Congenital afibrinogenemia is a rare autosomal recessive disorder characterized by bleeding that varies from mild to severe and by complete absence or extremely low levels of plasma and platelet fibrinogen. Although several mutations in the fibrinogen genes associated with dysfibrinogenemia and hypofibrinogenemia have been described, the genetic defects of congenital afibrinogenemia are largely unknown, except for a recently reported 11-kb deletion of the fibrinogen A-chain gene. Nevertheless, mutation mechanisms other than the deletion of a fibrinogen gene are likely to exist because patients with afibrinogenemia showing no gross alteration within the fibrinogen cluster have been reported. We tested this hypothesis by studying the affected members of two families, one Italian and one Iranian, who had no evidence of large deletions in the fibrinogen genes. Sequencing of the fibrinogen genes in the 2 probands detected 2 different homozygous missense mutations in exons 7 and 8 of the Bβ-chain gene, leading to amino acid substitutions Leu353Arg and Gly400Asp, respectively. Transient transfection experiments with plasmids expressing wild-type and mutant fibrinogens demonstrated that the presence of either mutation was sufficient to abolish fibrinogen secretion. These findings demonstrated that missense mutations in the Bβ fibrinogen gene could cause congenital afibrinogenemia by impairing fibrinogen secretion.
Fibrinogen is a 340-kd glycoprotein synthesized primarily by hepatocytes and secreted as a hexamer composed of 3 pairs of polypeptide chains (Aα, Bβ, and γ) (for review, see1). It has a trinodular structure with a central nodule (E-domain) that contains the N-terminus of each chain and 2 lateral globular domains (D-domains) that contain the C-terminus of Bβ- and γ-chains. The E-domain is linked to the 2 D-domains by a coiled-coil triple helix structure.2 The 3 chains are encoded by different genes, clustered in a region of approximately 50 kb on chromosome 4q28.3 Fibrinogen participates in hemostasis by forming the insoluble fibrin clot and mediating platelet aggregation.4 Moreover, fibrinogen is a class II protein involved in the acute phase response to injury and stress (for review, see5).
Congenital fibrinogen disorders include afibrinogenemia, hypofibrinogenemia, and dysfibrinogenemia, characterized by the complete absence or extremely low levels of plasma fibrinogen, by reduced amounts of plasma fibrinogen, or by the presence of dysfunctional fibrinogen molecules, respectively. Congenital afibrinogenemia (MIM 202 400) is a rare autosomal recessive disorder with a high rate of consanguinity within affected families, and it is characterized by bleeding manifestations that often start at birth with uncontrolled umbilical cord hemorrhages. Bleeding from mucosal membranes, hematomas, hemarthroses, and hemorrhages after trauma and surgery are relatively frequent.6-8 However, the disease usually can be controlled by fibrinogen replacement therapy.
Although several missense mutations in the 3 fibrinogen genes have been identified as the cause of dysfibrinogenemia, hypofibrinogenemia, or both,9 congenital afibrinogenemia, originally described in 1920,10 has been associated only with a recently reported homozygous 11-kb deletion of the Aα-chain gene.11 However, the existence of different genetic alterations in inherited afibrinogenemia was suggested by a previous study on 2 patients with afibrinogenemia who had no gross alterations within the fibrinogen cluster as revealed by Southern blot analysis.12
In this study 2 patients with afibrinogenemia, an Italian and an Iranian, each the offspring of a marriage between first cousins, were studied. Both had plasma fibrinogen levels that could not be measured by functional assay, but extremely low levels (between 0.05% and 0.5% of the average normal value) were detected by enzyme immunoassay. Two different homozygous missense mutations (Leu353Arg and Gly400Asp), neither of which has yet been described, were identified in exons 7 and 8 of the fibrinogen Bβ-chain gene. In vitro expression of the mutant proteins demonstrated that each mutation abolished fibrinogen secretion, thus causing the extremely reduced fibrinogen levels observed in the patients.
Methods
Materials
Full-length Aα, Bβ and γ cDNA, cloned in the expression vector pRSV-Neo, were kind gifts of Dr C. M. Redman (Lindsley F. Kimball Research Institute, New York Blood Center, New York, NY). Rabbit polyclonal antibodies to human fibrinogen were from DAKO (Copenhagen, Denmark). Oligonucleotides were purchased from Life Technologies (Inchinnan, Paisley, UK).
Coagulation tests
Fibrinogen was measured in plasma by a functional assay based on fibrin polymerization time using a commercial kit (Laboratoire Stago, Asnieres, France), and it was measured in plasma and washed platelets by enzyme immunoassay.13 The sensitivity of the functional assay was 5 mg/dL and that of the immunoassay was 0.02 mg/dL.
DNA extraction
All examined subjects signed informed consent. Genomic DNA was extracted from blood samples using the Nucleon BACC1 kit (Amersham Pharmacia Biotech, Uppsala, Sweden).
Linkage analysis
The polymorphic locus used for linkage analysis was the tetranucleotide repeat FGA-i3, located in intron 3 of the fibrinogen Aα-chain gene.14 Polymerase chain reaction (PCR) was performed on genomic DNA under standard conditions, and amplified products were separated using an automated 370A DNA sequencer (PE Biosystems, Foster City, CA). Genotyping was performed using the Genescan 3.1 software (PE Biosystems) and the LINKAGE package was used for linkage analysis.15 The average maximum expected LOD score was calculated using the SIMULATE software.16
Sequence analysis
DNA sequencing was performed on both strands, either directly on purified-PCR products or on plasmids using the Taq dye-deoxy terminator method and an automated 370A DNA sequencer (PE Biosystems). All primers used for sequencing were designed on the basis of known sequences of the fibrinogen genes and intergenic regions (Genbank accession numbers M64 982, M64 983, M10 014, and U36 478). Factura and Sequence Navigator software packages (PE Biosystems) were used for mutation detection.
Allele-specific oligonucleotide hybridization
20 ng PCR products, corresponding to regions in exon 7 or 8 of the fibrinogen Bβ-chain gene, were blotted onto nylon membranes (Amersham Pharmacia Biotech) and hybridized to 32P-labeled allele-specific oligonucleotides (ASO) according to standard procedures.17 Wild-type and mutant probes are listed in Table 1.
Oligonucleotide primers used in ASO hybridizations and in site-directed mutagenesis
| Primer . | Sequence . | Position* . | Location (FGB exon) . |
|---|---|---|---|
| FGB-L353† | 5′ TAATGCCCTCATGGATG 3′ | 7148-7164 | 7 |
| FGB-R353† | 5′ TAATGCCCGCATGGATG 3′ | 7148-7164 | 7 |
| FGB-G400† | 5′ AGACGGTGGTGGATGGT 3′ | 7907-7923 | 8 |
| FGB-D400† | 5′ AGACGGTGATGGATGGT 3′ | 7907-7923 | 8 |
| L353R-F‡ | 5′ CCGGTAATGCCCGCATGGATGGAGC 3′ | 7144-7168 | 7 |
| L353R-R‡ | 5′ GCTCCATCCATGCGGGCATTACCGG 3′ | 7168-7144 | 7 |
| G400D-F‡ | 5′ AAGAAGACGGTGATGGATGGTGGTA 3′ | 7903-7927 | 8 |
| G400D-R‡ | 5′ TACCACCATCCATCACCGTCTTCTT 3′ | 7927-7903 | 8 |
| Primer . | Sequence . | Position* . | Location (FGB exon) . |
|---|---|---|---|
| FGB-L353† | 5′ TAATGCCCTCATGGATG 3′ | 7148-7164 | 7 |
| FGB-R353† | 5′ TAATGCCCGCATGGATG 3′ | 7148-7164 | 7 |
| FGB-G400† | 5′ AGACGGTGGTGGATGGT 3′ | 7907-7923 | 8 |
| FGB-D400† | 5′ AGACGGTGATGGATGGT 3′ | 7907-7923 | 8 |
| L353R-F‡ | 5′ CCGGTAATGCCCGCATGGATGGAGC 3′ | 7144-7168 | 7 |
| L353R-R‡ | 5′ GCTCCATCCATGCGGGCATTACCGG 3′ | 7168-7144 | 7 |
| G400D-F‡ | 5′ AAGAAGACGGTGATGGATGGTGGTA 3′ | 7903-7927 | 8 |
| G400D-R‡ | 5′ TACCACCATCCATCACCGTCTTCTT 3′ | 7927-7903 | 8 |
Numbering according to Genbank accession number M64983.
Probes used for ASO hybridization.
Oligonucleotides used for site-directed mutagenesis.
FGB, fibrinogen Bβ-chain gene.
Site-directed mutagenesis
Leu353Arg and Gly400Asp mutations were introduced in the pRSV-Neo-Bβ plasmid by the Quick-change site-directed mutagenesis kit (Stratagene, La Jolla, CA), according to the manufacturer's instructions, using the oligonucleotides L353R-F, L353R-R, and G400D-F, and the oligonucleotide G400D-R, respectively (Table 1).
Cell cultures, transfections, and metabolic labeling
The human hepatoma cell line HepG2 was cultured in Dulbecco's modified Eagle's medium (DMEM) and Ham's F12 media (1:1, vol/vol), supplemented with 10% fetal calf serum. The African green monkey kidney cell line COS-1 was cultured in DMEM containing 10% fetal calf serum. Antibiotics (100 IU/mL penicillin and 100 μg/mL streptomycin) and glutamine (1%) were added to the media. Cells were grown at 37°C in a humidified atmosphere of 5% CO2 and 95% air. Using CELL FECTIN (Life Technologies), semiconfluent COS-1 cells were transfected with equimolar amounts (5 μg each) pRSV-Neo-Aα, pRSV-Neo-γ, and pRSV-Neo-Bβ plasmids, the latter either wild-type or mutant. Thirty-six hours after transfection, the cells were labeled by incubating each 100-mm dish for 3 hours with 5 mL methionine- and cysteine-free DMEM (ICN Biomedicals, High Wycombe, Berks, UK) supplemented with 150 μCil-[35S] methionine and 60 μCil-[35S] cysteine (Amersham Pharmacia Biotech), with 10% dialyzed fetal calf serum, 0.1 mg/mL heparin, and 1% glutamine. In pulse-chase experiments, cells were pulse labeled for 3 hours as described above and then chase incubated for various periods of time (30, 90, 180 minutes) with fresh medium containing 10 mmol/L methionine and 10 mmol/L cysteine.
Immunoprecipitation and sodium dodecyl sulphate– polyacrylamide gel electrophoresis
After metabolic labeling, culture media were collected and a protease inhibitor mixture was added (Complete; Roche, Basel, Switzerland). Five milliliters medium from each plate was centrifuged to remove cell debris and concentrated using a Centricon Plus-20 column (Millipore, Bedford, MA). Cells were washed 3 times with prechilled phosphate-buffered saline (PBS) and lysed for 1 hour on ice with lysis buffer containing 1 × PBS, 1% Triton X-100, and the protease inhibitor mixture. Cell lysates were centrifuged to remove cell debris. Rabbit antihuman fibrinogen antibodies, preadsorbed with protein G-Sepharose (Sigma Chemical, St. Louis, MO) at room temperature for 3 hours, were added to cell lysates and media and incubated for 4 hours on ice. Pellets were collected by centrifugation for 3 minutes and washed 3 times with lysis buffer. The immunoprecipitated proteins were released from protein G-Sepharose by boiling for 5 minutes in sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer. Samples were analyzed by SDS-PAGE according to Laemmli18 and were detected by autoradiography or by a STORM PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Results
Patient data
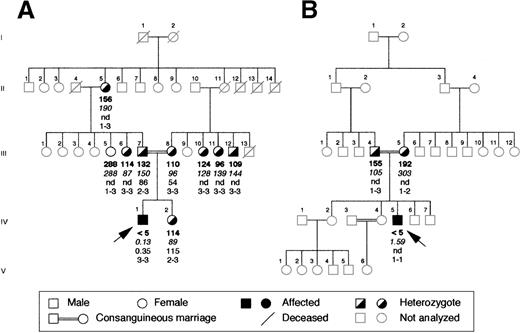
The proband of the Italian family was a 17-year old boy whose parents were first cousins (Figure 1A). The diagnosis of afibrinogenemia was made at birth because the patient had life-threatening bleeding from the umbilical cord, which rendered necessary transfusion with whole blood and fibrinogen concentrates. After that he had relatively mild symptoms, such as epistaxis and posttraumatic muscle hematomas. His parents, sister, and other family members available for study were asymptomatic. In the proband, plasma fibrinogen levels could not be measured by functional assay (less than 5 mg/dL; normal range, 160-400 mg/dL), and they were found to be very low by enzyme immunoassay (0.13 mg/dL; normal range, 160-400 mg/dL). Platelet levels of immunoreactive fibrinogen were also very low (0.35 × 109 platelets; normal range, 60-190 μg/109 platelets). His parents and sister had approximately half the normal levels of plasma fibrinogen and normal intraplatelet levels.
Family pedigrees of the 2 probands with congenital afibrinogenemia analyzed in this study.
(A) Pedigree of the Italian and (B) of the Iranian family. Plasma functional fibrinogen levels (mg/dL), immunoreactive fibrinogen levels (mg/dL), intraplatelet levels (μg/109 platelets) and haplotypes of the FGA-i3 tetranucleotide repeat marker (alleles numbered from the largest to the smallest) are indicated in this order below each symbol. Arrows indicate the probands. nd, not done.
Family pedigrees of the 2 probands with congenital afibrinogenemia analyzed in this study.
(A) Pedigree of the Italian and (B) of the Iranian family. Plasma functional fibrinogen levels (mg/dL), immunoreactive fibrinogen levels (mg/dL), intraplatelet levels (μg/109 platelets) and haplotypes of the FGA-i3 tetranucleotide repeat marker (alleles numbered from the largest to the smallest) are indicated in this order below each symbol. Arrows indicate the probands. nd, not done.
The proband of the Iranian family (Figure 1B) was a 24-year-old man, also born of a consanguineous marriage. He bled at birth from the umbilical cord and later during circumcision, and he was treated with whole blood and fresh-frozen plasma on both occasions. Subsequently, he suffered repeatedly from muscle hematomas and hemarthroses that occurred spontaneously or after minor trauma. His parents, 2 brothers, and 2 sisters were asymptomatic, and there was no history of bleeding in the other family members not investigated. In the proband, fibrinogen could not be measured by functional assay, and very low values (1.6 mg/dL) were detected by the more sensitive immunoassay. His father's plasma fibrinogen level was half the normal level, and his mother had a low borderline level of functional plasma fibrinogen and a normal level of immunoreactive fibrinogen. Platelet samples could not be obtained.
Mutational analysis
The absence of gross deletions in the fibrinogen cluster has been verified by Southern blot analysis and by long PCR amplification of each fibrinogen gene (data not shown). To verify whether the fibrinogen cluster was still a candidate for mutation screening, linkage analysis using the tetranucleotide repeat FGA-i3 marker located within the Aα-chain gene was performed in the Italian family. Evaluation of plasma levels of functional fibrinogen allowed us to classify phenotypically the 9 available members of this family as homozygous normal (more than 160 mg/dL) or heterozygous (less than 160 mg/dL) (Figure 1A). Linkage analysis resulted in an LOD score of 2.02 at θ = 0. This LOD score, close to the average maximum expected LOD score of 2.37 calculated for this pedigree, suggested the existence of linkage to the fibrinogen cluster. Because of the small number of available members, the Iranian pedigree could not be considered for this analysis. The proband received the same FGA-i3 allele from both parents, as expected for the affected offspring of a consanguineous marriage (Figure 1B).
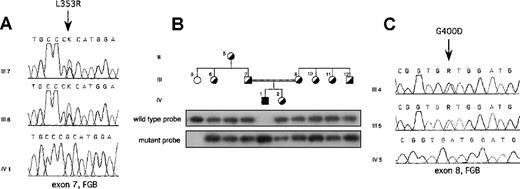
On the basis of these results in each proband, the entire coding region was sequenced, including exon–intron boundaries and approximately 300 bp of the promoter region of each fibrinogen gene. In the Italian proband, a single homozygous T-G transversion was found in exon 7 of the fibrinogen Bβ-chain gene at position 7156 (numbering according to Genbank accession number M64983) (Figure2A). This resulted in a missense mutation leading to a Leu-Arg substitution at position 353 (L353R). The proband's parents were heterozygous for the mutation (Figure 2A). All available family members were tested for the mutation by dot blot hybridization with ASO probes. Besides the proband's parents, the mutation was present in the heterozygous state in the proband's grandmother and in phenotypically heterozygous relatives (Figure 2B). Two hundred aploid genomes from unrelated persons in a northern Italian control population were also analyzed, and the mutation was absent. In the Iranian proband, a single G-A homozygous transition at position 7915 was identified in exon 8 of the fibrinogen Bβ-chain gene, replacing glycine with aspartate at position 400 (G400D) (Figure2C). This mutation was also present in the heterozygous state in the proband's parents (Figure 2C), but not in 200 unrelated Iranian aploid control genomes. Both amino acid substitutions are nonconservative and reside in the C-terminal globular portion of the Bβ-chain.
L353R and G400D mutations.
(A) Electropherograms showing the mutation identified in the Italian proband with afibrinogenemia. The T-G transversion leading to the L353R mutation, indicated by an arrow, was present in the heterozygous state in both parents. (B) ASO hybridization showing cosegregation of the L353R mutation and clinical phenotype in the Italian kindred. Family members were phenotypically classified as homozygous normal or heterozygote on the basis of plasma fibrinogen levels. Individuals within the pedigree are positioned above the corresponding lanes. Wild-type (FGB-L353) and mutant (FGB-R353) probes are listed in Table1. (C) Electropherograms showing the mutation identified in the Iranian proband with afibrinogenemia. The G-A transition leading to the G400D mutation, indicated by an arrow, was present in the heterozygous state in both parents. Family members are labeled as in Figure 1. FGB, fibrinogen Bβ-chain gene; K, G or T nucleotide; R, A or G nucleotide.
L353R and G400D mutations.
(A) Electropherograms showing the mutation identified in the Italian proband with afibrinogenemia. The T-G transversion leading to the L353R mutation, indicated by an arrow, was present in the heterozygous state in both parents. (B) ASO hybridization showing cosegregation of the L353R mutation and clinical phenotype in the Italian kindred. Family members were phenotypically classified as homozygous normal or heterozygote on the basis of plasma fibrinogen levels. Individuals within the pedigree are positioned above the corresponding lanes. Wild-type (FGB-L353) and mutant (FGB-R353) probes are listed in Table1. (C) Electropherograms showing the mutation identified in the Iranian proband with afibrinogenemia. The G-A transition leading to the G400D mutation, indicated by an arrow, was present in the heterozygous state in both parents. Family members are labeled as in Figure 1. FGB, fibrinogen Bβ-chain gene; K, G or T nucleotide; R, A or G nucleotide.
In vitro protein expression
To determine whether the L353R and G400D mutations affect fibrinogen assembly or secretion, mutant proteins were transiently expressed in COS-1 cells that do not express fibrinogen. The 2 point mutations were independently introduced by site-directed mutagenesis in a Bβ-chain expression plasmid (pRSV-Neo-Bβ)19 and checked by sequencing. Each mutated plasmid was transiently cotransfected in COS-1 cells, together with plasmids pRSV-Neo-Aα and pRSV-Neo-γ expressing the wild-type Aα- and γ-chains, respectively.20Transfections with equal amounts of wild-type and each of the mutated Bβ-chain cDNA were also performed, to mimic the heterozygous state. Transfected cells were incubated with l-[35S] methionine and l-[35S] cysteine. Fibrinogen expression was analyzed by immunoprecipitation using polyclonal antihuman fibrinogen antibodies and protein G-Sepharose and then by SDS-PAGE.
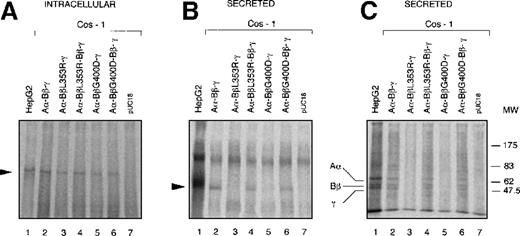
After SDS-PAGE under nonreducing conditions, a specific fibrinogen band was observed in cell lysates and culture media of COS-1 cells transfected with plasmids containing wild-type fibrinogen cDNA (Figures3A, 3B). The same band was also observed in untransfected control fibrinogen-expressing HepG2 cells (Figures 3A,3B). By contrast, in cells transfected with plasmids expressing L353R or G400D Bβ-chains, hexameric fibrinogen was detectable only in cell lysates (Figure 3A) but was undetectable in the corresponding culture media (Figure 3B). Cotransfection of wild-type and each of the mutated Bβ-chain cDNA restored fibrinogen secretion to the medium (Figure3B). Mock-transfected (pUC18) COS-1 cells showed only background bands in cell lysate and medium (Figures 3A, 3B). Immunoprecipitated proteins from the same culture media as in Figure 3B were also separated on SDS-PAGE under reducing conditions. The expected pattern of Aα, Bβ and γ-chains was observed in fibrinogen-containing media (Figure 3C).
Expression of wild-type and mutant fibrinogen chains in COS-1 cells.
Intracellular and secreted fibrinogen from COS-1 cells transfected with wild-type Aα and γ-chains together with wild-type and/or L353R or G400D Bβ, chains have been analyzed. Metabolic labeling, immunoprecipitation, and SDS-PAGE were carried out as described in “Materials and Methods.” (A) Immunoprecipitable proteins in cell lysates separated on 4% SDS-PAGE under nonreducing conditions. The arrowhead indicates the 340-kd fibrinogen complex. (B) Immunoprecipitable proteins in the culture media separated on 4% SDS-PAGE under nonreducing conditions. The arrowhead indicates the 340-kd fibrinogen complex. (C) Immunoprecipitable proteins in the culture media separated on 10% SDS-PAGE under reducing conditions. The positions of Aα, Bβ, and γ chains are indicated. The position of molecular weight markers (MW) in kd is denoted at the right-hand side of the figure.
Expression of wild-type and mutant fibrinogen chains in COS-1 cells.
Intracellular and secreted fibrinogen from COS-1 cells transfected with wild-type Aα and γ-chains together with wild-type and/or L353R or G400D Bβ, chains have been analyzed. Metabolic labeling, immunoprecipitation, and SDS-PAGE were carried out as described in “Materials and Methods.” (A) Immunoprecipitable proteins in cell lysates separated on 4% SDS-PAGE under nonreducing conditions. The arrowhead indicates the 340-kd fibrinogen complex. (B) Immunoprecipitable proteins in the culture media separated on 4% SDS-PAGE under nonreducing conditions. The arrowhead indicates the 340-kd fibrinogen complex. (C) Immunoprecipitable proteins in the culture media separated on 10% SDS-PAGE under reducing conditions. The positions of Aα, Bβ, and γ chains are indicated. The position of molecular weight markers (MW) in kd is denoted at the right-hand side of the figure.
To evaluate whether the observed absence of mutant fibrinogens in culture media resulted from an impairment of fibrinogen secretion or from increased intracellular or extracellular degradation, a pulse-chase experiment was performed. Transfected COS-1 cells were labeled for 3 hours with l-[35S] methionine and l-[35S] cysteine and were chase incubated with an excess of the corresponding unlabeled amino acids for various periods up to 3 hours. As shown in Figure4A, in lysates from cells transfected with the 3 wild-type cDNA, immunoprecipitated radioactive fibrinogen markedly declined at 90 minutes and disappeared at 180 minutes. By contrast, in lysates from cells transfected with plasmids expressing L353R or G400D Bβ-chains, hexameric fibrinogen persisted intracellularly up to 3 hours (Figure 4A). Secreted fibrinogen was present, at the analyzed chase-periods, in culture media from COS-1 cells cotransfected with Aα, Bβ, and γ wild-type cDNA, whereas it was absent in culture media from cells cotransfected with Aα and γ wild-type chains and either Bβ mutant chain expressing plasmids (Figure 4B).
Intracellular retention of L353R and G400D mutant fibrinogens.
COS-1 cells were incubated with l-[35S] methionine and l-[35S] cysteine for 3 hours and then chase incubated with 10 mmol/L l-methionine andl-cysteine for various periods of time up to 3 hours. At the specified chase periods (0, 30, 90, and 180 minutes), radioactive fibrinogen was immunoprecipitated from cell lysates and from the corresponding culture media, as described in “Materials and Methods.” Intracellular (A) and secreted (B) immunoprecipitable proteins were separated by 4% SDS-PAGE under nonreducing conditions. The arrowheads indicate the 340-kd fibrinogen complex. Intracellular and secreted fibrinogen at the end of the pulse period from control HepG2 cells was also analyzed. In both panels, pUC18 lane contains immunoprecipitable proteins at the end of the pulse period from COS-1 cells transfected with the unrelated pUC18 plasmid.
Intracellular retention of L353R and G400D mutant fibrinogens.
COS-1 cells were incubated with l-[35S] methionine and l-[35S] cysteine for 3 hours and then chase incubated with 10 mmol/L l-methionine andl-cysteine for various periods of time up to 3 hours. At the specified chase periods (0, 30, 90, and 180 minutes), radioactive fibrinogen was immunoprecipitated from cell lysates and from the corresponding culture media, as described in “Materials and Methods.” Intracellular (A) and secreted (B) immunoprecipitable proteins were separated by 4% SDS-PAGE under nonreducing conditions. The arrowheads indicate the 340-kd fibrinogen complex. Intracellular and secreted fibrinogen at the end of the pulse period from control HepG2 cells was also analyzed. In both panels, pUC18 lane contains immunoprecipitable proteins at the end of the pulse period from COS-1 cells transfected with the unrelated pUC18 plasmid.
Discussion
Inherited afibrinogenemia is a rare recessive bleeding disease characterized by fibrinogen deficiency in plasma and platelets. Even though, strictly speaking, the term should refer to the complete absence of fibrinogen, minute amounts of fibrinogen can sometimes be detected through sensitive immunoassays. Asymptomatic heterozygotes usually have approximately half the normal levels of functional and immunoreactive fibrinogen. The 2 probands studied here, both the offspring of consanguineous marriages, had unmeasurable, clottable fibrinogen and very low plasma levels of immunoreactive fibrinogen (0.13 mg/dL and 1.59 mg/dL). Genotypically heterozygous relatives had approximately half the normal fibrinogen levels and a good concordance between functional and immunologic values. The only notable exception was the mother of the Iranian proband, who had low borderline levels of functional plasma fibrinogen (192 mg/dL) with 50% higher immunoreactive fibrinogen (303 mg/dL). Some kindred afibrinogenemic putative carriers with normal fibrinogen levels have been described7; however, there is no clear explanation for the discrepancies between functional and immunologic test results except those that can be attributed to methodologic variability. In the Italian family, intraplatelet fibrinogen was also measured, and levels were normal in the 3 heterozygous members (III-7, III-8, and IV-2) studied. The intraplatelet fibrinogen level of the proband was very low (approximately 0.3% of the average normal level) but was less reduced than plasma fibrinogen levels (0.05%). This phenomenon may be explained by the ability of platelets actively to take up fibrinogen from plasma.21
Inherited fibrinogen deficiencies may have different causes, such as decreased protein synthesis, increased intracellular or circulatory degradation, defective assembly or secretion, or a combination of these defects. The only previously identified genetic basis of congenital hypofibrinogenemia and afibrinogenemia is a homozygous truncation of the Aα-chain gene. The deletion of 25% of the Aα-chain identified in fibrinogen Marburg was associated with mild hypofibrinogenemia (60 mg/dL, immunoassay),22 whereas in fibrinogen Otago, the more severe truncation of 56% of the Aα-chain was associated with severe hypofibrinogenemia (6 mg/dL, functional assay).23The truncation of 26% of the Aα-chain in fibrinogen Milano III was associated with normal circulating levels (260 mg/dL, functional assay).24 All these cases are also characterized by the presence of dysfunctional fibrinogen molecules and should therefore be defined as dysfibrinogenemia or hypo-disfibrinogenemia. The severest Aα-chain truncation is the 11-kb deletion recently identified by Neerman-Arbez et al,11 involving the whole gene except exon 1. In this case, fibrinogen was undetectable either by functional assay or immunoassay.
In this article, we report the identification of new pathogenetic defects underlying congenital afibrinogenemia. Two different missense mutations (L353R and G400D) were identified in the C-terminal globular portion of fibrinogen Bβ-chain (Figure5A). These nonconservative substitutions involve strictly conserved residues in a region highly conserved among vertebrates (Figure 5B). Inspection of the fibrinogen crystal structure2 showed that Leu353 and Gly400 were part of the same region of the protein, in the hydrophobic core of the Bβ-chain C-terminal region. Gly400 was solvent inaccessible, in contact with Phe262, Tyr269, Phe375, Tyr404, and Asn413. Similarly, Leu353 was buried beneath the protein surface, extensively interacting with the side chains of Tyr326, Val342, His370, Trp402, Ala410, and Trp437. The L353R and G400D mutations introduced bulky, charged side chains to these hydrophobic regions. Clearly, such drastic amino acid replacements would be expected to perturb the packing of the side chains in the Bβ-chain core and would likely affect the stability of the protein, which might be unable to fold correctly, with the ultimate result of impaired assembly and release of an active fibrinogen molecule.
Position of the L353R and G400D mutations in the structure of the fibrinogen Bβ chain.
(A) C-alpha trace of the AαBβγ trimer, outlining the position of Gly400 and Leu353, produced using the coordinates under the Protein Data Bank entry 1FZA. FGA, FGB, and FGG indicate the Aα, Bβ, and γ fibrinogen chains, respectively. N- and C-terminal residues of each chain are indicated. Light gray, black, and gray correspond to Aα, Bβ, and γ chains, respectively. (B) Multiple alignment of human, rat, bovine, xenopus, chicken, and lamprey fibrinogen Bβ chain in the region containing the 2 identified mutations. Identical amino acids are boxed. Positions of L353R and G400D mutations are indicated by arrows. Amino acid sequences were obtained from Swiss-Prot database (accession numbers: P02675 [human], P14480 [rat], P02676 [bovine], AAA85283 [xenopus], Q02020 [chicken], and FIBB_PETMA_2 [lamprey]) and numbered; the signal peptide is omitted. Secondary structures shown below the alignments (with cylinders representing α-helices and the arrow representing the β-strand) refer to the human protein, drawn from Spraggon et al2 data.
Position of the L353R and G400D mutations in the structure of the fibrinogen Bβ chain.
(A) C-alpha trace of the AαBβγ trimer, outlining the position of Gly400 and Leu353, produced using the coordinates under the Protein Data Bank entry 1FZA. FGA, FGB, and FGG indicate the Aα, Bβ, and γ fibrinogen chains, respectively. N- and C-terminal residues of each chain are indicated. Light gray, black, and gray correspond to Aα, Bβ, and γ chains, respectively. (B) Multiple alignment of human, rat, bovine, xenopus, chicken, and lamprey fibrinogen Bβ chain in the region containing the 2 identified mutations. Identical amino acids are boxed. Positions of L353R and G400D mutations are indicated by arrows. Amino acid sequences were obtained from Swiss-Prot database (accession numbers: P02675 [human], P14480 [rat], P02676 [bovine], AAA85283 [xenopus], Q02020 [chicken], and FIBB_PETMA_2 [lamprey]) and numbered; the signal peptide is omitted. Secondary structures shown below the alignments (with cylinders representing α-helices and the arrow representing the β-strand) refer to the human protein, drawn from Spraggon et al2 data.
It has been demonstrated, by deletion experiments, that the C-terminal half of the Bβ-chain, which is part of the D-domain, is not necessary for the assembly and secretion of fibrinogen.25Nevertheless the assembly of a misfolded protein could result in severe impairment of protein secretion. Numerous genetic diseases are known in which a single amino acid change can lead to failure of protein export,26 as evidenced in cystic fibrosis,27juvenile pulmonary emphysema,28 osteogenesis imperfecta,29 juvenile diabetes insipidus,30and hypercholesterolemia.31
To assess whether L353R and G400D mutations could be responsible for cases of congenital afibrinogenemia, the mutant Bβ-chains were expressed, together with the other 2 wild-type chains, in COS-1 cells. This cell line does not express fibrinogen and represents a widely used in vitro system for studying fibrinogen assembly and secretion by transient or stable transfections.20 Transfection experiments suggested that either mutation abolished fibrinogen secretion because mutant fibrinogen molecules were synthesized and assembled intracellularly but were undetectable in culture media. The observation that mutant fibrinogen molecules are assembled intracellularly might explain the minute amounts of protein measured in a patient's plasma using a sensitive immunoassay. This finding is consistent with the concept that missense mutations do not necessarily block protein production completely. The hypothesis that the main defect caused by these 2 mutations was at the secretion level was confirmed by the results of pulse-chase experiments showing the lack of cellular depletion of L353R and G400D mutant fibrinogens.
In conclusion, 2 different homozygous missense mutations were identified in the same region of the Bβ-chain in 2 patients with afibrinogenemia. Transfection experiments in COS-1 cells with plasmids expressing the mutant proteins demonstrated, for the first time, that these mutations can abolish fibrinogen secretion without affecting the protein synthesis. These results demonstrated that congenital afibrinogenemia can be caused by point mutations that impair the normal fibrinogen secretion pathway.
Acknowledgments
We thank Dr C. Redman for kindly providing Aα-, Bβ-, and γ-chain-expressing pRSV-Neo plasmids. We thank Dr A. Mattevi for helpful discussion on the structural effect of mutations on the protein folding and Drs A. Clivio, C. Pinter, and B. Pedrotti for helpful suggestions on expression and immunoprecipitation experiments. We also thank the members of both families for their participation in this study.
Supported by the Ministero dell'Università e della Ricerca Scientifica e Tecnologica (MURST 60%) and by IRCCS Maggiore Hospital, Milan, Italy.
Reprints:Maria Luisa Tenchini, Dipartimento di Biologia e Genetica per le Scienze Mediche, via Viotti, 3/5-20133 Milano, Italy; e-mail: marialuisa.tenchini@unimi.it.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

![Fig. 4. Intracellular retention of L353R and G400D mutant fibrinogens. / COS-1 cells were incubated with l-[35S] methionine and l-[35S] cysteine for 3 hours and then chase incubated with 10 mmol/L l-methionine andl-cysteine for various periods of time up to 3 hours. At the specified chase periods (0, 30, 90, and 180 minutes), radioactive fibrinogen was immunoprecipitated from cell lysates and from the corresponding culture media, as described in “Materials and Methods.” Intracellular (A) and secreted (B) immunoprecipitable proteins were separated by 4% SDS-PAGE under nonreducing conditions. The arrowheads indicate the 340-kd fibrinogen complex. Intracellular and secreted fibrinogen at the end of the pulse period from control HepG2 cells was also analyzed. In both panels, pUC18 lane contains immunoprecipitable proteins at the end of the pulse period from COS-1 cells transfected with the unrelated pUC18 plasmid.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/95/4/10.1182_blood.v95.4.1336.004k16_1336_1341/6/m_bloo00416004w.jpeg?Expires=1767786666&Signature=diZJLMVFx6-5te4pORgr4owUIVUrFhk4Jaj5nkelWSBS0Ibgu8jjoJkpMUQc~-CbDXBCLzKMynQo2G2qAiqpsyV6NM3fvXp3OnvnhXhssVglv~KSF6a5z71-43mQCCfgJ4d5Dg1PAuZAYZwpsS-NPfJDgS7fP4hWV22Awk2DLXR5gyyL9uEPje9V~pOHnpNuo1mvqtHLP81gC6ang6b9aNa7sOy1kQtKkKxHWJSMTa0RoK4Xc~NRVWETF1YYE7hBAEInayoFT9gre0Gby0q4J8exe4BjyVTi6p9eWC9JtqM4YS4wP14l7MzJRN9A25zm9miH-UWb98t2p6sOx3aYiw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 5. Position of the L353R and G400D mutations in the structure of the fibrinogen Bβ chain. / (A) C-alpha trace of the AαBβγ trimer, outlining the position of Gly400 and Leu353, produced using the coordinates under the Protein Data Bank entry 1FZA. FGA, FGB, and FGG indicate the Aα, Bβ, and γ fibrinogen chains, respectively. N- and C-terminal residues of each chain are indicated. Light gray, black, and gray correspond to Aα, Bβ, and γ chains, respectively. (B) Multiple alignment of human, rat, bovine, xenopus, chicken, and lamprey fibrinogen Bβ chain in the region containing the 2 identified mutations. Identical amino acids are boxed. Positions of L353R and G400D mutations are indicated by arrows. Amino acid sequences were obtained from Swiss-Prot database (accession numbers: P02675 [human], P14480 [rat], P02676 [bovine], AAA85283 [xenopus], Q02020 [chicken], and FIBB_PETMA_2 [lamprey]) and numbered; the signal peptide is omitted. Secondary structures shown below the alignments (with cylinders representing α-helices and the arrow representing the β-strand) refer to the human protein, drawn from Spraggon et al2 data.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/95/4/10.1182_blood.v95.4.1336.004k16_1336_1341/6/m_bloo00416005y.jpeg?Expires=1767786666&Signature=k6UVYrGEEsKYClNJbgza8t1tGChzhZREyp6~f~SI402y0aO~5hshmcR2Lozwrb~aXZQVF7gWYguU9wmF56ipaZAXrfnSvaFZyktBs~hjsR37SD5MWp8D9n6VR2e0pl3r~VV9HJUfyNsUHS47SMzc42mOcWUE~0k530thE0R~t5jylipayjDM7QRqvH4bmNE4OPOSKVo5~9DtuT4FWpGE7vA9pZRCEihA0stAnnEnwGpcFTkcOrkP2ctvZQF6dZkhD-msiTK~p7eIU2U8jyir3wjU4KyD7KmUDSN1LU5UWOT0w63eJwArB1NFsSeFaa7cWtTxRIBjm3AUIk4BxEbVlg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal