In this study, we wished to determine whether familial chronic lymphocytic leukemia of B-cell phenotype (CLL) shares with sporadic B-CLL the same immunoglobulin (Ig) heavy chain variable region (VH) gene usage and occurrence of somatic mutation, to gain insight into the pathogenetic relatedness of these epidemiologically distinct forms of CLL. We therefore analyzed the expressed Ig heavy chain genes in 23 cases (11 families) of familial CLL, and compared these results with data previously reported for sporadic CLL. In addition, we assessed the relationship of the occurrence of somatic mutation to several clinical and phenotypic features. The distribution of V genes among these cases was similar to that observed in sporadic CLL: VH3 > VH1 > VH4. Thirteen of the 23 cases (57%) showed germ line VH gene sequences, whereas somatic mutations were detected in 10 cases (43%). The average mutation frequency of these latter 10 cases of was 6.7% (ranging from 1.7% to 8.8%), and evidence of antigen selection was noted in 6. Intraclonal variation, followed by clonal evolution and the appearance of a second clone over a 20-year period was observed in 1 case, suggesting that mutations can continue to accumulate after neoplastic transformation. The presence of somatic mutations correlated with age at presentation, low white blood cell (WBC) count, and low fluorescence intensity of surface CD5, and the potential significance of these relationships is discussed. Our data indicate that familial and sporadic B-CLL display a similar pattern of immunoglobulin gene usage and frequency of somatic mutation, and are consistent with a common ontogeny and immunogenetic origin for these 2 epidemiologically distinct forms of CLL.
Chronic lymphocytic leukemia of B-cell phenotype (CLL) is the most common leukemia in the adult population in the United States and Europe.1 This is a clinically indolent neoplasm and results from an accumulation of CD5+ neoplastic B-cells with a low proliferative rate. The normal counterpart of B-CLL is believed to be the B-1a cell,2 which accounts for 5% to 30% of the normal circulating B-cell population, and is characterized by surface expression of IgM/IgD and CD5, and the ability to produce autoantibodies.3 CLL has been reported to show nonrandom usage of VH region families and genes, an observation hypothesized to be related to the B-cell subset of origin, and to the peculiar antibody-mediated autoimmunity sometimes accompanying this leukemia.3 4
Traditionally, CLL has been viewed as a homogeneous disease, with its characteristic immunophenotype and indolent clinical course. However, more recent cytogenetic and immunogenetic studies have challenged this viewpoint. Several different cytogenetic abnormalities have been reported, each with somewhat different accompanying clinical and/or phenotypic features.5,6 Trisomy 12 and 11q23 deletions have been associated with a shorter median survival,7-12 whereas cases with 13q14 deletions appear to have a more favorable outcome.12
Further evidence for heterogeneity in CLL comes from investigations of the expressed immunoglobulin genes. Although early studies of the IgH gene in CLL failed to demonstrate somatic mutation,13-16more recent analyses have shown that a substantial (20%-50%) percentage of cases show somatic mutations, suggesting that CLL may arise from either pregerminal center naive B-cells, or germinal center exposed B-cells.3,17-20 These studies also suggest that the presence or frequency of somatic mutation correlates with immunophenotype,20 the usage of variable region gene (VH) families18 and the presence of specific chromosomal abnormalities.19 Fais et al18 reported somatic mutation in 50% of the IgM+ expressing cases, with the highest incidence noted in the VH3 family, compared with VH1 and VH4. Oscier et al19 noted that cases with trisomy 12 lacked somatic mutation, whereas cases with 13q14 deletion showed significant levels of somatic mutation. The existence of CLL lymphocytes having undergone somatic mutation suggests that a subset of CLL is derived from memory B cells that have passed through the germinal center stage of B-cell differentiation. Taken together, the above observations indicate that CLL is more heterogeneous than previously thought and may develop either from ontogenically naive B lymphocytes or from a more mature antigen-exposed memory B cells.
Familial aggregation of CLL (familial CLL) has been observed more frequently than in any other type of leukemia, and immunogenetic factors related to HLA type have been implicated in its development.21-23 Besides familial and genetic predisposition, environmental factors may also play a role in this clustering of cases. Although familial CLL is indistinguishable from sporadic CLL in its morphology and immunophenotype, the pathogenetic relationship between these epidemiologically distinct forms of CLL has not been clearly established. There are virtually no cytogenetic or molecular genetic studies comparing familial CLL with sporadic CLL. Likewise, studies of immunoglobulin gene usage and somatic mutation of immunoglobulin genes are limited in familial CLL, for comparison to sporadic CLL.24 25 To gain insight into the pathogenetic relationship between familial CLL and sporadic CLL, we wished to study the spectrum of immunoglobulin heavy chain gene usage and the presence or absence of somatic mutation in familial CLL. Because of the genetic overlay to familial CLL, we hypothesized that it may be a more restricted disease with regard to its immunoglobulin gene usage or ontogenic development, as reflected by presence or absence of somatic mutation.
We therefore investigated the immunoglobulin gene usage and pattern of somatic mutation in 23 patients from 11 families with familial CLL and compared these findings with those reported in sporadic CLL. Furthermore, we examined the relationship between the presence or absence of somatic mutation in the expressed IgH genes, and several clinical and immunophenotypic features. We found a pattern of immunoglobulin gene usage and somatic mutation similar to that reported for sporadic CLL, suggesting that familial CLL displays a similar heterogeneity. We also found an unexplained inverse correlation between CD5 antigen density and the presence of somatic mutation.
Materials and methods
Patients/samples
Twenty-three individuals with familial B-CLL from 11 families (a minimum of 2 members [first degree relatives] per family) were selected on the basis of the availability of stored frozen specimens. The diagnosis of B-CLL was confirmed in all affected individuals by using morphologic and immunophenotypic criteria, and the patients were subsequently followed in the Family Studies Section Clinic, National Cancer Institute (NCI) in an IRB-approved protocol. All analyzed samples were collected before any therapy was given, with the exception of patient 2a. Two samples from 1988 and 1998 were available from 1 member of an affected family (case 2a). This patient received 1 course of chlorambucil at the time of his second sample.
Peripheral blood mononuclear cells (PBMCs) were obtained from heparinized peripheral blood by Ficoll-Isopaque gradient density centrifugation (GIBCO BRL, Gaithersburg, MD), then treated with ACK lysing buffer (Quality Biological, Gaithersburg, MD) to remove contaminating red blood cells (RBCs), and resuspended in freezing media (RPMI 1640 + 20% FCS + 7.5% DMSO). Frozen aliquots were stored in liquid nitrogen.
Immunophenotypic analysis
Blood samples were washed with phosphate-buffered saline (PBS) and 10% fetal calf serum, then double labeled using the following monoclonal antibodies: CD5 (Leu 1), CD19 (Leu 12), CD20 (Leu 16), CD23 (Leu 20) (BDIS, San Jose, CA), kappa and lambda (BDIS, San Jose, CA; Caltag, Burlingame, CA; DAKO, Carpinteria, CA). RBCs were removed by 1 × FACS-lyse (BDIS, San Jose, CA) after labeling. Immunophenotyping was performed by flow cytometry using a FACS scan (BDIS, San Jose, CA).
Isolation of RNA and first-strand cDNA
Total cellular RNA was isolated from cryo-preserved mononuclear cell suspensions using TRIZOL reagent (GIBCO BRL, Gaithersburg, MD) according to the manufacturer's instructions. After DNase I (GIBCO BRL, Gaithersburg, MD) treatment, 1 to 5 μg total RNA were incubated with 50 μmol of random hexamer primers (PE, Foster City, CA) for 10 minutes at 70°C. After cooling on ice, the reaction mixture was added to a final volume of 20 μL containing 200 U of Super Script II, 1 × First Strand Buffer (50 mmol/L Tris-HCl, pH 8.3, 75 mmol/L KCl, 3 mmol/L MgCl2), 10 mmol/L of DTT, 20 U of RNase inhibitor, and 0.5 mmol/L of each dNTP (GIBCO BRL, Gaithersburg, MD). The reaction was performed for 10 minutes at 25°C followed by 50 minutes at 42°C. Subsequently, the enzyme was inactivated for 15 minutes at 70°C.
Polymerase chain reactions
One microliter of cDNA was amplified using GeneAmp System 2400 (PE, Foster City, CA) with a 50 pmol specific upstream primer corresponding to 1 of the 6 human VH family leader sequences (VHL1, 5′-CCATGGACTGGACCTGGAGG-3′; VHL2, 5′-ATGGACATACTTTGTTCCAGC-3′; VHL3, 5′-CCATGGAGTTTGGGCTGAGC-3′; VHL4, 5′-ATGAAACACCTGTGGT TCTT-3′; VHL5, 5′-ATGGGGTCAACCGCCATCCT-3′; VHL6, 5′-ATGTCTGTCT CCTTCCTCAT-3′)26 and a 50 pmol downstream primer (JHa, 5′-ACCTGAGGAGACGGTGACC-3′)27corresponding to a consensus sequence at the 3′ end of the J region in a 50 μL volume. The polymerase chain reaction (PCR) contained 1 × PCR buffer (10 mmol/L Tris-HCl, pH 8.3, 50 mmol/L KCl), 1.5 mmol/L MgCl2 (for VHL1, VHL3, and VHL5) or 2.0 mmol/L MgCl2 (for VHL2, VHL4, and VHL6), 0.2 mmol/L of each dNTP, 1.25 U Taq polymerase (PE, Foster City, CA) mixed with TaqStart™antibody (Clontech, Palo Alto, CA). First, 1 cycle of amplification was performed under the following conditions: denaturation at 95°C for 7 minutes, annealing at 65°C for 4 minutes, and extension at 72°C for 1 minute. The next 40 cycles of amplification were at 95°C for 1 minute, 61°C for 30 seconds, and at 72°C for 1 minute. The reaction was completed with extension at 72°C for 6 minutes. The PCR products were analyzed on a 3% NuSieve GTG agarose gel (FMC, Rockland, ME) and visualized by staining with ethidium bromide.
Three colonies were sequenced for each case if no mismatches between VH sequences were found. If mismatches among the 3 clones were identified, at least 2 additional colonies were sequenced.
Cloning and sequencing of PCR products
After excision of the PCR products from an agarose gel, DNA was isolated using a QIAEX DNA extraction kit (Qiagen, Santa Clarita, CA). The recovered DNA was ligated into the pCR2.1 vector, following the manufacturer's instruction (TA cloning kit; Invitrogen, San Diego, CA). Three to 5 colonies were picked at random and grown overnight in 3 mL LB (Quality Biological, Gaithersburg, MD) medium. Recombinant plasmids were purified by DNA affinity columns (Qiagen, Valencia, CA) and selected by restriction analysis. The double-stranded plasmid was sequenced in an automatic DNA sequencer (PE Applied Biosystems 377 × l Automated DNA; PE Applied Biosystems, Foster City, CA) using the BigDye™ Terminator Cycle Sequencing Ready Reaction kit (PE, Foster, CA), following manufacturer's instruction.
Analysis of VH gene usage and mutations
Sequences obtained from each sample were compared with germ line sequences in the V Base sequence directory (I.M. Tomlinson, MRC Center for Protein Engineering, Cambridge, UK) using MacVector 6.0 sequence analysis software (Oxford Molecular Group, Campbell, CA), and the closest sequence was assigned. Attribution of the D segments was based on the identification of at least 6 consecutive bases without mismatches. The nomenclature proposed by Corbett et al28was adopted. We considered a VH gene sequence to be mutated if it had equal or more than 1.5% sequence alterations when compared with the published germ line sequence. Mutated cases had between 5 and 26 nucleotide differences. Nonmutated cases had 0 or 1 nucleotide differences from published germ line VH gene sequences.
The probability that somatic mutations in the rearranged VH genes resulted from antigen selection was analyzed according to the method of Chang and Casali,29 taking into account the inherent susceptibility of VH complementarity-determining regions (CDRs) to mutations. A binomial probability model was used:P = {n!/[k!(n − k)!]} × qk × (1 − q)n−k, where n is the total number of observed mutations, k is the number of observed R (replacement) mutations in the CDRs or framework regions (FRs), andq is the probability that an R mutation will localize to the CDRs or FRs (q = CDR rel × CDR Rf. or FRrel × FR Rf). The inherent susceptibility of R mutations of the CDRs and FRs (CDR Rf and FR Rf, respectively) is calculated for each of the identified germ line genes and it is based on any single nucleotide change that gives rise to an amino acid replacement, whereas substitutions that result in stop codons are excluded. CDR rel and FR relrepresented the relative sizes of the CDRs and FRs.
Statistical analysis
The relationship between age, gender, WBC count, Rai stage, duration of disease from diagnosis, the intensity of surface antigens, and the presence of somatic mutations was examined using a variety of approaches, including Mann-Whitney U test (nonparametric) and independent sample t tests. Multiple linear regression was used to examine the effects of several independent variables, but this analysis was limited by the small number of subjects. The relationship between the presence of somatic mutation and the fluorescence intensity of CD5, CD20, and CD45 on the leukemia cells was analyzed by similar methods. A 2-sided P value of <.05 was considered significant.
Results
Clinical features
The clinical features of 23 cases of familial B-CLL are summarized in Table 1. The age, WBC count, Rai stage, and duration of disease correspond to the date when the patients were referred to NIH. One individual (2a) was evaluated at 2 time points with a 10-year interval and the Rai stage remained unchanged. Except for 1 family (no 4) with 3 affected individuals, there were 2 affected individuals in each family. In all families but 1, the patients were siblings. Thirteen of the 23 cases were male, with an age ranging from 40 to 75 years (median, 61 years). The WBC count ranged from 6.1 to 148.8 × 103 /μL (median, 45.2 × 103/μL), and Rai stage at presentation are listed in Table 1. The duration of disease ranged from less than 1 year to 20 years.
Clinical features of the patients
| Family No. . | Relation . | Age . | WBC Count (×103/uL) . | Rai Stage . | Duration of Disease (y)* . | Mutations of VH Genes† . |
|---|---|---|---|---|---|---|
| 1a | Mother | 75 | 41.6 | 0 | 1 | − |
| 1b | Son | 56 | 34.3 | 0 | 1 | − |
| 2a | Sister | 73 | 30.7 | II | 20 | + |
| 2b | Brother | 59 | 33.6 | IV | 10 | + |
| 3a | Sister | 62 | 21.9 | I | <1 | + |
| 3b | Sister | 61 | 22.1 | I | 4 | + |
| 4a | Sister | 74 | 16.5 | I | 4 | + |
| 4b | Sister | 75 | 77.4 | I | 18 | + |
| 4c | Brother | 72 | 16.1 | IV | 2 | + |
| 5a | Brother | 54 | 6.1 | I | 5 | − |
| 5b | Sister | 51 | 124.0 | 0 | 4 | − |
| 6a | Brother | 56 | 27.7 | I | 1 | − |
| 6b | Brother | 53 | 42.5 | I | 3 | − |
| 7a | Brother | 75 | 8.0 | 0 | 9 | + |
| 7b | Brother | 72 | 13.3 | I | <1 | + |
| 8a | Brother | 50 | 27.9 | 0 | 3 | + |
| 8b | Brother | 64 | 70.9 | II | 2 | − |
| 9a | Sister | 40 | 95.8 | I | 3 | − |
| 9b | Brother | 43 | 58.0 | I | 1 | − |
| 10a | Sister | 57 | 148.8 | I | 8 | − |
| 10b | Brother | 45 | 97.1 | I | 1 | − |
| 11a | Brother | 65 | 15.9 | I | <1 | − |
| 11b | Sister | 62 | 8.8 | 0 | <1 | + |
| Family No. . | Relation . | Age . | WBC Count (×103/uL) . | Rai Stage . | Duration of Disease (y)* . | Mutations of VH Genes† . |
|---|---|---|---|---|---|---|
| 1a | Mother | 75 | 41.6 | 0 | 1 | − |
| 1b | Son | 56 | 34.3 | 0 | 1 | − |
| 2a | Sister | 73 | 30.7 | II | 20 | + |
| 2b | Brother | 59 | 33.6 | IV | 10 | + |
| 3a | Sister | 62 | 21.9 | I | <1 | + |
| 3b | Sister | 61 | 22.1 | I | 4 | + |
| 4a | Sister | 74 | 16.5 | I | 4 | + |
| 4b | Sister | 75 | 77.4 | I | 18 | + |
| 4c | Brother | 72 | 16.1 | IV | 2 | + |
| 5a | Brother | 54 | 6.1 | I | 5 | − |
| 5b | Sister | 51 | 124.0 | 0 | 4 | − |
| 6a | Brother | 56 | 27.7 | I | 1 | − |
| 6b | Brother | 53 | 42.5 | I | 3 | − |
| 7a | Brother | 75 | 8.0 | 0 | 9 | + |
| 7b | Brother | 72 | 13.3 | I | <1 | + |
| 8a | Brother | 50 | 27.9 | 0 | 3 | + |
| 8b | Brother | 64 | 70.9 | II | 2 | − |
| 9a | Sister | 40 | 95.8 | I | 3 | − |
| 9b | Brother | 43 | 58.0 | I | 1 | − |
| 10a | Sister | 57 | 148.8 | I | 8 | − |
| 10b | Brother | 45 | 97.1 | I | 1 | − |
| 11a | Brother | 65 | 15.9 | I | <1 | − |
| 11b | Sister | 62 | 8.8 | 0 | <1 | + |
Expressed in years from time of diagnosis.
Mutations of immunoglobin heavy-chain variable region (VH): +, presence of somatic mutations; −, absence of somatic mutations.
Immunophenotypic analysis and clonality
All cases had a typical immunophenotype characterized by monotypic surface immunoglobulin, CD5, CD19, and CD23 expression. Two- color FACS analysis revealed coexpression of CD5 in more than 99% of the CD19+ B cells, except for cases 5b, 7b, and 11b, in which there were some residual normal B cells (1.0%, 4% and 1.1%, respectively). All neoplastic B cells expressed CD23, κ or λ. Clonality was verified by PCR using cDNA generated from PBMC and the VH family specific primers. PCR products from all cases showed a single predominant band corresponding to 1 of 6 VH families. The 3 cases containing 1% or more normal Bcells, 5b, 7b, and 11b, showed some additional faint bands presumably originating from this population (data not shown).
VH, D, and JH gene usage by familial B-CLL
A summary of the data are shown in Table2. All VH families, except VH6, were observed at least once in these cases of familial B-CLL. VH3, VH1, and VH4 families were expressed at the highest frequency (37.5%, 33.3%, and 20.8%, respectively), with VH2, VH4, VH5, and VH7 each occurring only once. In 4 families, identical usage of the VH family was recognized in at least 2 cases, ie, VH3 in family nos 3, 4, and 7, VH1 in family no 6. However, only the 2 cases of family no 6 used the same VH gene (VH1-69/DP-10). In the other cases, different VH genes were used. Comparison with the germ line JH gene segments showed the preferential usage of JH4b, followed by JH6b and JH5b (43.5%, 30.4%, and 13.0%, respectively). D segments were attributed in 70% of cases.
Mutation analysis of the familial B-CLL
| Family . | VH Family . | VH Gene . | % Identity . | n . | CDR1 + CDR2 . | FR1 + FR2 + FR3 . | D Gene . | JH Gene . | Sequence Pattern . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Observed R/S (Exp. R) . | Inherent R:S . | P . | Observed R/S (Exp. R) . | Inherent R:S . | P . | ||||||||
| 1a | VH4 | 4-34 | 100 | 0 | 0/0 | nd | nd | 0/0 | nd | nd | D3-10 | JH4b | germ line |
| 1b | VH3 | 3-13 | 99.7 | 1 | 1/0 (0.2) | 3.23 | nd | 0/0 (0.6) | nd | nd | D2-2 | JH5b | germ line* |
| 2a/1('88) | VH1 | 1-02 | 91.8 | 24 | 8/0 (4.4) | 4.34 | 0.035# | 7/9 (13.9) | 2.99 | 2.92 × 10−3† | D6-19 | JH5b | intraclonal mutation |
| 2a/2('88) | VH1 | 1-02 | 93.2 | 20 | 6/0 (3.6) | 4.34 | 0.085 | 7/7 (11.6) | 2.99 | 0.021† | D6-19 | JH5b | intraclonal mutation |
| 2a/3('88) | VH1 | 1-02 | 91.2 | 26 | 8/0 (4.7) | 4.34 | 0.051 | 10/8 (15.1) | 2.99 | 0.021† | D6-19 | JH5b | intraclonal mutation |
| 2a′/1('98) | VH1 | 1-02 | 91.2 | 26 | 8/0 (4.7) | 4.34 | 0.051 | 10/8 (15.1) | 2.99 | 0.021† | D6-19 | JH5b | intraclonal mutation |
| 2a′/2('98) | VH7 | 7-4-1 | 92.4 | 22 | 3/2 (3.9) | 3.63 | 0.209 | 8/9 (12.9) | 2.94 | 0.023† | D2-15 | JH4b | intraclonal mutation |
| 2b | VH4 | 4-61 | 94.6 | 16 | 6/3 (3.0) | 4.03 | 0.042# | 4/3 (8.9) | 2.61 | 0.010† | D3-22 | JH4b | somatic mutation |
| 3a | VH3 | 3-73 | 91.8 | 24 | 9/1 (4.4) | 3.08 | 0.015# | 10/4 (13.6) | 2.99 | 0.057 | D1-7 | JH4b | somatic mutation |
| 3b | VH3 | 3-74 | 93.2 | 20 | 6/0 (3.6) | 3.90 | 0.079 | 9/5 (11.4) | 2.80 | 0.097 | D3-9 | JH5a | somatic mutation |
| 4a | VH4 | 4-34 | 93.8 | 18 | 6/1 (3.3) | 4.50 | 0.065 | 4/7 (10.2) | 2.73 | 2.65 × 10−3† | D3-3 | JH5a | somatic mutation |
| 4b | VH3 | 3-30 | 91.2 | 26 | 4/7 (4.6) | 3.87 | 0.200 | 9/6 (14.9) | 2.85 | 0.011† | N/A | JH4b | somatic mutation |
| 4c | VH3 | 3-30 | 99.7 | 1 | 0/0 (0.2) | nd | nd | 1/0 (0.6) | 2.85 | 0.574 | D6-25 | JH4b | germ line* |
| 5a | VH1 | 1-69 | 99.7 | 1 | 0/1 (0.2) | nd | nd | 0/0 (0.6) | nd | nd | D2-2 | JH6b | germ line* |
| 5b | VH5 | 5-51 | 100 | 0 | 0/0 | nd | nd | 0/0 | nd | nd | N/A | JH6b | germ line |
| 6a | VH1 | 1-69 | 100 | 0 | 0/0 | nd | nd | 0/0 | nd | nd | D2-2 | JH6b | germ line |
| 6b | VH1 | 1-69 | 100 | 0 | 0/0 | nd | nd | 0/0 | nd | nd | D2-2 | JH6b | germ line |
| 7a | VH3 | 3-30 | 94.6 | 16 | 2/4 (2.9) | 4.05 | 0.242 | 6/4 (9.2) | 2.85 | 0.056 | N/A | JH4b | somatic mutation |
| 7b | VH3 | 3-7 | 92.2 | 23 | 8/1 (4.3) | 4.94 | 0.032# | 11/3 (13.2) | 2.84 | 0.108 | N/A | JH4b | somatic mutation |
| 8a | VH1 | 1-24 | 98.3 | 5 | 5/0 (0.9) | 4.08 | 1.94 × 10−4# | 0/0 (2.9) | nd | nd | N/A | JH6b | somatic mutation |
| 8b | VH3 | 3-9 | 100 | 0 | 0/0 | nd | nd | 0/0 | nd | nd | D3-16 | JH4b | germ line |
| 9a | VH4 | 4-39 | 100 | 0 | 0/0 | nd | nd | 0/0 | nd | nd | N/A | JH5b | germ line |
| 9b | VH1 | 1-02 | 100 | 0 | 0/0 | nd | nd | 0/0 | nd | nd | D4-17 | JH6b | germ line |
| 10a | VH1 | 1-02 | 100 | 0 | 0/0 | nd | nd | 0/0 | nd | nd | D6-19 | JH4b | germ line |
| 10b | VH4 | 4-39 | 100 | 0 | 0/0 | nd | nd | 0/0 | nd | nd | D1-26 | JH3b | germ line |
| 11a | VH3 | 3-30 | 100 | 0 | 0/0 | nd | nd | 0/0 | nd | nd | D6-19 | JH6b | germ line |
| 11b | VH2 | 2-70 | 96.3 | 11 | 5/0 (2.1) | 4.05 | 0.031# | 5/1 (6.3) | 2.90 | 0.177 | D6-6 | JH4b | somatic mutation |
| Family . | VH Family . | VH Gene . | % Identity . | n . | CDR1 + CDR2 . | FR1 + FR2 + FR3 . | D Gene . | JH Gene . | Sequence Pattern . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Observed R/S (Exp. R) . | Inherent R:S . | P . | Observed R/S (Exp. R) . | Inherent R:S . | P . | ||||||||
| 1a | VH4 | 4-34 | 100 | 0 | 0/0 | nd | nd | 0/0 | nd | nd | D3-10 | JH4b | germ line |
| 1b | VH3 | 3-13 | 99.7 | 1 | 1/0 (0.2) | 3.23 | nd | 0/0 (0.6) | nd | nd | D2-2 | JH5b | germ line* |
| 2a/1('88) | VH1 | 1-02 | 91.8 | 24 | 8/0 (4.4) | 4.34 | 0.035# | 7/9 (13.9) | 2.99 | 2.92 × 10−3† | D6-19 | JH5b | intraclonal mutation |
| 2a/2('88) | VH1 | 1-02 | 93.2 | 20 | 6/0 (3.6) | 4.34 | 0.085 | 7/7 (11.6) | 2.99 | 0.021† | D6-19 | JH5b | intraclonal mutation |
| 2a/3('88) | VH1 | 1-02 | 91.2 | 26 | 8/0 (4.7) | 4.34 | 0.051 | 10/8 (15.1) | 2.99 | 0.021† | D6-19 | JH5b | intraclonal mutation |
| 2a′/1('98) | VH1 | 1-02 | 91.2 | 26 | 8/0 (4.7) | 4.34 | 0.051 | 10/8 (15.1) | 2.99 | 0.021† | D6-19 | JH5b | intraclonal mutation |
| 2a′/2('98) | VH7 | 7-4-1 | 92.4 | 22 | 3/2 (3.9) | 3.63 | 0.209 | 8/9 (12.9) | 2.94 | 0.023† | D2-15 | JH4b | intraclonal mutation |
| 2b | VH4 | 4-61 | 94.6 | 16 | 6/3 (3.0) | 4.03 | 0.042# | 4/3 (8.9) | 2.61 | 0.010† | D3-22 | JH4b | somatic mutation |
| 3a | VH3 | 3-73 | 91.8 | 24 | 9/1 (4.4) | 3.08 | 0.015# | 10/4 (13.6) | 2.99 | 0.057 | D1-7 | JH4b | somatic mutation |
| 3b | VH3 | 3-74 | 93.2 | 20 | 6/0 (3.6) | 3.90 | 0.079 | 9/5 (11.4) | 2.80 | 0.097 | D3-9 | JH5a | somatic mutation |
| 4a | VH4 | 4-34 | 93.8 | 18 | 6/1 (3.3) | 4.50 | 0.065 | 4/7 (10.2) | 2.73 | 2.65 × 10−3† | D3-3 | JH5a | somatic mutation |
| 4b | VH3 | 3-30 | 91.2 | 26 | 4/7 (4.6) | 3.87 | 0.200 | 9/6 (14.9) | 2.85 | 0.011† | N/A | JH4b | somatic mutation |
| 4c | VH3 | 3-30 | 99.7 | 1 | 0/0 (0.2) | nd | nd | 1/0 (0.6) | 2.85 | 0.574 | D6-25 | JH4b | germ line* |
| 5a | VH1 | 1-69 | 99.7 | 1 | 0/1 (0.2) | nd | nd | 0/0 (0.6) | nd | nd | D2-2 | JH6b | germ line* |
| 5b | VH5 | 5-51 | 100 | 0 | 0/0 | nd | nd | 0/0 | nd | nd | N/A | JH6b | germ line |
| 6a | VH1 | 1-69 | 100 | 0 | 0/0 | nd | nd | 0/0 | nd | nd | D2-2 | JH6b | germ line |
| 6b | VH1 | 1-69 | 100 | 0 | 0/0 | nd | nd | 0/0 | nd | nd | D2-2 | JH6b | germ line |
| 7a | VH3 | 3-30 | 94.6 | 16 | 2/4 (2.9) | 4.05 | 0.242 | 6/4 (9.2) | 2.85 | 0.056 | N/A | JH4b | somatic mutation |
| 7b | VH3 | 3-7 | 92.2 | 23 | 8/1 (4.3) | 4.94 | 0.032# | 11/3 (13.2) | 2.84 | 0.108 | N/A | JH4b | somatic mutation |
| 8a | VH1 | 1-24 | 98.3 | 5 | 5/0 (0.9) | 4.08 | 1.94 × 10−4# | 0/0 (2.9) | nd | nd | N/A | JH6b | somatic mutation |
| 8b | VH3 | 3-9 | 100 | 0 | 0/0 | nd | nd | 0/0 | nd | nd | D3-16 | JH4b | germ line |
| 9a | VH4 | 4-39 | 100 | 0 | 0/0 | nd | nd | 0/0 | nd | nd | N/A | JH5b | germ line |
| 9b | VH1 | 1-02 | 100 | 0 | 0/0 | nd | nd | 0/0 | nd | nd | D4-17 | JH6b | germ line |
| 10a | VH1 | 1-02 | 100 | 0 | 0/0 | nd | nd | 0/0 | nd | nd | D6-19 | JH4b | germ line |
| 10b | VH4 | 4-39 | 100 | 0 | 0/0 | nd | nd | 0/0 | nd | nd | D1-26 | JH3b | germ line |
| 11a | VH3 | 3-30 | 100 | 0 | 0/0 | nd | nd | 0/0 | nd | nd | D6-19 | JH6b | germ line |
| 11b | VH2 | 2-70 | 96.3 | 11 | 5/0 (2.1) | 4.05 | 0.031# | 5/1 (6.3) | 2.90 | 0.177 | D6-6 | JH4b | somatic mutation |
CDR = Complementarity-determining region; FR = frame work region; R = number of detected R mutations; S = number of detected S mutations; inherent R:S = mutation ratio of the quotient of total possible R to total possible S mutations (Chang and Casali29); P = probability;
, statistically significant (P < .05);
, case showing one nucleotide alteration; N/A = D region not assigned (Corbett et al28).
Mutation pattern in familial B-CLL
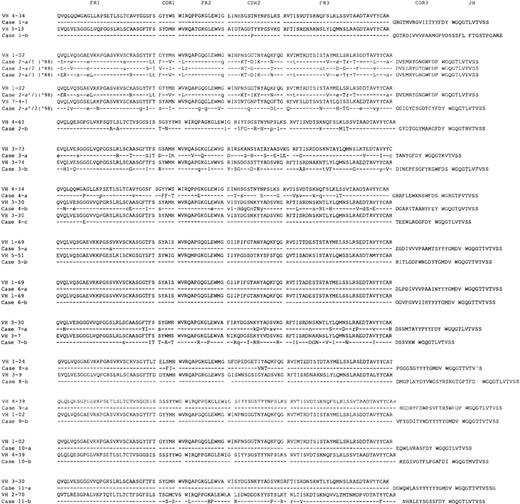
The mutation analysis data are summarized in Table 2, and the deduced amino acid sequences of the IgH (VDJ) gene are depicted in Figure 1. The similarity of the VH genes to the closest germ line genes is shown as percentage identity and ranged from 91.2% to 100% (Table 2). Thirteen of the 23 cases showed 99% to 100% homology to their closest germ line genes. The others showed differences of more than 5 nucleotides (ranging from 5 to 26). It cannot be excluded that some of these nucleotide differences derive from PCR errors or represent germ line polymorphisms, especially, in those cases showing 1 nucleotide difference. However, since most, if not all, of VH germline gene segments have been identified,30 these nucleotide differences were considered to be the result of somatic mutations.
Deduced amino acid sequences of the immunoglobulin gene regions (VH-DH-JH) used in the cases of familial CLL.
In each case, the top sequence represents the closest germ line VH genes. The DH and JH segments of the closest germ line are shown. The individual FR and CDR are indicated according to the V BASE sequence directory (I.M. Tomlinson, MRC Center for Protein Engineering, Cambridge, UK). Replacement mutations are shown by uppercase letters and silent mutations by lowercase letters. Identities are represented by dashes.
Deduced amino acid sequences of the immunoglobulin gene regions (VH-DH-JH) used in the cases of familial CLL.
In each case, the top sequence represents the closest germ line VH genes. The DH and JH segments of the closest germ line are shown. The individual FR and CDR are indicated according to the V BASE sequence directory (I.M. Tomlinson, MRC Center for Protein Engineering, Cambridge, UK). Replacement mutations are shown by uppercase letters and silent mutations by lowercase letters. Identities are represented by dashes.
Analysis of the distribution of somatic mutations in each sequence was based on a binomial distribution model of Chang and Casali.29 In 6 of the 10 cases carrying mutations (2a and b, 3a, 7a, 8a, and 11b), there was evidence of antigen selection (P < .05) as demonstrated by a higher number of replacement mutations than expected in the CDRs (see Table 2). With regard to the FR regions, a scarcity of replacement mutations reached a level of significance in 4 cases (2a and b, 4a and b) suggesting that these were under negative selective pressure. A lower than expected number of replacement mutations in the framework has been suggested to be the more critical parameter in determining whether a VH gene has been subjected to antigen selection.31 We examined VH gene family concordance within kindreds but failed to observe a significant excess.
Intraclonal mutations in family 2
We were able to analyze 2 samples collected over a 10-year interval (1988 and 1998) from case 2a (Figure 1). Six independent colonies were screened from the 1988 sample. All displayed rearrangements of the same VH1 family (VH1-02/DP-75), and used the same D and JH region (D6-19 and JH5b, respectively). However, sequence variations were detected among the 6 colonies. Three clones formed 1 identical set (a/1), 2 formed a second identical set (a/2), and the last clone (a/3) had yet a third distinct, although related, sequence. The subsequent specimen, obtained 10 years later in 1998, contained 2 distinct sequences. 1 (a′/1) was nearly identical to the a/3 sequence variant from the 1988 sampling, with the exception of 2 additional mutations present in CDR2. The second sequence was a completely different rearrangement, using a VH7 family gene (VI-4), and D and JH region genes, D2-15 and JH4b, respectively. These 2 subclones were verified by sequencing multiple colonies from separate RT-PCR reactions. No additional intraclonal variations were detected. These findings are consistent with the occurrence of intraclonal variations in the 1988 biopsy and clonal evolution in 1998 (a′/1), and suggest the emergence of a new clone as shown by the a′/2 sequence. Consistent with the molecular data, flow analysis of the 1998 specimen suggested a bimodal distribution of CD5 positive B cells (data not shown). Because intraclonal variation is highly unusual in CLL, it is important to reemphasize that the morphologic and phenotypic features of this patient's disease were fully consistent with CLL (CD5+, CD23+).
Correlation of somatic mutations with clinical and phenotypic parameters
The age at presentation, gender, Rai stage, WBC count, and duration of disease were examined in relation to the presence or absence of somatic mutations. Subjects with somatic mutations tended to be older 66.8 versus 54.9, P = .005 and have lower WBC counts 25.1 versus 63.6, P = .015 in univariate analysis. The Rai stage, duration since presentation, and gender distributions by mutation status were similar. Small numbers limited the multivariate analysis.
We also examined the relationship between somatic mutation and the relative density of expression of several surface markers (CD45, CD20, and CD5), as measured by fluorescence intensity. For this analysis, the relative antigenic density of the tumor cells was normalized to the average fluorescence intensity of the particular antigen as expressed on residual normal cells in the specimen (CD20 and CD45 on B cells and CD5 on T cells). By normalizing the flourescence intensities in this way, we could compare 1 case with another, eliminating fluorescence differences attributable to the particular analysis. Of the 3 markers studied, only the relative intensity of CD5 expression was significantly (inversely) related to the presence of somatic mutations (Mann-Whitney, 2-tailed, P = .02, analysis of varianceP = .012).
Discussion
B-cell chronic lymphocytic leukemia has traditionally been thought of as a homogeneous disease because of its rather uniform morphology and immunophenotype. Recent cytogenetic and molecular biologic analyses have challenged this view. It has become apparent that cases of CLL may contain distinct and recurring cytogenetic abnormalities with up to 30% of cases with trisomy 12,9,32 45% with del 13q24,11 and another 20% with deletions of 11q23.11 Moreover, analysis of the expressed immunoglobulin genes in CLL indicate that as many as 30% to 50% of cases show evidence of somatic mutation, with a high percentage of these showing evidence of having undergone antigen selection.18 These data suggest that sporadic CLL develops from either a CD5+ naive B cells or from a more mature CD5+ B cell that has undergone somatic mutation and antigen selection. This recently recognized heterogeneity in sporadic CLL led us to ask whether the familial variant of CLL would show the same heterogeneity with respect to the expressed immunoglobulin gene as that seen in sporadic CLL, or whether familial CLL would show a more restricted profile.
Our study indicates that familial CLL is indistinguishable from sporadic CLL with respect to immunoglobulin VH gene usage and the occurrence of somatic mutation. The majority of cases in this study used either VH1, VH3, or VH4, at frequencies of 33.3%, 37.5%, and 20.8%, respectively. This is very similar to the VH gene usage frequencies reported by Fais et al18 for cases of IgM+ sporadic CLL (28.1%, 37.5%, and 30.1% for VH1, VH3, and VH4, respectively). Compared with the pattern of VH family usage in adult peripheral blood derived CD5 positive B cells reported by Brezinschek et al,33 both sporadic CLL and familial CLL show higher than expected usage of VH1 genes, and lower than expected usage of VH3 genes. However, contrary to a recent report by Rosenquist et al,34 who found a very high usage of the VH1 family member VH1-69 in sporadic CLL (6/9), only 2 of our 8 VH1 family cases expressed VH1-69. VH1-69 has been associated with autoimmune manifestations in sporadic CLL, and although the 2 VH1-69 expressing cases in this study have not displayed autoimmune manifestations, it is interesting that these 2 patients were members of the same family. Consistent with data reported by Johnson et al,35 both VH1-69 expressers used JH6b. In contrast to the relatively low usage of VH1-69, 4 of 8 (50%) of our familial CLL cases used VH1-02. Two of these were from the same family, and the remaining 2 from different additional families. In sporadic CLL, VH1-02 was reported to occur in 4 of 18 (22%) IgM+ cases.18 This is higher than the incidence reported in “normal” peripheral CD5+ B cells, but half the incidence found in the familial CLL cases. Whether this is a statistical anomaly of our cases or a true bias in gene usage will require the examination of additional familial cases. Interestingly, VH1-02 has also been reported in autoimmune disease, although less commonly than VH1-69.36
The presence of somatic mutations was demonstrated in 43% of cases (10/23) and evidence for antigen selection was noted in 60% of these (6/10). These percentages are again similar to those reported for sporadic CLL,18 and suggest that, like sporadic CLL, familial CLL is heterogeneous and may arise from pregerminal center cells or postgerminal center cells. Although early studies of small numbers of cases of sporadic CLL showed minimal numbers of somatic mutations in sporadic CLL,13,14 more recent larger surveys have demonstrated a greater than 5% mutation rate in up to 32% of IgM+ CLL.3,18-20 In the study of Fais et al,18a hierarchy of mutation was seen with VH3 > VH4 > VH1. This same hierarchy of mutation was also seen in our cases of familial CLL.
The majority of cases with mutated VH genes showed evidence of antigenic selection. This was determined by the method of Chang and Casali.29 According to these authors, replacement mutations higher than expected by chance alone reflects a positive pressure by antigen, and conversely, replacement mutations lower than that expected by chance alone indicate a negative pressure by antigen. Using this formulation, we found that 60% of the mutated cases (25% of our total cases) showed evidence of antigen selection. This percentage is similar to that reported for sporadic CLL by Fais et al18 who found evidence for antigenic stimulation in 20% of their total cases, using the same method.
Although we used the method of Chang and Casali to estimate antigen selection, recent data from Dorner et al31 suggest that this formula may not accurately assess antigen selection. Dorner et al found that peripheral blood B cells possess similar ratios of replacement to silent mutations in the CDRs of unexpressed alleles and expressed alleles, suggesting that the analysis of CDR regions cannot provide information regarding antigen selection. On the contrary, these investigators found that the most consistent difference between the expressed and unexpressed alleles was the conservation of framework regions in the expressed allele, suggesting that there was selective pressure to maintain the immunoglobulin structure only in the expressed allele. These data suggest that the assessment of antigen selection should focus on mutations occurring only within the framework regions. Because the Chang and Casali formulation considers mutations occurring in both the CDRs and the framework regions, this method for assessing antigen selection may need modification. However, because all the comparable literature in sporadic CLL have used the Chang and Casali method, and because we wished to compare the molecular features of Ig usage and somatic mutation with the published literature regarding sporadic CLL, we chose to use the same model, realizing that it may not be providing a completely accurate estimate of antigen selection.
One case of familial CLL (patient 2) showed evidence of intraclonal variation and clonal evolution occurring over a 10-year period. In this case, 2 samples acquired 10 years apart were analyzed. The first sample showed a single rearrangement involving VH1 (VH1-02/D6-19/JH5b). Several distinct but related subclones were identified within this sample (intraclonal variation). The second sample contained a sequence that was similar to 1 of the VH1-02 variants, but with 2 additional mutations located within CDR2 (clonal evolution), and a second unrelated sequence using VH7 (VI-4/D2-15/JH4b). It is unclear whether this second sequence represents a completely new CLL clone or whether it represents a further rearrangement within the original tumor. The presence of intraclonal variation in this case was unique and contrasts with recent studies that failed to demonstrate intraclonal diversification in sporadic B-CLL.18,37 38 This case suggests that in rare instances, mutations in familial CLL may continue to occur after the neoplastic transformation.
Specific cytogenetic lesions in sporadic CLL have been associated with the presence or absence of somatic mutation. CLL cases carrying 13q14 deletions showed high levels of mutations compared with cases with trisomy 12.19 Unfortunately, the material used in this study was cryopreserved, and cytogenetic analyses had not been performed as part of the initial investigation of these patients. Therefore, we were unable to compare the cytogenetic features of familial CLL with sporadic CLL, and confirm this correlation for the familial CLL cases.
Although this is one of the larger series of molecularly characterized familial CLL, the number of cases is still relatively small to draw clinical correlations from the data. Nonetheless, we attempted to correlate several clinical and phenotypic features with the presence or absence of mutations. Of the characteristics assessed, the age at presentation, WBC and CD5 fluorescence intensity showed a significant relationship to the presence or absence of mutation.
Patients with mutated VH genes tended to be older and had significantly lower peripheral blood white cell counts than did patients without somatic mutations. It is interesting to speculate that there may be a tendency in older individuals for the leukemia to originate from an antigen exposed postgerminal center cell, and in younger individuals from a naive pregerminal center B cell. The difference in the number of circulating neoplastic cells could reflect differences in homing properties or alternatively, it could reflect differences in homeostatic properties of pregerminal and postgerminal center B cells that ultimately determine the levels of leukemia cells attainable in the blood.
We did not have a sufficiently large enough sample or a long enough follow-up to compare survival in the mutated and nonmutated patients. In light of recent studies by Hamblin et al39 and Damle et al40 indicating that mutated cases have much better survival than nonmutated cases, it will be important to follow these patients over time to see if this clinical correlation with mutation status holds up in the familial patients.
We also found a significant inverse correlation between the fluorescence intensity of CD5 and the presence of somatic mutation. Recently Pospisil et al41 42 reported that CD5 has a low affinity binding capacity for the Ig heavy chain framework sequence, as well as its traditional receptor molecules. These investigators suggested that the interaction between CD5 and the FR structures in the VH genes might affect the maintenance and selective expansion of B cells using specific VH genes, and conceivably play a role in the growth of transformed cells such as in CLL. It is interesting to speculate that these mutations could affect CD5 binding and thereby alter the proliferative capacity of the leukemic clone. Preliminary experiments to investigate this possibility are underway in our laboratories.
Acknowledgments
We thank Patricia H. Carter for sample preparation and immunophenotyping results and Laura Fontaine, RN, for invaluable assistance with patient management and specimen organization.
Reprints:Mark Raffeld, Laboratory of Pathology, National Cancer Institute, Building 10, Room 2N110, 9000 Rockville Pike, Bethesda, MD 20892; e-mail: mraff@box-m.nih.gov.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The US government retains nonexclusive, royalty-free license to any copyright covering this article.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal