Mice deficient in intercellular adhesion molecule-1 (ICAM-1), lacking membranous ICAM-1, show a normal development but abnormalities of inflammatory and immune functions. Although the membrane-bound form of ICAM-1 is not detectable in the mutant strain, circulating ICAM-1 (cICAM) is present in serum from ICAM-1-deficient mice in similar amounts as in serum from wild-type mice. These findings were confirmed in vitro by flow cytometric analysis of lipopolysaccharide-stimulated spleen cells, and cICAM-enzyme-linked immunosorbent assay analysis of supernatants of cultured spleen cells. To analyze for the source of cICAM-1, spleen cell RNA was isolated and ICAM-1 RNA was amplified by reverse transcriptase-polymerase chain reaction using primers binding in the 5′ and 3′ untranslated regions. Different fragments were cloned and sequenced. In wild-type RNA the common 5 domain form of ICAM-1 was identified. In RNA from ICAM-1 mutant mice only 3 smaller fragments were found. Sequencing these fragments identified 3 alternatively spliced isoforms of ICAM-1, lacking 2 or 3 extracellular domains. However, in all spliced fragments the transmembrane domain was included. Therefore, we postulate that circulating forms of ICAM-1 are generated by proteolytic cleavage of membranous ICAM-1. The data indicate that the expression of membranous ICAM-1 and the appearance of circulating forms in serum are independently regulated mechanisms.
Intercellular adhesion molecule-1 (ICAM-1), a member of the immunoglobulin (Ig) superfamily, consists of 5 extracellular Ig-like domains, a transmembrane domain, and a short cytoplasmic region. ICAM-1 is important in adhesion of leukocytes and endothelial leukocyte migration through binding to lymphocyte function-associated antigen (LFA)-1.1-3 ICAM-1 also plays an important role in various immune responses4 and it can provide a costimulatory signal in T-cell activation.5
The ICAM-1 gene consists of 7 exons. Exon 1 encodes the signal sequence, exons 2 to 6 encode the Ig-like domains 1 to 5, respectively, and exon 7 encodes the transmembrane domain and the short cytoplasmic tail.6 Two different ICAM-1-deficient mice have been generated by homologous recombination of a neomycin resistance gene into distinct exons of the ICAM-1 gene—one with the neomycin gene inserted in exon 47 and the other in exon 5.8 Both mice had normal development, but showed abnormalities of inflammatory and immune functions, that is, resistance to lethal effects of high doses of lipopolysaccharide (LPS)7 or protection against ischemic renal injury.9 Although residual ICAM-1 expression could be detected in different tissues of exon 5 mutant mice, no expression of ICAM-1 was identified in exon 4 mutant mice.7 10
Circulating forms of ICAM-1 (cICAM-1) have been detected in normal human serum11,12 and elevated levels were described in various inflammatory and immune disorders.11-18 However, the pathophysiologic function and the molecular origin of cICAM-1 are still under investigation. To approach the function of cICAM-1, a recombinantly produced soluble form of ICAM-1 (rICAM-1) was used in in vitro and in vivo experiments. We described an immunomodulatory function of rICAM-1 in the inhibition of autoimmune T-cell proliferation.19 Recently, we could show an immunomodulatory potential of rICAM-1 by reducing leukocyte adherence after ischemia/reperfusion20 and by inhibiting insulitis and the development of type I diabetes in nonobese diabetic (NOD) mice.21
The molecular origin of cICAM-1 is currently unclear. Two mechanisms are discussed by which serum ICAM-1 can be generated. Evidence for proteolytic cleavage of membrane-integrated human ICAM-1 has been reported by Budnik et al.22 In contrast, Wakatsuki et al23 identified an alternatively spliced form of human ICAM-1 messenger RNA (mRNA) lacking the transmembrane domain.
Here we describe cICAM-1 in ICAM-1-deficient mice. Because ICAM-1–exon 4 mutant mice do not express ICAM-1 on the membrane, this mouse strain seems to be a good model for studying the natural source of cICAM-1. In a reverse transcriptase-polymerase chain reaction (RT-PCR) approach different ICAM-1 isoforms could be detected in mRNA of ICAM-1 mutant mice. Interestingly, the transmembrane spanning region was detected in all mRNAs identified in these mice. Therefore, we postulate that cICAM-1 is produced via proteolytic cleavage.
Materials and methods
Animals
Wild-type C57BL/6 mice (Bomholtgard, Ry, Denmark) and ICAM-1 mutant mice7 were bred and maintained in our animal facilities. ICAM-1 mutant mice, originally on a mixed genetic background out of C57BL/6 and DBA mice, were backcrossed to the C57BL/6 background. Animals were screened by PCR for the H-2b tissue type using specific oligonucleotides: D17-Mit102, -Mit31, -Mit32, -Mit34, and -Mit83, which were purchased from Research Genetics (Huntsville, AL). Animals were kept under pathogen-free conditions and had free access to food and tap water. For the different experiments female and male mice were taken at ages of 14 to 19 months.
Antibodies
Antimurine ICAM-1 antibody YN1/1.7.4 was purchased from ATCC (Rockville, MD) and was purified as described before.24Further antibodies directed to murine ICAM-1 (3E2-biotin, 3E2-phycoerythrin [PE]) and antimurine CD45-fluorescein isothiocyanate (FITC) were obtained from Pharmingen (San Diego, CA). Streptavidin-horseradish peroxidase (HRP) was purchased from Zymed (San Francisco, CA).
cICAM-1 enzyme-linked immunosorbent assay (ELISA)
Serum and supernatants were analyzed for cICAM-1 concentration in 2 different “sandwich” ELISAs using monoclonal antibodies directed to the first extracellular domain of murine ICAM-1. In the first assay (ELISA-1), YN1/1.7.4 was plated at 30 μg/mL (50 μL/well) on flat bottom 96-well microtiter plates (Nunc, Wiesbaden-Bieberich, Germany) and allowed to adsorb for 1 hour at 37°C. The plates were then washed and blocked with phosphate-buffered saline (PBS)/2% bovine serum albumin (BSA) for 1 hour at 37°C. Standard and samples were diluted in PBS/1% BSA and were incubated in duplicate for 1 hour at 37°C. Biotinylated 3E2 (1:250 in PBS/1% BSA) was used as detection antibody (30 minutes, 37°C) followed by incubation with Streptavidin-HRP (1:1000 in PBS/1% BSA, 30 minutes, 37°C). Staining was performed with ABTS and hydrogen peroxide (Zymed) in citrate buffer and adsorption was measured by 405 nm. The second ELISA (ELISA-2) was a commercial murine ICAM-1 ELISA (Endogen ELISA, Biozol, Eching, Germany), which was used according to the supplier's instructions.
mRNA analysis
Total RNA was isolated from spleen tissue by homogenization in Trizol reagent (Gibco BRL, Karlsruhe, Germany). Determination of specific mRNA was performed by RT-PCR on 5 μg RNA as previously described.25 Specific oligonucleotides were designed in the untranslated regions (UTR) of the ICAM-1 complementary DNA (cDNA): 5′ UTR primer 5′-TAC CTG CAC TTT GCC CTG GCC CTG CAA TG-3′ and 3′ UTR primer 5′-GTC CTC TTA GCT CTG AGG CTA CAA GTG TGC-3′ (Gibco BRL). After a denaturation step for 5 minutes at 95°C, product amplification was performed by 35 cycles of 1 minute 95°C, 1 minute 63°C, and 2 minutes 72°C. Finally, a 7-minute incubation at 72°C completed the PCR. PCR products were resolved on 1.5% agarose gels and blotted on Hybond-N nitrocellulose (Amersham, Braunschweig, Germany). Blots were hybridized with a digoxygenin (DIG)-labeled ICAM-1–exon 2 primer (5′-GGG CCT GGT GAT GCT CAG GTA-3′) according to the supplier's instructions (DIG oligonucleotide tailing kit, Boehringer Mannheim, Mannheim, Germany). PCR products were cloned into the pCR2.1-TOPO vector (Invitrogen, Groningen, the Netherlands) and 2 or 3 clones of each fragment were sequenced on an ABM PRISM sequencer (Perkin Elmer, Weiterstadt, Germany) according to the manufacturer's instructions.
Size exclusion chromatography
Sera from wild-type and ICAM-1 mutant mice (500 μL each), and a mixture of both, were diluted 2 times in PBS, and filtered through 0.2-μm filters. The samples were applied to a Superdex-200 column with PBS as the mobile phase at a flow rate of 0.7 mL/min. Collected fractions (0.5 mL) were analyzed for the presence of cICAM-1 by ELISA-1. The column was calibrated with molecular weight markers for gel filtration, which ranged from 12 to 200 kd (Sigma).
Spleen cell isolation
Spleen cells were isolated by passing spleens through a sterile strainer. Cells were washed in Hanks' buffered saline (HBSS; Life Technologies, Eggenstein, Germany) and incubated in 2 volumes 10 mM KHCO3, 155 mM NH4Cl, 0.1 mM EDTA, pH 7.4, for 15 minutes on ice. Cells were washed again with HBSS and were resuspended in RPMI 1640 (Life Technologies) supplemented with 10% fetal calf serum (FCS) (Life Technologies) and 0.08% 2-mercaptoethanol (Merck, Darmstadt, Germany).
Spleen cell cultivation
Isolated spleen cells were cultured (5 × 105cells/well) in 48-well microtiter plates (Nunc) for 3 days (37°C, 5% CO2). Supernatants were removed at different time points (1 hour, 3 days) and analyzed for cICAM-1 by ELISA-1.
Fluorescence-activated cell sorter (FACS) analysis
Spleen cells (2 × 106 cells/well) were stimulated for 18 hours with 100 U/mL interferon-gamma (IFN-γ) and 10 ng/mL LPS (Sigma, Deisenhofen, Germany) in 300 μL RPMI/10% FCS (37°C, 5% CO2) in 24-well microtiter plates (Nunc). Cells were washed 3 times with PBS (pH 7.5) and fixed for 15 minutes with 100 μL PBS/1% formaldehyde (Merck). After another 2 wash steps, cells were incubated with 5 μL rabbit serum (Dako, Hamburg, Germany), 15 μL FITC-conjugated anti-CD45 and 15 μL PE-conjugated anti-ICAM-1 in a final volume of 100 μL for 30 minutes at 4°C. Cells were washed twice, resuspended in PBS, and analyzed by flow cytometry using a FACScan cytometer. In each analysis 10 000 cells were examined.
Septic shock
Wild-type and ICAM-1 mutant mice (10 in each group) were treated intraperitoneally with a single dose (40 mg/kg) of LPS (Sigma). All animals were monitored during the experiment for signs of morbidity. Blood samples were taken from the retro-orbital plexus with heparinized capillaries at indicated time points. Animals were killed at the end of the experiment. Serum from collected blood samples was obtained by a centrifugation step (5 minutes, 12 000g) and cICAM-1 concentrations were determined in duplicate by ELISA-2.
Statistics
The immunoreactivity of ICAM-1 in serum of LPS-treated mice was expressed as the mean ± SEM. Student t tests were performed to determine the significance of the increase in ICAM-1 immunoreactivity between means of different time points after treatment.
Results
ICAM-1-deficient mice produce soluble ICAM-1
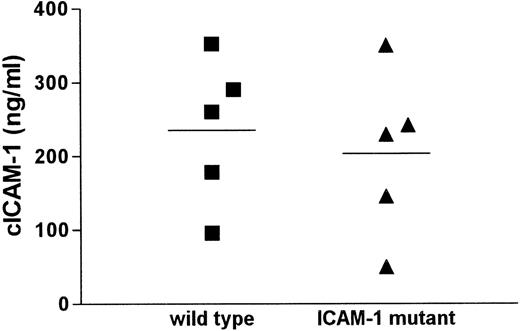
Serum from mice deficient for ICAM-1, initially used as a negative control in cICAM-1 ELISA (ELISA1), surprisingly showed cICAM-1 immunoreactivity. This result was confirmed by a different cICAM-1 ELISA (ELISA-2) with different monoclonal antibodies. All antibodies used in the different ELISAs recognize epitopes within the first extracellular domain of ICAM-1. To characterize this observation we compared different serum samples from wild-type and ICAM-1-deficient mice in cICAM-1 ELISA-1. The cICAM-1 levels showed a similar distribution in both mouse strains (Figure1). The mean cICAM-1 value in the mutant strain (203 ng/mL) did not differ significantly from the mean level in the control strain (235 ng/mL). In contrast, serum from mice with the ICAM-1 disruption in exon 5 (kindly provided by Prof Dr K. Scharffetter-Kochanek, Cologne, Germany) contained cICAM-1 protein of about 40 ng/mL, approximately 70% less than in wild-type mice (data not shown), which is in accordance with an earlier report.26
cICAM-1 concentrations in serum of wild-type and ICAM-1 mutant mice.
Sera from 5 C57BL/6 wild-type and 5 ICAM-1–exon 4 mutant mice were assayed for cICAM-1 by ELISA-1 in duplicate. Mean values are indicated by lines.
cICAM-1 concentrations in serum of wild-type and ICAM-1 mutant mice.
Sera from 5 C57BL/6 wild-type and 5 ICAM-1–exon 4 mutant mice were assayed for cICAM-1 by ELISA-1 in duplicate. Mean values are indicated by lines.
Stimulated spleen cells of ICAM-1 mutant mice do not have ICAM-1 surface expression, but produce cICAM-1
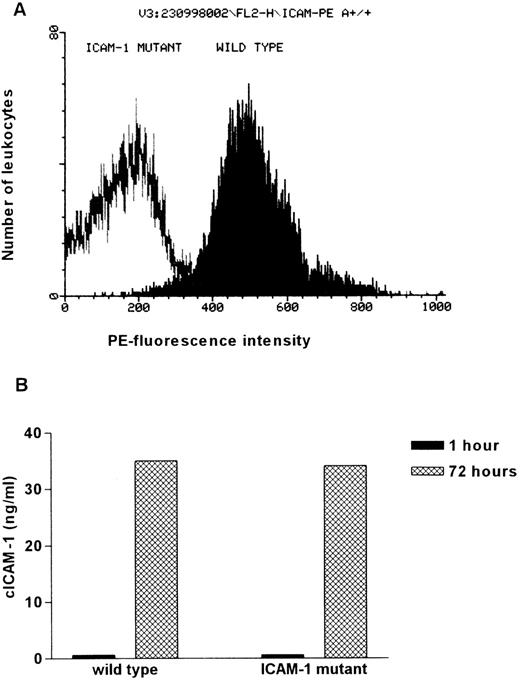
To confirm the absence of residual ICAM-1 expression in ICAM-1 mutant mice, isolated spleen cells of ICAM-1-deficient and wild-type mice were examined for ICAM-1 expression by FACS analysis. The ICAM-1 on the cell surface was determined by incubation with a PE-conjugated anti-ICAM-1 antibody. Whereas 92% of the IFN-γ and LPS-stimulated leukocytes from wild-type animals show high PE-fluorescence intensity, only background staining was observed in ICAM-1 mutant mice. This indicates that ICAM-1 expression is not detectable on leukocytes of ICAM-1 mutants (Figure 2A). Similar results were obtained from FACS analysis of peripheral blood mononuclear cells (data not shown).
Comparison of membrane integrated and circulating ICAM-1 in wild-type and ICAM-1–exon 4 mutant mice.
(A) Visualization of the ICAM-1 expression on leukocytes isolated from wild-type and ICAM-1–exon 4 mutant mice. Isolated spleen cells were stimulated with 100 U/mL IFN-γ and 10 ng/mL LPS for 18 hours to increase ICAM-1 expression. Cells were incubated with FITC-conjugated anti-CD45 (1:7) and PE-conjugated anti-ICAM-1 (1:7). In each measurement PE-fluorescence intensity of 10 000 FITC-positive cells was determined. (B) cICAM-1 production by in vitro cultured spleen cells. Freshly isolated spleen cells (5 × 105 cells/well) were cultured for 3 days in RPMI supplemented with 10% FCS and 0.08% β-mercaptoethanol. Supernatant (150 μL) was taken after 1 hour and at the end of cultivation. cICAM-1 concentrations were determined by ELISA-2 in duplicate.
Comparison of membrane integrated and circulating ICAM-1 in wild-type and ICAM-1–exon 4 mutant mice.
(A) Visualization of the ICAM-1 expression on leukocytes isolated from wild-type and ICAM-1–exon 4 mutant mice. Isolated spleen cells were stimulated with 100 U/mL IFN-γ and 10 ng/mL LPS for 18 hours to increase ICAM-1 expression. Cells were incubated with FITC-conjugated anti-CD45 (1:7) and PE-conjugated anti-ICAM-1 (1:7). In each measurement PE-fluorescence intensity of 10 000 FITC-positive cells was determined. (B) cICAM-1 production by in vitro cultured spleen cells. Freshly isolated spleen cells (5 × 105 cells/well) were cultured for 3 days in RPMI supplemented with 10% FCS and 0.08% β-mercaptoethanol. Supernatant (150 μL) was taken after 1 hour and at the end of cultivation. cICAM-1 concentrations were determined by ELISA-2 in duplicate.
Despite the absence of membranous ICAM-1, cultivated spleen cells from ICAM-1 mutant mice are able to produce cICAM-1. After 3 days of cultivation, ICAM-1 concentrations in the supernatants were determined by ELISA-2. After 1 hour of incubation no cICAM-1 was detected and after 3 days of cultivation concentrations of 34 ng/mL (mutant) and 35 ng/mL (wild type) were measured (Figure 2B). Thus, cICAM-1 can be identified in leukocyte cultures from ICAM-1 mutants, although no membrane integrated ICAM-1 is detectable.
RT-PCR reveals a number of ICAM-1 splice products, none of them coding for a soluble molecule
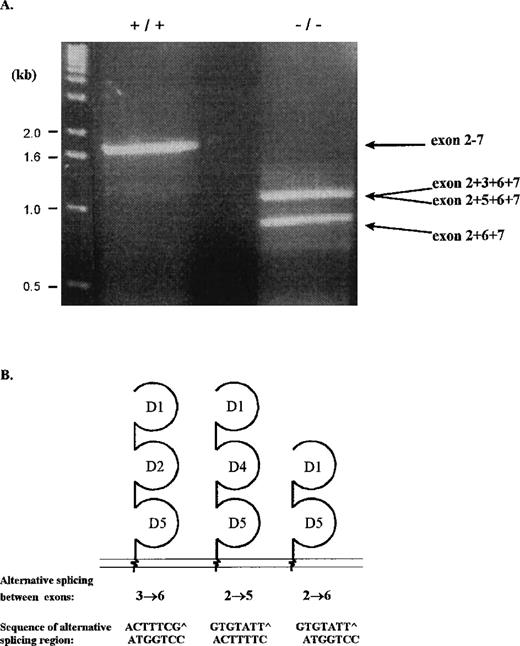
To analyze for the source of cICAM-1 in serum and in spleen cell supernatants in wild-type and ICAM-1 mutant mice, spleen cell RNA was isolated and analyzed by RT-PCR. Two specific primers both based in the UTRs of the murine ICAM-1 gene were used for the amplification. PCR products were resolved on agarose gels (Figure3A). In wild-type mice a 1.8-kb fragment was detected, whereas in ICAM-1-deficient mice bands of 1.2 and 0.9 kb were observed. Further analysis of the bands via Southern blot preparation, which was hybridized with a DIG-labeled oligonucleotide specific for exon 2, revealed that the fragments contained the coding region for at least the first extracellular domain of ICAM-1 (data not shown). PCR fragments were cloned and sequenced for further identification. The 1.8-kb fragment from wild-type RNA corresponded to the common form of ICAM-1. The 1.2-kb band from ICAM-1-deficient mice consisted of 2 fragments with slightly different lengths, representing 2 alternatively spliced isoforms of ICAM-1. The larger 1.26-kb fragment is spliced between exons 3 and 6, retaining the coding sequence for extracellular domains 1, 2, and 5. The smaller 1.20-kb fragment is spliced between exons 2 and 5, retaining the coding sequence for extracellular domains 1, 4, and 5. The 0.9-kb fragment represented an isoform spliced between exons 2 and 6, encoding the extracellular domains 1 and 5 (Figure 3B). However, all isoforms contained exon 7, which encodes the transmembrane region, indicating that no alternatively spliced form coding for cICAM-1 was identified.
RT-PCR analysis of alternatively spliced ICAM-1 RNA isoforms.
RT-PCR was performed on spleen RNA from wild-type and ICAM-1 mutant mice using 5′UTR and 3′UTR primers. (A) RT-PCR products resolved on a 1.5% agarose gel. For identification, fragments were transferred to a nitrocellulose membrane, which was hybridized with an exon 2-specific probe. Positive fragments were cloned and sequenced. Indicated on the diagram are short sequences of the exons joining the alternatively spliced sites. (B) Schematic representation of the ICAM-1 isoforms of ICAM-1–exon 4 mutant mice. The isoforms contain 2 or 3 extracellular domains, a transmembrane domain, and a cytoplasmic region.
RT-PCR analysis of alternatively spliced ICAM-1 RNA isoforms.
RT-PCR was performed on spleen RNA from wild-type and ICAM-1 mutant mice using 5′UTR and 3′UTR primers. (A) RT-PCR products resolved on a 1.5% agarose gel. For identification, fragments were transferred to a nitrocellulose membrane, which was hybridized with an exon 2-specific probe. Positive fragments were cloned and sequenced. Indicated on the diagram are short sequences of the exons joining the alternatively spliced sites. (B) Schematic representation of the ICAM-1 isoforms of ICAM-1–exon 4 mutant mice. The isoforms contain 2 or 3 extracellular domains, a transmembrane domain, and a cytoplasmic region.
Circulating ICAM-1 in ICAM-1 mutant mice differs in molecular weight from that in wild-type mice
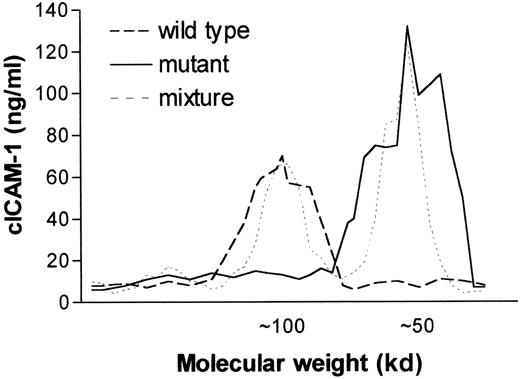
To analyze the molecular weight of the circulating adhesion molecule, serum from wild-type or ICAM-1 mutant mice was separated by gel filtration chromatography. ICAM-1 immunoreactivity in the different fractions was analyzed by cICAM-1 ELISA-1. cICAM-1 in control mice appeared to have a molecular weight of about 100 kd, whereas the molecular weight of cICAM-1 in mutant mice was estimated at 50 kd (Figure 4). These data are in accordance with the RT-PCR analysis in which in wild-type mice a larger fragment was identified than in mutants.
Comparison of cICAM-1 in wild-type and mutant C57BL/6 mice by fast protein liquid size exclusion chromatography.
Sterile filtered serum (500 μL) from wild-type (+/+), ICAM-1–exon 4 mutant mice (-/-) and a mixture of +/+ and -/-serum were chromatographed on a Superdex-200 column in separate runs. Collected fractions of 0.5 mL were analyzed for the presence of cICAM-1 by ELISA-1. The molecular weight of proteins in the different fractions was estimated from the elution profile of marker proteins of molecular weight in the range of 12 to 200 kd.
Comparison of cICAM-1 in wild-type and mutant C57BL/6 mice by fast protein liquid size exclusion chromatography.
Sterile filtered serum (500 μL) from wild-type (+/+), ICAM-1–exon 4 mutant mice (-/-) and a mixture of +/+ and -/-serum were chromatographed on a Superdex-200 column in separate runs. Collected fractions of 0.5 mL were analyzed for the presence of cICAM-1 by ELISA-1. The molecular weight of proteins in the different fractions was estimated from the elution profile of marker proteins of molecular weight in the range of 12 to 200 kd.
Regulation of cICAM-1 in ICAM-1 mutant mice is comparable to that in wild-type animals
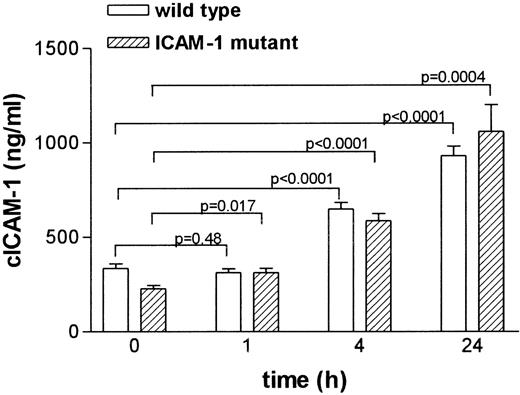
To study the regulation of cICAM-1 in vivo, wild-type and ICAM-1 mutant mice were treated with LPS. Serum samples were taken at different time points and analyzed for cICAM-1 by ELISA-2 (Figure 5). Mean cICAM-1 levels showed a significant increase after 4 and 24 hours in wild-type mice as well as in ICAM-1-deficient mice.
Circulating ICAM-1 concentrations in serum from LPS-treated mice.
Mice were given a single dose of 40 mg/kg of LPS intraperitoneally. Blood samples were taken from the retro-orbital plexus with heparinized capillaries before (0 hour) and at indicated times after LPS injection. At different time points, different groups of animals were used. Serum ICAM-1 concentrations were analyzed by ELISA-2 in duplicate. Results are expressed as the mean (± SEM) ICAM-1 immunoreactivity.
Circulating ICAM-1 concentrations in serum from LPS-treated mice.
Mice were given a single dose of 40 mg/kg of LPS intraperitoneally. Blood samples were taken from the retro-orbital plexus with heparinized capillaries before (0 hour) and at indicated times after LPS injection. At different time points, different groups of animals were used. Serum ICAM-1 concentrations were analyzed by ELISA-2 in duplicate. Results are expressed as the mean (± SEM) ICAM-1 immunoreactivity.
Discussion
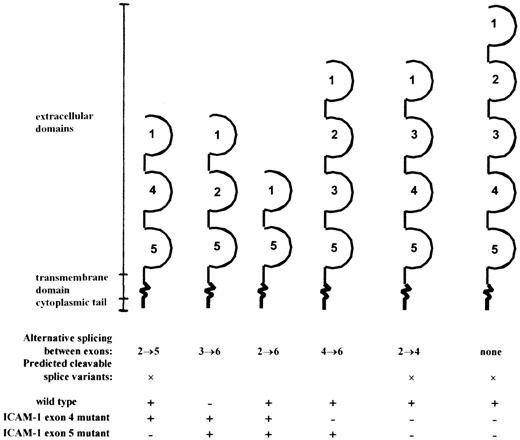
During the last few years, 2 different ICAM-1-deficient mouse strains have been generated. Previous studies of ICAM-1–exon 5 mutant mice showed a residual expression of membranous ICAM-1 in the selected tissue.10 In contrast, no background expression of ICAM-1 was found in the ICAM-1-deficient mice used in this study, which have the ICAM-1 gene disrupted in exon 4.7 Despite the absence of membranous ICAM, we show here that the ICAM-1 immunoreactivity level in serum of ICAM-1 mutants is detectable and even comparable to the ICAM-1 immunoreactivity level in serum of wild-type mice. We detected 3 alternatively spliced ICAM-1 isoforms in RNA of ICAM-1-deficient mice. The products spliced between exons 2 and 6, (2→6), 3→6, and 2→5 encode ICAM-1 proteins with the Ig-like domains 1 + 5, 1 + 2 + 5, and 1 + 4 + 5, respectively (Figure 6). Similar splicing of the ICAM-1 gene has been reported for the ICAM-1–exon 5-deficient mouse,10 in which splice products 2→6, 3→6, and 4→6 were shown. Both mutant strains differ in cICAM-1 levels. We detected a similar serum ICAM-1 level in wild-type and ICAM-1–exon 4-deficient mice, whereas Komatsu et al26 described a 70% reduction of plasma cICAM-1 in exon 5-deficient mice. Because different isoforms were identified in both mutant strains (see Figure 6), we can assume that splice variant 2→5 is responsible for the increase in cICAM-1 levels in exon 4-deficient mice. In a similar way we may argue that the 4→6 fragment is responsible for the residual ICAM-1 in some tissues of exon 5-deficient mice.
Overview of the different splice variants encoded by the mRNA detected in wild-type, ICAM-1–exon 4, and ICAM-1–exon 5 mutant mice.
Extracellular domains are indicated: +, detected; −, not detected.
Overview of the different splice variants encoded by the mRNA detected in wild-type, ICAM-1–exon 4, and ICAM-1–exon 5 mutant mice.
Extracellular domains are indicated: +, detected; −, not detected.
The molecular weight of the ICAM-1 2→5 splice variant, encoding 256 extracellular amino acids would be around 50 kd, and of the common 5-domain form, encoding 457 extracellular amino acids, about 100 kd, taking the glycosylation of the first domain into account.27 This supports that the ICAM-1 2→5 splice variant and the common form are the main molecular source of cICAM-1 in mutant and wild-type mice, respectively.
Several splice variants have been described in wild-type mice10; however, these were not detected in unstimulated spleen cells as presented in this report. LPS stimulation of these cells resulted in a weak transcription of 2 splice variants (data not shown). Furthermore, in the earlier study different splice variants have been found in lung and thymus, suggesting activation and tissue-specific splicing of the ICAM-1 gene.
Alternative splicing has been postulated as a possible mechanism to explain the generation of cICAM-1 in human tissue.23 In our approach to identify cICAM mRNA, the presence of the first domain in each isoform was confirmed by hybridizing with a probe directed to exon 2. Using this sensitive approach, it is unlikely we have failed to detect any splice variant unless expressed at a very low level. Interestingly, all our and previously identified alternatively spliced forms of murine ICAM-1 contain exon 7, which encodes the transmembrane spanning domain. We have sequenced several clones of all alternatively spliced fragments to exclude that a frame shift with a consecutive stop codon would be responsible for an alternatively spliced source of cICAM-1, as was reported for human cICAM-1.23 No frame shift was identified. Previously, it has been shown that all spliced variants can be expressed in Chinese hamster ovary cells and detected by ICAM-1-specific monoclonal antibodies.10 These findings of membrane retention of the ICAM-1 variants promote a mechanism other than alternative splicing. Proteolytic cleavage of membrane integrated ICAM-1 or ICAM-1 splice variants seems more likely to be the mechanism for the generation of cICAM-1, previously postulated for human keratinocytes.22 Indeed, in a recent study proteolytic cleavage of ICAM-1 by human neutrophil elastase has been shown.28 Direct evidence for proteolytic cleavage of membrane integrated adhesion molecules has already been described for L-selectin.29 30 On binding of a leukocyte L-selectin is cleaved within seconds. If the protease recognition site is destroyed, no cleavage and consequently no rolling is detectable. Taken together, we support the view that proteolytic cleavage, but not alternative splicing, is the molecular mechanism for the generation of cICAM-1.
An explanation for the differences in cICAM-1 expression in the 2 ICAM-1 knockout models can be that some of the different ICAM-1 splice variants get cleaved and others do not. The common form and the 2→5 splice variant are presumably cleaved, whereas the 4→6 variant remains in the membrane. We postulate that the protease responsible for cleavage has an Ig-domain specificity. The 2→5 splice product and the common form contain the Ig-like domains 4 and 5. In contrast, the combination of domains 4 and 5 is lacking in splice variants of exon 5 mutants, because the coding region for domain 4 is disrupted in these mice. Therefore, the recognition site for the postulated protease to generate cICAM-1 seems to involve parts of domain 4 and 5.
Even though the total amount of cICAM-1 in ICAM-1-deficient mice is not increased, the variants might have different binding avidities to its receptors as reported for different ICAM-1 splice variants in cell-binding assays.10 Therefore, it has to be considered that pathophysiologic abnormalities in ICAM-1-deficient mice as increased body weight31 or septic shock7 might be related to the cICAM-1 variants in addition to the missing ICAM-1 expression. During the last few years we have described an immunomodulatory potential of soluble rICAM-1.19,21,32,33Recently, we showed that injection of rICAM-1 can prevent leukocyte adherence after ischemia/reperfusion injury.20Interestingly, ICAM-1 mutant mice are more resistant to ischemia/reperfusion injury than wild-type mice.34 35 This might support a therapeutic potential for rICAM-1 or different spliced circulating forms of ICAM-1 in immune-mediated diseases. However, in rICAM-1- treated mice no increase in body weight was found, indicating that obesity of ICAM-1-deficient mice might be related more to the deficiency of membrane-bound ICAM-1 than to the presence of mutant forms of cICAM-1. Further experiments are needed to clarify the (patho-) physiologic role of circulating ICAM-1 in ICAM-1-mutant mice.
Acknowledgments
We would like to thank Prof Dr J-C. Gutierrez-Ramos and Prof Dr K. Scharffetter-Kochanek for providing the different strains of ICAM-1-deficient mice, Bettina Schulte for excellent technical assistance, Dr B. Schwippert for performing FACS analysis, and Dr Markus Walz for sequencing analysis.
Supported by a grant from the Deutsche Forschungsgemeinschaft (Ma 1260/4), the “Bennigsen-Foerder” grant from the Minister für Wissenschaft und Forschung des Landes Nordrhein-Westfalen, Düsseldorf, and institutional funds provided by the Bundesminister für Gesundheit, Bonn, and the Minister für Wissenschaft und Forschung des Landes Nordrhein-Westfalen, Düsseldorf.
Reprints:Priv.-Doz. Dr Stephan Martin, Diabetes Research Institute at the Heinrich-Heine-Universität Düsseldorf, Auf ‘m Hennekamp 65, D-40225 Düsseldorf, Germany.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal