic groups. It would be of interest to know whether Africans with congenital anhaptoglobinemia have ever been found to have antibodies against Hp or to have experienced transfusion reactions.
Recently, it has been reported that haptoglobin gene-knockout mice showed a higher susceptibility during acute hemolysis compared with control mice, although the clearances of free plasma hemoglobin in these mice and in control mice were not significantly different.28 These mice also had a small reduction in postnatal viability. These results suggest that individuals homozygous for the Hpdel allele may be at a disadvantage during acute hemolysis.
Undesired reactions caused by blood transfusion have focused on the hemolytic immune reactions with red blood cells. However, less attention has been paid to nonhemolytic reactions after blood transfusion or infusion of human blood products. Anaphylactic reactions to blood and blood components in patients with IgA deficiency, or with fourth component of complement (C4) deficiency, have been reported to be associated with antibodies to IgA, and to C4, respectively.29-32 IgA anaphylactic reactions have occurred in persons who were IgA-deficient and in whom class-specific anti-IgA was detected in the serum. Determination of serum concentrations of Hp could be used for initial screening of patients at risk for anti-Hp transfusion reaction. However, the measurement of plasma Hp concentration is not sufficient for determination of true anhaptoglobinemia, since plasma Hp concentration is known to decrease to below detectable levels in certain pathologic states such as hemolysis and liver dysfunction.3 In addition, certainly the incidence of anhaptoglobinemia in many populations is so low that it would not be cost-effective to do such screening in the interest of preventing what would be a very rare incidence of transfusion reactions.
The incidence of IgA deficiency in Japanese was reported to be about 1/30 000, which is lower than those in Europeans (1/2 500).33,34 However, the incidence of anhaptoglobinemia appears to be much higher than that of IgA deficiency in Asian populations. More attention should be paid on haptoglobin deficiency and anti-haptoglobin antibody as the cause of transfusion-related anaphylactic reactions in East Asians. Accordingly, our simple PCR method to detect the Hpdelis recommended as a reliable method to prevent anaphylactic transfusion reactions in individuals having a risk to produce anti-Hp antibody by blood transfusions. If patients who are shown to be homozygous for the haptoglobin deletion need to be transfused with red cells, it would be necessary to meticulously wash the red cells before transfusion to avoid an immunologic transfusion reaction.30
Acknowledgments
We thank all members of the 2 families for consenting to the study and giving blood samples. We also thank Dr Ernette D. du Toit (Department of Immunology, Medical School, Cape Town, South Africa) and Dr Doo-Sung Kim (Korean Red Cross Central Blood Center, Seoul, Korea) for providing blood samples. We thank Osaka Red Cross Blood Center for collecting blood samples of members of NK family.
Supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Sports, and Culture of Japan, and a grant from the Uehara Memorial Foundation.
Nucleotide sequence data reported in this article have been deposited in the DDBJ/EMBL/GenBank nucleotide sequence databases with the accession numbers AB025320 (the junction region of the Hpdel) and AB025321 (the 5′ flanking region of the Hp). The accession number for the 5′ flanking region of the Hp gene of New World monkey (Ateles geoffroyi) is U04852.
Reprints:Hiroshi Kimura, Department of Forensic Medicine, Kurume University School of Medicine, Kurume 830-0011, Japan; e-mail: hkimura@med.kurume-u.ac.jp.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal