A substantial body of published data suggests activation of lineage-specific genes in multipotential hemopoietic cells before their unilineage commitment. Because the behavior and plasticity of cells isolated in vitro away from microenvironmental constraints exercised in vivo may be altered, one wonders whether similar findings can be observed in a physiologic setting in vivo. We used a transgenic mouse model harboring human micro LCR together with β promoter sequences as a transgene to examine activation of lineage-specific programs in vivo. By using LacZ as a reporter, we had the ability to detect, quantitate, and select live cells with different levels of LacZ activation. We found strong expression of LacZ by X-gal staining in 2 lineages—erythroid and megakaryocytic. Activation in the latter was a novel finding not previously observed when similar transgenes were used. We also found activation of μLCR-βpro at low levels in progenitor cells of granulocytic-macrophagic, erythroid, or megakaryocytic lineage detected by in vitro assays, suggesting activation before commitment to a specific lineage pathway. In particular, the expression of LacZ was graded among progenitors, so that in a proportion of them activation occurred only after commitment to erythroid or megakaryocytic lineage. In addition, we found quantitative reduction in LacZ expression between fetal liver and bone marrow-derived cells, the basis of which is unclear. Collectively our data provide in vivo evidence supporting the view that lineage-specific genes are expressed in a graded fashion in pluripotential cells before their irreversible unilineage commitment.
Functional hemopoietic cells of different lineages are constantly replenished by a pool of lineage-committed progenitor cells, which in turn are generated on demand by multipotent stem cells. The ability of the latter cells to become differentiated, together with their ability to self-renew, constitutes the unique fundamental function of stem cells.1 Understanding the molecular processes that underlie both the self-renewal process and the differentiation decisions in the hemopoietic system is not only of biologic significance, it has implications in transplantation, gene therapy, and disease pathogenesis (ie, leukemias). Therefore, parameters that influence the decision-making process by stem cells are under intense investigation. Based on numerous cellular or molecular studies conducted thus far, it has been suggested that commitment of adult hemopoietic stem cells to a specific lineage is an intrinsic, stochastic process,2-10 though the possibility of external influence at some stem cell–progeny level is still actively debated.11 Early data generated by Ogawa et al4on the fates of 2 daughter cells separated in vitro have suggested that lineage selection is accomplished not by acquisition of new markers but by sequential restriction of potentialities of multipotent cells in a stochastic fashion. Other variations of this theory suggest that the restriction of multipotentiality is accomplished not stochastically but in a predetermined, hierarchical fashion.12 These views have gained new life by recent molecular analyses of early multipotential cells.5-9 The goal of these studies was to attempt to analyze the molecular expression profiles of multipotential or uncommitted cells to provide a glimpse at the molecular basis of lineage commitment. Initial studies were conducted in cell lines thought to represent multipotential cells, especially the ones capable of responding to physiologic stimuli with downstream differentiation to more than 1 lineage.13-15 Various genes with distinctive lineage affiliations were found to be present in these cells at the chromatin level and at the transcriptional level of these genes. Furthermore, it was found that the expression and/or transcription of both of these genes is either accentuated or is down-regulated, depending on the lineage pathway chosen. Thus, commitment to nonerythroid (ie, monocytic or lymphocytic) pathway shuts down expression of the epo-receptor gene or of the erythroid transcription factor GATA-1, whereas a dramatic up-regulation of these genes is seen with erythroid differentiation.
However, many of the cell lines studied continuously proliferate in vitro, unlike true stem cells, and show spontaneous downstream differentiation to limited lineage pathways.16 Therefore, many of the data addressing characteristics of primitive hemopoietic cells at the population level may simply represent differences among subpopulations of more differentiated cells in these cell lines. These concerns were greatly alleviated when single cells from cell lines were tested by sensitive reverse transcription–polymerase chain reaction (RT-PCR) approaches.9 These important studies showed that low levels of transcription of lineage-affiliated genes, such as globin or myeloperoxidase, are coexpressed within individual cells. The survey of genes investigated included lineage-affiliated transcription factors, such as GATA-1, growth-factor receptors (GM-CSF, kit, M-CSF, or Epo receptors), or lineage-specific genes (globin). In general, the expression levels were low, and no lineage-affiliated proteins were detected in these studies. In contrast to expectations, transcriptional factors characteristic of a single lineage and lineage-specific genes (ie, globin, GATA-1, or epo-R) were not expressed in tandem in these studies. Most important, coactivation of genes of multiple lineages within single cells was also found when primary CD34+ cells were examined instead of FDCP cells.
Several arguments can be raised with these previously published data. They concern cells adapted to in vitro conditions, cells that are continuously proliferating and that have potential activation of certain molecular pathways. These cells may have options that may not be exercised in vivo. In addition, primary cells, because of methods of isolation or of conditions under which they are kept in vitro to ensure viability, may not be subjected to negative constraints exercised in vivo by the hemopoietic microenvironment and as a result may display a certain state of activation in vitro. Because the hemopoietic microenvironment cannot be faithfully imitated in vitro, it is unclear whether the results can be directly extrapolated in vivo. Furthermore, in other studies17 18 lineage-affiliated genes were not found to be activated in early cells, though the sensitivity of methods used may have been the problem in these studies.
Because the priming of lineage-specific genes in precommitted cells has been thus far only an in vitro phenomenon, in the current study we explored whether it could also be observed in vivo. To do so, we used a transgenic murine model in which a μLCR-driven β-globin promoter, together with LacZ as a reporter, was used as a transgene. We found that progenitors of all lineages displayed low levels of μLCR-βpro-LacZ activation with subsequent full activation in 2 lineages, erythroid and megakaryocytic. Moreover, though we detected priming of lineage-affiliated genes (μLCR-βpro) in all types of progenitor cells in vivo, this priming appeared graded and was not detected in a small proportion of progenitors. In aggregate, our data provided insight into mechanisms of lineage commitment operating in a physiologic setting.
Materials and methods
Synthesis of μLCR-β promoter LacZ construct
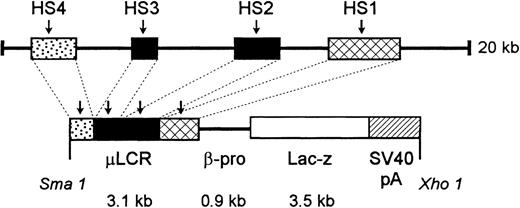
An 865-bp HpaI-ClaI human β-globin gene promoter fragment was isolated from pSP73 βm.19 This fragment was made blunt-ended by filling the 5′ single-stranded tails withEscherichia coli DNA polymerase I Klenow fragment. It was then ligated into XmnI-digested and phosphatased (calf intestinal alkaline phosphatase) pGNA, a plasmid containing a promoterlessLacZ gene,20 producing pβ-proLacZ. To produce the final construct, a 3.1-kb μLCR fragment was released from a modified LCR,21 made blunt-ended, and ligated into pGNA βpro-LacZ that had been digested with XbaI and made blunt-ended, creating pμLCR-βpro-LacZ. A 7.5-kb fragment containing μLCR-βpro-LacZ was released from plasmid sequence by XmaI/XhoI double digestion. After purification from agarose gel, the fragment was used for microinjection. (See Figure1.)
Diagram of construct used to generate μLCR-βpro-LacZ mice.
See “Materials and Methods” for details.
Diagram of construct used to generate μLCR-βpro-LacZ mice.
See “Materials and Methods” for details.
Generation of μLCR-βpro-LacZ transgenic mice
The 7.5-kb μLCR-βpro-LacZ was used to microinject fertilized mouse oocytes and subsequently was transferred to pseudopregnant females. Positive founder (F0) mice were identified by Southern slot blots with a probe directed to β-promoter sequence (865 bp). Briefly, DNA was isolated from tail biopsies, and 50 μL was precipitated, resuspended in 0.1 mol/L NaOH, 2 mol/L NaCl, heat-denatured, and slot-blotted to nitrocellulose (Schleicher and Schuell, Keene, NH). Six positive founders were bred with nontransgenic mice, and the resultant progeny were screened for integrity of the construct. Transgenic mice carrying correct integration with high X-gal red cell expression were used for breeding.
FACS-Gal assay
Bone marrow or fetal liver cell suspensions were prepared, subjected to ammonium chloride lysis to eliminate mature red cells, labeled with anti-TER119 antibody, and then subjected to VarioMacs (Miltenyi-Biotec, Auburn, CA) separation to remove TER119-positive nucleated erythroid cells. More than 80% of fetal liver cells and 30% to 40% of bone marrow cells were removed by this separation. The resultant TER119-negative cells were labeled with fluorescein di-β-D-galactoside (FDG; Molecular Probes, Eugene, OR) as described by Fiering et al.22 Briefly, cells to be assayed were suspended in buffered medium at a concentration of 107/mL and warmed to 37°C. An equal volume of FDG at 2 mmol/L in H2O was also warmed to 37°. The prewarmed cells and FDG were rapidly mixed together and immediately returned to the water bath for 1 minute. The tube was then placed on ice, 9 mL isotonic medium was added, and the mixture was held on ice for 30 to 60 minutes until it was sorted using a FACStar Plus (Becton Dickinson, San Jose, CA). As a positive control, cells from Rosa-β-geo mice,23in which all hemopoietic cells are LacZ positive, were used. In selected studies, FDG staining was carried out in lineage-depleted cells previously labeled with antibodies to CD34, kit, or Sca−1. All antibodies were purchased from Pharmingen (San Diego, CA).
X-gal assay
Mononuclear cells from bone marrow, fetal liver, or blood were rinsed in phosphate-buffered saline (PBS), fixed for 5 minutes on ice in a fixative solution of 1% formaldehyde and 0.5% glutaraldehyde in PBS, rinsed in detergent buffer (PBS, 0.02% NP40, 0.01% deoxycholate, 2 mmol/L MgCl2), and stained at 37°C in the dark for 3 to 4 hours. Staining solution contained 1 mg/mL X-gal [5-bromo-4-chloro-3-indolyl-β-D-galacto-pyranoside], 5 mmol/L K3Fe(CN)6, 5 mmol/L K4Fe(CN)6, and 1 mmol/L EGTA (ethylene glycol-bis(β-aminoethyl ether) N, N, N', N'-tetraacetic acid) in detergent buffer.24 Frozen sections were fixed and stained similarly. Whole organs were fixed for 30 minutes, processed as above, and stained overnight. Plasma clots were flattened on a slide with filter paper, fixed for 5 minutes, and stained as above for exactly 1 hour, rinsed, fixed in 3% glutaraldehyde, rinsed in distilled H20, then stained with benzidine and hematoxylin as previously described.25
Colony-forming unit-C assays
Colony-forming unit (CFU)-C assays were performed using a mixture containing 30% fetal bovine serum (FBS; Intergen, Purchase, NY), 1% bovine serum albumin (BSA; Intergen, Purchase, NY), 0.1 mmol/L 2-mercaptoethanol (Sigma, St. Louis, MO), 5 U/mL recombinant human erythropoietin (EPO; Genetics Institute, Cambridge, MA), 10% vol/vol mouse IL-3 culture supplement (Collaborative Biomedical Products, Bedford, MA), 5% pokeweed mitogen spleen cell-conditioned medium, 100 ng/mL recombinant murine stem cell factor (Peprotech, Rocky Hill, NJ), and 1.2% methyl cellulose (Fisher Scientific, Fairlawn, NJ). Plasma clot cultures contained the above mixture (without methyl cellulose), 2 mmol/L CaCl2, 0.25 U/mL bovine thrombin (Sigma), and 10% bovine citrated plasma (Animal Technologies, Tyler, TX). Medium components were made up in Iscove's modified Dulbecco's medium (Mediatech, Hernden, VA). Cells were plated as previously described26 and counted after 7 to 8 days.
Colony-forming unit-S assays
Fetal liver or bone marrow mononuclear cells from the LacZmice were subjected to red cell lysis, labeled with TER119 antibody followed by antirat IgG-phycoerythrin, then stained with FDG and sorted using a Becton Dickinson FACStar Plus. 5 × 104sorted FDG(+) or FDG(−) cells were then injected into the tail veins of sublethally irradiated SCID/NOD recipient mice. (Because of histocompatability problems, no syngeneic mice could be used.) Mice were killed 8 or 12 days later, their spleens were excised, and CFU-S were counted in situ or individually dissected out of the spleens, fixed, and stained with X-gal. Some spleens were mounted in OCT embedding medium (Tissue-Tek, Torrance, CA), and frozen sections were then fixed and stained with X-gal to assess positivity of CFU-S.
Results
LacZ activation in the developing fetus
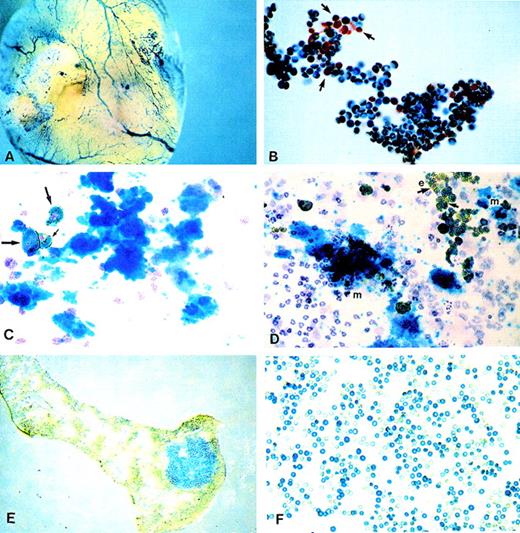
Histochemical X-gal staining was carried out using either whole embryos or yolk sacs or using cell suspensions prepared from yolk sac, fetal blood, or fetal liver. Whole embryo staining revealed LacZexpression in blood vessels and hemopoietic organs (yolk sac, fetal liver, and bones; Figure2A). This pattern of expression, restricted to hemopoietic cells and organs, was reminiscent of results published previously when similar constructs were used.27 28 Circulating embryonic or definitive erythroblasts and red cells present in blood at days 11 to 12 days post coitus were heavily labeled. However, white cells present in the same samples were not labeled (Figure 2B). Cell preparations from fetal liver also showed heavy labeling obscuring even the presence of white, unlabeled cells. High proportions of erythroblasts in these preparations were responsible for the intense labeling. To secure the identity of many small cells labeled with LacZ, we used a double labeling approach in which benzidine staining was combined with X-gal staining. All erythroid cells were benzidine positive, and the combination of gold (from benzidine staining) and green (for X-gal staining) gave these cells a deep bronze appearance (Figures 2C, 2D).
LacZ activation as revealed by histochemical X-gal staining.
(A) Whole embryo staining by X-gal. Yolk sac blood vessels were intensely stained. (B) Embryonic blood stained for X-gal showing that white cells (shown by arrows) are negative. (C, D) Plasma clots stained for benzidine, X-gal, and counterstained with hematoxylin. (C) One megakaryocytic colony is shown. Also present is a group of 3 benzidine-positive and X-gal-positive erythroid cells (arrows) and scattered white cells negative for X-gal. (D) Two megakaryocytic (m) colonies and 1 erythroid (e) colony (right upper corner) are seen, along with scattered white cells. Note the presence of positive and negative X-gal erythroid cells (arrows). (E) Spleen section with an X-gal-positive colony (CFU-S). (F) Red cells from a SCID/NOD mouse transplanted with FDG(+)/TER119(−) fetal liver cells at 5 weeks after transplantation. Note the larger size of X-gal(+) fetal red cells.
LacZ activation as revealed by histochemical X-gal staining.
(A) Whole embryo staining by X-gal. Yolk sac blood vessels were intensely stained. (B) Embryonic blood stained for X-gal showing that white cells (shown by arrows) are negative. (C, D) Plasma clots stained for benzidine, X-gal, and counterstained with hematoxylin. (C) One megakaryocytic colony is shown. Also present is a group of 3 benzidine-positive and X-gal-positive erythroid cells (arrows) and scattered white cells negative for X-gal. (D) Two megakaryocytic (m) colonies and 1 erythroid (e) colony (right upper corner) are seen, along with scattered white cells. Note the presence of positive and negative X-gal erythroid cells (arrows). (E) Spleen section with an X-gal-positive colony (CFU-S). (F) Red cells from a SCID/NOD mouse transplanted with FDG(+)/TER119(−) fetal liver cells at 5 weeks after transplantation. Note the larger size of X-gal(+) fetal red cells.
LacZ activation in differentiated erythroid and megakaryocytic cells
To test the presence of X-gal activity among differentiated cells of several lineages, peripheral blood cells or bone marrow mononuclear cells from adult mice were labeled in suspension by the X-gal technique, as described in the “Materials and methods” section. After staining, cytocentrifuge preparations were made to score individual cells. Among peripheral blood cells, the only cells with X-gal activity were the red cells. The proportion of positive red cells among 26 mice tested was between 84% and 100%, and among positive cells there were visible differences in the intensity of staining. Timed incubation for X-gal (between 15 minutes and 2 hours) exaggerated these differences. In 1 sample the proportion of X-gal-positive cells was approximately 60% at 15 minutes, but virtually all cells were positive at 2 hours. In bone marrow preparations, erythroblasts at all maturation stages were positive, and the majority appeared strongly positive. In addition to erythroid cells, megakaryocytes on the smears were positive, again with great differences in intensity of staining.
LacZ activation in BFUe and CFU-meg-derived progeny
To ensure that only differentiated erythroid or megakaryocytic cells at all stages of maturation were positive, we cultivated cell suspensions from yolk sac, fetal liver, or adult bone marrow in semi-solid (methyl cellulose or plasma clot) media and tested the X-gal positivity in the in vitro progeny of all clonogenic progenitors. Methyl cellulose culture plates were evaluated in situ for all types of colonies present on the basis of morphologic criteria and then were stained with the X-gal for evaluation of total “blue” colonies versus “non-blue” colonies. Plasma clots were evaluated after they were fixed and stained as described in “Materials and Methods.” The proportion of erythroid bursts, especially those of mixed erythroid and megakaryocytic colonies, was higher in yolk sac or fetal liver than in adult bone marrow (Ery/Meg colonies: yolk sac, 8.7 ± 1.8/105 cells plated; fetal liver, 3.9 ± 1.8/105; adult bone marrow, less than 0.01). The total number of blue colonies was sometimes slightly lower than the total number of erythroid/megakaryocytic colonies (data not shown). This difference was as high as 5% in fetal samples and 30% in adult samples. In other words, more than 92% of erythroid and megakaryocytic colonies were strongly positive for X-gal in fetal samples, whereas in adult samples an average of 67% of the colonies were strongly X-gal positive. Whether colonies scored as negative were totally negative or whether some weakly positive cells were present was not assessed because these methylcellulose counts were performed only under the dissecting microscope. In flattened plasma clot cultures, in addition to apparently negative bursts, bursts displaying mixed positive and negative cells were clearly present (Figure 2D), especially in adult bone marrow cells. Heterogeneity in staining was also seen in megakaryocytic colonies in the same cultures.
FDG(+) cells include progenitors of all lineages
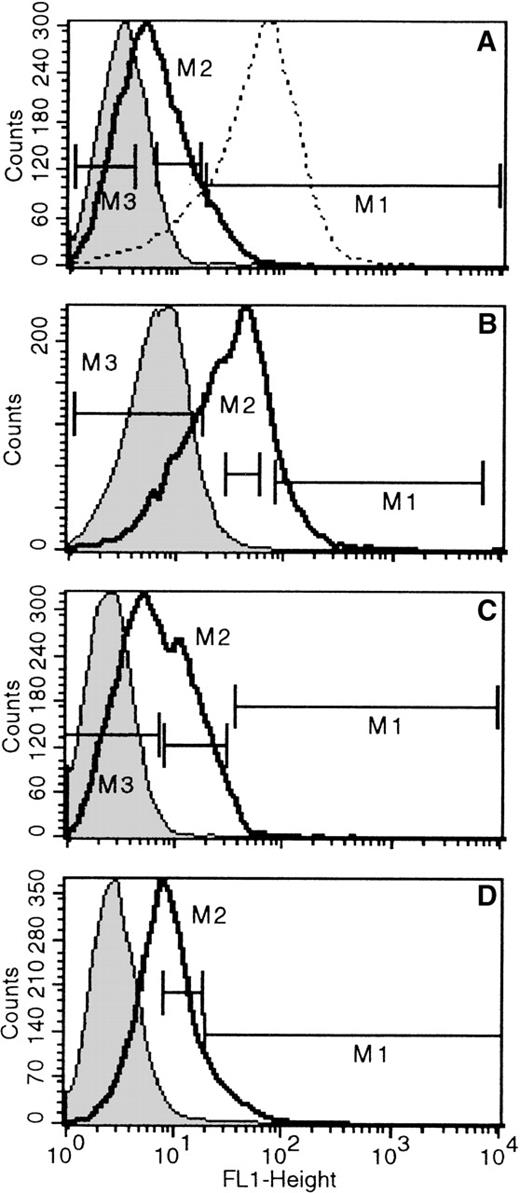
In the previous sections we described that differentiated erythroid and megakaryocytic cells in vivo, and the ones generated in vitro by committed progenitors in culture were positive in X-gal staining. To test whether these positive cells were generated after terminal commitment to these 2 lineages or whether the activation ofLacZ was already present in precommitted cells, we depleted bone marrow or fetal liver cells of erythroid cells (by removing TER119+ cells) and labeled the resultant TER119(−) cells with the fluorescent LacZ substrate FDG, as described in “Materials and Methods.” FDG labeling is a highly sensitive way to assess even low levels of LacZ activation. FDG-stained bone marrow cells from nontransgenic mice were used as negative control samples. Positive controls were FDG-stained bone marrow cells from Rosa-β-geo mice in which all cells are X-gal positive except red cells. In fetal livers TER-119(+) cells displayed similar positivity to Rosa-β-geo cells, and these were also used as positive controls. The proportion of FDG(+) cells among TER119(−) fetal liver samples was very high (see Table 1) and showed a broad spectrum of positivity (only 2% to 13% of cells were FDG(−); Table 1, Figure3). By contrast, the proportion of FDG(+) cells among adult bone marrow/TER119(−) cells was much lower; in 4 bone marrow samples, the proportion of FDG(−) cells was 36% to 75% (Figure4). In bone marrow samples of 2 newborns, proportions of FDG(+) and TER119(−) cells were 50.2% and 34.2%. Not only was the proportion of FDG(+)/TER119(−) cells lower in bone marrow samples when compared with fetal liver cells, the mean fluorescence intensity (Figure 3 vs Figure 4) was also much lower. After the distribution of FDG positivity was determined, populations highly positive for FDG, displaying similar positivity with Rosa-β-geo controls (Figure 3A), or populations with intermediate FDG positivity and populations totally negative for FDG were sorted and subjected to clonogenic cultures to determine the types of colonies present in these fractions. Results from these analyses are listed in Tables 1 and 2. Three fetal liver samples and 4 bone marrow samples were analyzed. From fetal liver samples, clearly the majority of colonies generated were among the FDG(+) fractions (considering the frequency among plated cells and the frequency of FDG(+) among total cells). Most interesting, not only the erythroid/megakaryocytic progenitors but also the majority of myeloid progenitors, CFU-GM, were found in these positive fractions, though progeny of the latter was negative for X-gal in vitro. Independent evaluation of X-gal positive (blue) versus X-gal negative (non-blue) colonies secured these findings (ie, number of blue colonies was similar to the number of erythroid and megakaryocytic colonies). In addition to informative findings from FDG(+) cells, the types of colonies generated from FDG(−) cells were also of interest. Erythroid bursts with X-gal(+) progeny were also generated from these fractions. Although LacZ positivity was not detected in progenitors, the gene was not irreversibly silenced; its progeny displayed full activation of LacZ during erythroid differentiation. Highly positive FDG fractions in 1 liver sample (Figure 3A) generated virtually only CFUe. The degree of positivity in this fraction was equivalent to that of TER119(+) cells or the Rosa-β-geo cells (Figure 3A). Therefore, it may not be surprising that this fraction showed only late erythroid progenitors and precursors.
CFU generated from TER-119(−) fetal liver cells separated according to FDG positivity
| Sample . | Liver A: CFU-C/105 * . | Liver B: CFU-C ± SEM/105 . | Liver C: CFU-C ± SEM/105 . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % of Total . | CFU-E . | BFU-E . | Mk . | GM . | % of Total . | BFU-E . | Mk . | GM . | % of Total . | BFU-E . | Mk . | GM . | |
| Pre-sort | 1200 | 150 | 580 | 1700 ± 80 | 730 ± 71 | 940 ± 96 | 5067 ± 536 | 1200 ± 252 | 7700 ± 874 | ||||
| FDGhi | 0.9 | (1029) | 0 | 0 | 0 | 19.6 | 89 ± 21 | 202 ± 9.5 | 330 ± 30 | 18.6 | 5830 ± 830 | 1750 ± 320 | 4650 ± 920 |
| FDGmed | 96.2 | 5952 | 128 | 2016 | 57.3 | 733 ± 240 | 131 ± 35 | 988 ± 156 | |||||
| FDGneg | 2.2 | 507 | 216 | 343 | 13.2 | 523 ± 73 | 383 ± 40 | 1146 ± 94 | 5.5 | 4123 ± 420 | 2411 ± 490 | 7300 ± 780 | |
| Sample . | Liver A: CFU-C/105 * . | Liver B: CFU-C ± SEM/105 . | Liver C: CFU-C ± SEM/105 . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % of Total . | CFU-E . | BFU-E . | Mk . | GM . | % of Total . | BFU-E . | Mk . | GM . | % of Total . | BFU-E . | Mk . | GM . | |
| Pre-sort | 1200 | 150 | 580 | 1700 ± 80 | 730 ± 71 | 940 ± 96 | 5067 ± 536 | 1200 ± 252 | 7700 ± 874 | ||||
| FDGhi | 0.9 | (1029) | 0 | 0 | 0 | 19.6 | 89 ± 21 | 202 ± 9.5 | 330 ± 30 | 18.6 | 5830 ± 830 | 1750 ± 320 | 4650 ± 920 |
| FDGmed | 96.2 | 5952 | 128 | 2016 | 57.3 | 733 ± 240 | 131 ± 35 | 988 ± 156 | |||||
| FDGneg | 2.2 | 507 | 216 | 343 | 13.2 | 523 ± 73 | 383 ± 40 | 1146 ± 94 | 5.5 | 4123 ± 420 | 2411 ± 490 | 7300 ± 780 | |
Plating efficiencies per 105 cells plated from each fraction.
FACS histograms of TER119(−) cells from 3 (day 14) fetal livers stained for FDG.
The 3 fetal livers are A, B, and C. Negative controls are indicated by shaded histograms. Solid, bold lines represent FDG positivity in fetal liver TER119(−) cells, and dashed lines show FDG positivity in Rosa-β-geo cells used as positive controls. Negative controls are non-FDG-stained cells (shaded peak) or FDG-stained nontransgenic fetal liver cells (dashed line in Figure 3C; both peaks are virtually superimposable). Populations sorted for culture are indicated (M1 = FDG high; M2 = FDG med; M3 = FDG neg).
FACS histograms of TER119(−) cells from 3 (day 14) fetal livers stained for FDG.
The 3 fetal livers are A, B, and C. Negative controls are indicated by shaded histograms. Solid, bold lines represent FDG positivity in fetal liver TER119(−) cells, and dashed lines show FDG positivity in Rosa-β-geo cells used as positive controls. Negative controls are non-FDG-stained cells (shaded peak) or FDG-stained nontransgenic fetal liver cells (dashed line in Figure 3C; both peaks are virtually superimposable). Populations sorted for culture are indicated (M1 = FDG high; M2 = FDG med; M3 = FDG neg).
FACS histograms of 4 bone marrow TER119(−) cells, after staining with FDG.
Fractions were sorted and cultured, as in Figure 3. Negative controls are shaded, and the dotted line peak in #1 represents FDG positivity of TER119(+) cells used as a positive control. Labels A-D correspond respectively to sample numbers 1-4 of bone marrows used for culture in Table 2. The proportions of FDG(−) cells were: A (#1), 75.2%; B (#2), 35.6%; C (#3), 50%; and D (#4), 69%.
FACS histograms of 4 bone marrow TER119(−) cells, after staining with FDG.
Fractions were sorted and cultured, as in Figure 3. Negative controls are shaded, and the dotted line peak in #1 represents FDG positivity of TER119(+) cells used as a positive control. Labels A-D correspond respectively to sample numbers 1-4 of bone marrows used for culture in Table 2. The proportions of FDG(−) cells were: A (#1), 75.2%; B (#2), 35.6%; C (#3), 50%; and D (#4), 69%.
CFU-C generated from TER119(−) bone marrow cells separated according to FDG positivity
| . | CFU ± SEM/105Cells . | ||||
|---|---|---|---|---|---|
| BFUE . | CFU-Mk . | CFU-GM . | Total CFC . | Total CFC “blue” . | |
| Bone marrow #1* | |||||
| Pre-sort | 10 ± 1.8 | 16 ± 2.3 | 306 ± 7.6 | 332 ± 8.7 | 8 ± 0.7 |
| FDG ++ (9.0) | 15 ± 2.1 | 3 ± 0.6 | TC† | 7 ± 0.5 | |
| FDG +/− (35.0) | 64 ± 4.7 | 31 ± 3.4 | 411 ± 29.3 | 506 ± 9.2 | 61 ± 4.8 |
| FDG neg (32.0) | 17 ± 2.1 | 3 ± 0.2 | 56 ± 1.8 | 76 ± 3.8 | 13 ± 1.1 |
| Bone marrow #2* | |||||
| Pre-sort | 28 ± 2.1 | 24 ± 1.7 | 258 ± 3.8 | 320 ± 5.1 | 32 ± 1.8 |
| FDG ++ (10.5) | 31 ± 1.8 | 29 ± 2.3 | 733 ± 35.3 | 793 ± 40.2 | 33 ± 3.2 |
| FDG + (37.0) | 210 ± 5.9 | 64 ± 4.6 | 1270 ± 40.8 | 1543 ± 50.2 | 202 ± 7.5 |
| FDG neg (27.0) | 35.5 | 17.5 ± 2.8 | 587 ± 39.7 | 640 ± 35.7 | 36 ± 4.6 |
| Bone marrow #3* | |||||
| Pre-sort | (No Epo used in these cultures.) | 350 ± 19 | |||
| FDG ++ (2.0) | 380 ± 39 | ||||
| FDG + (39.0) | 248 ± 21 | ||||
| FDG neg (55.0) | 114 ± 17 | ||||
| Bone marrow #4* | |||||
| Pre-sort | 179 ± 3.0 | 45 ± 3.0 | |||
| FDG ++ (13.3) | 603 ± 7.5 | 165 ± 5.4 | |||
| FDG +/− (46.0) | 112 ± 5.0 | 0 | |||
| . | CFU ± SEM/105Cells . | ||||
|---|---|---|---|---|---|
| BFUE . | CFU-Mk . | CFU-GM . | Total CFC . | Total CFC “blue” . | |
| Bone marrow #1* | |||||
| Pre-sort | 10 ± 1.8 | 16 ± 2.3 | 306 ± 7.6 | 332 ± 8.7 | 8 ± 0.7 |
| FDG ++ (9.0) | 15 ± 2.1 | 3 ± 0.6 | TC† | 7 ± 0.5 | |
| FDG +/− (35.0) | 64 ± 4.7 | 31 ± 3.4 | 411 ± 29.3 | 506 ± 9.2 | 61 ± 4.8 |
| FDG neg (32.0) | 17 ± 2.1 | 3 ± 0.2 | 56 ± 1.8 | 76 ± 3.8 | 13 ± 1.1 |
| Bone marrow #2* | |||||
| Pre-sort | 28 ± 2.1 | 24 ± 1.7 | 258 ± 3.8 | 320 ± 5.1 | 32 ± 1.8 |
| FDG ++ (10.5) | 31 ± 1.8 | 29 ± 2.3 | 733 ± 35.3 | 793 ± 40.2 | 33 ± 3.2 |
| FDG + (37.0) | 210 ± 5.9 | 64 ± 4.6 | 1270 ± 40.8 | 1543 ± 50.2 | 202 ± 7.5 |
| FDG neg (27.0) | 35.5 | 17.5 ± 2.8 | 587 ± 39.7 | 640 ± 35.7 | 36 ± 4.6 |
| Bone marrow #3* | |||||
| Pre-sort | (No Epo used in these cultures.) | 350 ± 19 | |||
| FDG ++ (2.0) | 380 ± 39 | ||||
| FDG + (39.0) | 248 ± 21 | ||||
| FDG neg (55.0) | 114 ± 17 | ||||
| Bone marrow #4* | |||||
| Pre-sort | 179 ± 3.0 | 45 ± 3.0 | |||
| FDG ++ (13.3) | 603 ± 7.5 | 165 ± 5.4 | |||
| FDG +/− (46.0) | 112 ± 5.0 | 0 | |||
Numbers in parentheses represent the gated portion sorted expressed as percentage of total cells (see example in Figure 4, #2). Note that the totals are less than 100.
Too crowded to count.
Results with bone marrow samples were qualitatively similar to fetal liver samples, but there were significant quantitative differences. As illustrated by FACS profiles in Figure 4, the overall positivity of FDG(+) fractions in bone marrow were approximately 25% to 65% compared to fetal liver values of 87% to 98%. Fetal liver samples also displayed higher fluorescence intensity (compare Figures 3 and 4). In samples from both bone marrow and fetal liver, the majority of colonies of all types were generated from FDG(+) (either intermediate or high positivity) rather than FDG(−) fractions. A feature that was not infrequent in bone marrow samples was the appearance of mixed positivity (by X-gal staining) among erythroid or megakaryocytic colonies. Totally negative colonies were also seen. These mixed and negative colonies were either absent or extremely rare in cultures from fetal liver samples.
CFU-S12 are generated from transplantation of FDG(+) cells
One bone marrow and one fetal liver sample were used for CFU-S12 studies (Table 3). The samples were simultaneously labeled for TER119-PE and for FDG–FITC. After the TER119(+) cells were excluded, the population was divided into FDG(+) and FDG(−) cells. After 5 × 104 cells [FDG(+) or FDG(−)] were injected into each mouse, CFU-S12 colonies were counted in whole spleens before or after X-gal staining. Forty colonies were counted in 5 mice given FDG(−) cells from the bone marrow sample. Four mice given FDG(+) cells generated 39 colonies. (In the 5th mouse, colonies were too confluent for accurate counting.) Because the SCID/NOD mice were only sublethally irradiated, CFU-S12 in these mice generated from FDG(+) or FDG(−) cells could represent just endogenous CFU-S. Spleens were then subjected to X-gal staining. The diffuse green color in these spleens (because of the presence of red cells and erythroblasts matured from CFU-S8) obscured the true positivity of CFU-S. Therefore, a different approach was used in the next experiment. From 1 fetal liver sample, only the FDG(+) cells were injected into 8 mice. Four mice were killed at day 8, and CFU-S8 were counted (there were 36 CFU-S, and most appeared blue after staining of frozen spleen sections with X-gal; Figure 2E). From 4 mice killed at day 12, 26 colonies were counted, and 17 of these were individually dissected to be X-gal stained separately in vitro. 70% of colonies individually stained showed more than 20% positive cells in each. (Colonies with less than 20% positive cells were considered negative.) Spleen sections stained for X-gal (Figure 2E) were also prepared. These sections showed either fairly uniform X-gal positivity (Figure 2E) or scattered positive cells among each CFU-S colony. Thus, in these experiments, we showed that CFU-S, like CFC, were present among FDG(+) cells, though the fraction of total CFU-S12 could not be calculated with accuracy (because of the nature of recipient mice).
CFU-Ss from FDG(+) or (−) fractions given to SCID/NOD mice
| Sample . | Mice . | CFU-S12 . |
|---|---|---|
| Bone marrow (ter119-) | ||
| FDG(+) | 1 | TC3-150 |
| 2 | 12 | |
| 3 | 8 | |
| 4 | 7 | |
| 5 | 12 | |
| Mean ± SEM | 9.8 ± 1.32 | |
| FDG(−) | 1 | 3 |
| 2 | 6 | |
| 3 | 10 | |
| 4 | 10 | |
| 5 | 11 | |
| Mean ± SEM | 8 ± 1.52 | |
| Fetal liver (Ter119-)3-151 | ||
| FDG(+) | 1 | 6 |
| 2 | 11 | |
| 3 | 9 | |
| 4 | 0 | |
| Mean ± SEM | 6.5 ± 2.4 |
| Sample . | Mice . | CFU-S12 . |
|---|---|---|
| Bone marrow (ter119-) | ||
| FDG(+) | 1 | TC3-150 |
| 2 | 12 | |
| 3 | 8 | |
| 4 | 7 | |
| 5 | 12 | |
| Mean ± SEM | 9.8 ± 1.32 | |
| FDG(−) | 1 | 3 |
| 2 | 6 | |
| 3 | 10 | |
| 4 | 10 | |
| 5 | 11 | |
| Mean ± SEM | 8 ± 1.52 | |
| Fetal liver (Ter119-)3-151 | ||
| FDG(+) | 1 | 6 |
| 2 | 11 | |
| 3 | 9 | |
| 4 | 0 | |
| Mean ± SEM | 6.5 ± 2.4 |
Too crowded to count.
In a separate fetal liver experiment, a total of 17 CFU-S12 colonies were individually dissected and stained for X-gal. All colonies had positive cells. Colonies with >20% of cells X-gal(+) were considered positive (10 of 17).
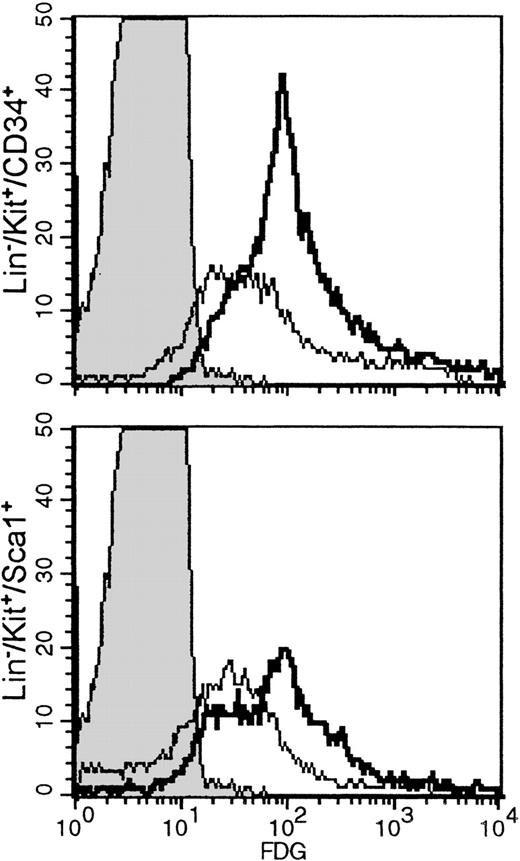
Hemopoietic cell subsets with a stem cell phenotype are FDG positive
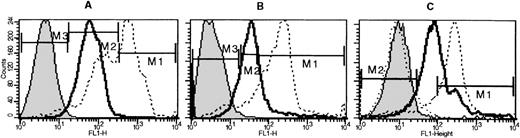
Histocompatibility problems precluded long-term repopulation experiments using FDG(+) or FDG(−) cells. Using a surrogate approach, we evaluated the LacZ activation in subsets of stem cells previously shown to include long-term repopulating cells, such as Lin−/kit+/CD34+ or Lin−/Sca−1+/kit+. One fetal liver sample and 1 bone marrow sample from a newborn were used for these studies. The proportion of Lin−/kit+/34+ subset in fetal liver was 11.4%, whereas the proportion of Lin−/Sca−1+/kit+ cells was 15.7%. The proportions in newborn bone marrow were 3.5% and 7.1%, respectively. However, the great majority of either Lin−/Sca−1+/kit+ and Lin−/kit+/CD34+ in both samples were FDG(+) [Figure 5], suggesting thatLacZ activation may indeed start early at the multipotential cell level, possibly in a proportion of long-term repopulating cells. Our experiments thus far do not allow any conclusions regarding the level of activation in long-term repopulating cells. The latter can only be reliably tested with transplantations of FDG(+) stem cell subsets in histocompatible recipients. Nevertheless, we have evaluated SCID/NOD mice that received FDG(+) cells 5 weeks after transplantation. Two such mice showed approximately 50% of their red cells X-gal (+) (Figure 2F). In addition, bone marrow from 1 mouse was cultured in plasma clots to assess the presence of X-gal (+) colonies. This marrow yielded a total of 229 ± 19 CFU-C/105plated cells. The total erythroid bursts were 51 ± 4, CFU-Meg were 20 ± 3, CFU-MixEry + Meg were 7 ± 2, and CFU-GM were 151 ± 18/105 cells plated. Fifty-five colonies were blue—ie, approximately 73% of the total erythroid and megakaryocytic colonies were X-gal (+).
Lineage-depleted fetal liver cells or newborn bone marrow cells labeled with anti-CD34 and anti-kit or anti-kit and anti-Sca−1, followed by FDG labeling.
Histograms shown are from gated populations that were either Lin−/kit+/34+ or Lin−/kit+/Sca−1+. Negative controls (indicated by shaded peaks) are non-FDG-labeled fetal liver Lin− cells. In fetal liver (bold lines), the proportion of FDG(+) cells in the Lin−/kit+/34+ subset was 93.3%, and the Lin−/kit+/Sca−1+ was 79.8%, whereas in bone marrow (thin lines) they were 72.6% and 51.6%, respectively.
Lineage-depleted fetal liver cells or newborn bone marrow cells labeled with anti-CD34 and anti-kit or anti-kit and anti-Sca−1, followed by FDG labeling.
Histograms shown are from gated populations that were either Lin−/kit+/34+ or Lin−/kit+/Sca−1+. Negative controls (indicated by shaded peaks) are non-FDG-labeled fetal liver Lin− cells. In fetal liver (bold lines), the proportion of FDG(+) cells in the Lin−/kit+/34+ subset was 93.3%, and the Lin−/kit+/Sca−1+ was 79.8%, whereas in bone marrow (thin lines) they were 72.6% and 51.6%, respectively.
Discussion
Although previous data showing simultaneous expression of lineage-affiliated genes in multipotential cells from established cell lines or in primary human CD34+ cells are compelling,9 the concept of activation of lineage-restricted genes in cells before their overt unilineage commitment has not been tested in an in vivo setting. We elected to explore this concept in vivo using transgenic mice, carrying the LCR-driven β-promoter as a transgene. Our choice of this transgenic model was deemed particularly appropriate because LCR chromatin status and β-globin gene transcripts have been used as lineage-specific probes for in vitro studies that addressed issues of lineage commitment, and both were detected in multipotential cells before their final commitment to a specific lineage.10Furthermore, because of the use of LacZ as a reporter in our model, we had the opportunity not only to detect expression at the single-cell level but also to grade quantitatively the levels of expression at the cellular level. This was possible through the use of 2 assays for LacZ expression, the FACS-gal assay and the X-gal histochemical assay, which differ in sensitivity by more than 100-fold.22 Using the FACS-gal assay we detected positive cells that could be negative by the histochemical assay, the X-gal assay. Finally, because of extensive prior experience, it was known that transgene activation using a number of similar constructs27 28 is tissue specific with no evidence of ectopic or inappropriate expression in nonhemopoietic tissues.
Erythroid and megakaryocytic cells display high levels of LacZ activity
As expected from previous studies using similar constructs,24, 27,28 we found significant expression of LacZ on erythroid cells. However, in contrast to previously reported data, we found X-gal-positive cells among cells of megakaryocytic lineage. Megakaryocytes generated in vivo or induced from CFU-Meg during clonogenic cultures in vitro were X-gal positive. Of particular interest, bipotent erythroid megakaryocytic progenitors giving rise to mixed erythroid/megakaryocytic colonies were frequently encountered in yolk sac and fetal liver samples. Thus, by using this particular transgene, we uncovered in vivo a molecular pathway of differentiation that is active in early developmental stages but is less frequent in adult bone marrow samples.29 Sharing of certain transcription factors or of certain surface antigens between erythroid and megakaryocytic cells has been emphasized by Papayannopoulou et al29 and the sources cited for that study and could be the basis of activation of β-globin in megakaryocytic cells. In view of the high levels of LacZ expression in megakaryocytic cells, it was of great interest to test whether endogenous murine β-globin mRNA could also be detected in positively selected CD41+ cells (FACS sorted from in vitro Tpo-stimulated bone marrow cells), which included megakaryocytic cells at different levels of development. Using dilutions equivalent to approximately 13 CD41+ cells, we were able to detect β-globin mRNA by PCR (Asano H, unpublished data). Because CD41 is already expressed by a significant proportion of erythroid progenitors (BFUe),29 it may be argued that the PCR results stemmed only from the presence of erythroid progenitors in our samples. However, no EKLF transcripts were detected in the same preparations, making it unlikely that β-globin in adult erythroid cells is dissociated from EKLF activation.30 Rather, murine β-globin transcripts could be present in cells before their definitive commitment to erythroid lineage or in nonerythroid-committed (ie, Meg) cells and could provide added assurance that results with LacZ in our mice had physiologic relevance and did not result from the presence of the artificial construct. Although β-globin transcripts could be detected, no protein was detectable by immunofluorescence. This suggests that in the absence of a proper regimen of transcriptional factors, including EKLF, no efficient translation or protein accumulation occurs.
Variegation in μLCR-βpro-LacZ expression
Position effects or a variegated expression of μLCR-βpro transgene were expected and were seen in our studies, as they were in other studies using similar constructs.27, 28,31 For this reason, only lines with high LacZ expression in red cells (80% to 100% positive red cells) were selected for study. In addition to variation in the proportion of positive cells, 3 additional features in our transgenic animals were consistent with the graded rather than the binary model of LCR-dependent position effects 27 32: (1) the red cells of adult mice displayed significant variation in intensity of staining, which was particularly evident when positivity in cells was evaluated kinetically from 15 minutes to 2 hours; (2) the proportion of FACS-gal(+)/TER119(−) (nonerythroid) cells was higher in FL samples than in bone marrow-derived samples (Tables 1, 2) (in the latter, the mean fluorescence intensity was also lower than in fetal liver-derived cells [Figures 3, 4]); (3) the distribution of FDG activity among positive cells was broad, both in fetal liver and bone marrow-derived samples, consistent with varying levels ofLacZ among cells within a given sample.
The differences in quantitative expression of LacZ between fetal liver cells and bone marrow cells deserve further comment. It was previously suggested that LacZ expression is increasingly silenced with age.31 Indeed, significant differences in the proportion of X-gal positivity were seen when total nucleated cells from fetal liver cells were compared with those from adult bone marrow. This could partially be explained by the higher proportion of erythroid cells and erythroid progenitors in fetal liver compared with bone marrow samples. However, additional reasons, related neither to the proportion of erythroid progenitors nor to age per se, may be at play. Newborn and adult bone marrow samples differed from fetal liver samples. It is thus possible that activation of LacZis more efficient in fetal liver cells or in cells harboring a fetal rather than an adult cellular environment. The combination of transcriptional factors within fetal liver progenitor cells, in a quantitative or a qualitative sense, may be more conducive to the LCR-β-promoter activation than bone marrow-derived progenitors. Alternatively, cell cycling differences between fetal liver and bone marrow cells33 may play a role. The situation is reminiscent of differences detected between primitive and definitive progenitors in LCR-driven LacZ expression.24 A repressive effect of LacZ on LCR was seen in definitive (fetal liver) progenitors but not in primitive (yolk sac) progenitors in the above study.24 Further experiments using other transgenes may shed some light on this issue.
In vivo activation of μLCR-βpro-LacZ in progenitor cells
The full activation (ie, X-gal positivity) of LCR-β promoter in differentiated erythroid and megakaryocytic cells, as described in this article, could be explained by 2 different hypotheses. First, the activation can be at or after commitment to erythroid lineage, megakaryocytic lineage, or both. Second, the activation could be before commitment to erythroid/nonmegakaryocytic lineage but silenced later in other, nonerythroid/nonmegakaryocytic lineages. The expectations from these 2 hypotheses are distinct. According to the first hypothesis, which assumes expression of LacZ after commitment to erythroid/megakaryocytic lineage, if FACS-gal(+) progenitors exist, these should give rise only to erythroid colonies, megakaryocytic colonies, or both. By contrast, according to the second hypothesis, which assumes expression of LacZ before commitment, positivity is expected in all types of committed progenitors. Results of cultures of FDG(+) cells generating all types of progenitor cells, especially CFU-GM, are highly compatible with the second hypothesis. Indeed, the great majority of all progenitors was recovered in the FDG(+) fraction. However, because both X-gal(+) BFUe-derived colonies and X-gal(+) CFU-Meg-derived colonies were grown from FDG(−) progenitors, the data further implied that activation of LCR-β promoter is not uniformly activated before erythroid commitment in all progenitors and that it may not be occurring in a fraction of erythroid or other progenitor cells. The distribution of scattered rather than uniform X-gal cell positivity within erythroid bursts derived from FDG(−) progenitors suggests that activation occurs after commitment in a proportion of BFUe, as proposed in previous studies using only X-gal staining in colonies from adult mice with low levels of LacZ expression in red cells.28 Collectively, our data suggest that although priming of lineage-restricted genes can be detected in vivo in all types of progenitor cells, a spectrum of molecular phenotypes is seen, indicating a gradual and progressive final commitment to a specific lineage. Alternatively, one may suggest that this low level of activation in progenitor cells is of a transient nature alternating with states of no activation at any given time (“see-saw” phenomenon). Our data of LacZ activation in μLCR-βpro-LacZ mice share certain similarities with data in mice in which LacZ was linked to SCL promoter.34Because SCL is a transcription factor essential for primitive and definitive hemopoiesis, its activation in all types of progenitors was not surprising. However, full activation was found only within differentiated erythroid and megakaryocytic cells, similar to our findings.
The importance of the current data is that progenitor cells in vivo, before their final commitment, display priming of lineage-specific genes under physiologic conditions without any putative in vitro perturbations of their behavior. Such priming in hemopoietic progenitors does not represent ectopic expression of LCR-βpro-LacZ; extensive prior experience with several similar constructs did not show activation in other than hemopoietic organs. Rather, it reflects functionally incompetent levels of expression, but the requirements for the transition from the “primed” state to the state of “full activation” are entirely unclear. Our data seem to support more the stochastic rather than the instructive model of differentiation. However, they do not preclude the fact that external influences, as envisioned by the instructional model, can be exercised at some level downstream of the initial activation by simply altering threshold levels of transcriptional regulators, of growth factors, etc, and by influencing the lineage pathway chosen. We cannot explain why the levels of activation were higher in fetal liver cells rather than adult progenitor cells. Whether this is a peculiarity of the LCR or the LacZ construct, or both, is unclear. Nevertheless, we believe our in vivo data in aggregate, especially those in fetal liver, strengthen and complement prior in vitro data on lineage commitment decisions.
Acknowledgments
We thank Dr H. Asano for performing the PCR in CD41+ cells and Gina Alvino and Tyler Kimbrough for creating the μLCR-βpro-LacZconstruct. We also thank Margaret Oppenheimer and Domino Hawks for their expert secretarial assistance.
Supported by National Institutes of Health grants DK30852 and HL46557.
Reprints:Thalia Papayannopoulou, Division of Hematology, University of Washington, Box 357710, Seattle, WA 98195-7710; email: thalp@u.washington.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal