Single cell studies aimed at clarifying the nature and clonality of Hodgkin and Reed-Sternberg (HRS) cells of classical Hodgkin's disease (HD) have so far produced conflicting results. Using an improved single cell procedure, the HRS cells of 25 patients with nodular sclerosing HD lacking B- and T-cell antigens, with and without Epstein-Barr virus infection, were analyzed for the presence of immunoglobulin (Ig) gene rearrangements. One patient with HD developed follicular lymphoma 2 years later. Both lymphomas originated from a common precursor identified as a germinal center B cell. The data show that all but one of the investigated cases harbored rearranged Ig genes, which were clonal in all instances and carried a high load of somatic mutations. The Ig coding capacity was preserved in 18 of the 24 cases (75%) with rearrangements. However, expression of Ig messenger RNA was not detectable in the HRS cells with the exception of Ig kappa light chain expression in some tumor cells of 1 case. The lack of Ig gene transcription in HRS cells was confirmed by analyzing the HD cell lines L428 and KM-H2 in transient transfection experiments. An Ig promoter/enhancer reporter construct showed virtually no activity in these cells compared to 5 control B-cell lines. We conclude that (1) classical HD is a B-cell lymphoma in most instances, (2) HRS cells are clonal without any exception, (3) they are derived from germinal center B-cells that (4) mostly lack crippling mutations but (5) have consistently lost their Ig gene transcription ability, due to functional defects in the Ig gene regulatory elements.
Hodgkin's disease (HD), one of the most frequent lymphomas in Western countries, is characterized by scattered large atypical cells residing in a complex admixture of inflammatory cells.1 Two different biologic entities have been recognized within HD: nodular lymphocyte predominance HD (LPHD) and classical HD comprising nodular sclerosis, mixed cellularity, and lymphocyte depletion.2 Whereas the nature of the tumor cells of LPHD is regarded as clarified,3-5 the cellular origin of the atypical cells associated with classical HD designated as Hodgkin and Reed-Sternberg (HRS) cells is less clear. Immunophenotypical studies demonstrated the lymphocyte activation antigen CD30 on HRS cells and this pointed toward a lymphocytic origin for these cells.6 However, B- and T-cell antigens were absent in the majority of the cases, thus drawing their lymphocytic nature into doubt. Attempts to confirm or disprove a lymphocytic origin for the HRS cells at the genomic level by Southern blot or polymerase chain reaction (PCR) analysis for the detection of clonal antigen receptor gene rearrangements in whole tissue DNA extracts have been largely unsuccessful.7-11
The development of techniques for the isolation of single cells from histologic sections raised the possibility of directly analyzing immunoglobulin (Ig) chain gene rearrangements in individual HRS cells. However, the results obtained have so far provided more confusion than clarification. In 1 study of this kind, all 12 cases investigated revealed no Ig rearrangements12 whereas in the studies by Rajewsky's group, all but 1 of the classical HD cases represented the outgrowth of a dominant B-cell clone13,14 The results by our group15 showed clonal as well as polyclonal rearrangements, whereas Delabie et al16 described exclusively polyclonally rearranged HRS cells. Interestingly, additional polyclonal Ig rearrangements were also encountered in 2 of the cases included in the study of Rajewsky and coworkers14and also in a recent study by Deng et al.17 The detailed analysis of the clonally rearranged HRS cells disclosed that they carried high loads of somatic Ig mutations indicating that they originate from germinal or postgerminal center B cells. Moreover, with the exception of only 1 case, Rajewsky's group did not detect potentially functional rearrangements in any HRS cell. It was suggested that this disruption of the coding capacity is responsible for the absence of Ig expression described in classical HD.14
To clarify the confusing findings concerning the clonality of the HRS cells, we applied an improved single cell isolation procedure and PCR to the investigation of 25 patients with classical HD. We restricted our approach to the analysis of cases of nodular sclerosis lacking expression of B- and T-cell antigens, and with and without Epstein-Barr virus (EBV) infection, because the most discordant data were obtained from cases of this main histotype of classical HD.15 16 Our results demonstrate that all HRS cells harbored exclusively monoclonal Ig rearrangements. Surprisingly, the majority of the HRS cell-derived Ig rearrangements proved to be functional. Therefore, we extended our study to the expression of Ig-specific messenger RNA (mRNA) in primary HRS cells and to the transcription activity of Ig genes in the HD lines L428 and KM-H2. We found absence of Ig transcripts in all cases with the exception of IgLκ in some HRS cells in a single case and a highly reduced Ig transcription activity in the transfected HD cell line cells. This demonstrates that the nonexpression of Ig in HRS cells is not caused by crippling mutations but by defective Ig gene-specific transcription machinery.
Materials and methods
Tissues
Samples of frozen tissue from 25 patients with classical HD of nodular sclerosing type were selected from the files of the Institute of Pathology at the Free University of Berlin. The phenotype of the HRS cells was determined by immunohistochemical staining with a panel of antibodies and detection by the APAAP technique.18 In addition, several reactive lymph nodes were selected as controls.
Isolation of single cells
Frozen sections, 7 μ thick, immunostained for CD30 (Ber-H2), were used for the isolation of individual HRS cells. For control purposes, sections of the same cases and from the reactive lymph nodes were immunostained for the β-chain of the T-cell receptor (βF1) and for CD20 (L26), respectively, and used for the isolation of B and T cells. Single cells were extracted from tissue sections and transferred into PCR tubes with a minimal volume of buffer covering the section (0.05-0.1 μL) by hydraulic micromanipulators. After the isolation of each cell an aliquot of approximately 1 μL was drawn from the buffer to serve as a negative control.
Single cell PCR and sequence analysis
After treatment of the isolated cells with proteinase K (1 hour, 55°C), the DNA was subjected to PCR for the detection of rearranged variable heavy chain (VH) or variable kappa light chain (VLκ) genes, respectively. For amplification of VH rearrangements, a full-nested PCR was performed as previously described, using family-specific framework (FW) 1 and FW2 primer sets.3 For amplification of VLκrearrangements the first round of amplification was carried out with family-specific VLκ primers as described by Kanzler et al.14 For reamplification we designed a new set of VLκ primers capable of binding in FW1 region downstream of the first primer set. In contrast to previous assays, this allows for the detection of all VLκ rearrangements in only 1 reamplification. Both VLκ primer sets were used in conjunction with 2 nested sets of JLκprimers.14
Resulting PCR products were isolated from the polyacrylamide gels by elution and directly sequenced on an automated DNA sequencer (PE/Applied Biosystem, 377A, Weiterstadt, Germany) by the chain termination technique using fluorescence labeled ddNTPs (BigDye; Applied Biosystems). The sequences obtained were compared to the corresponding germline VH and VLκ segments (VBASE)19 to determine the rearranged V segment and to demonstrate the number of somatic mutations. In addition, the sequences were compared to each other and to our own database to establish their clonal relationship, to detect intraclonal diversities, and to rule out contamination with rearrangements previously amplified in our laboratory. Furthermore, the sequences were translated into amino acids to calculate the ratio of replacement and silent mutations and to detect disruptions of the coding capacity.
Antigen selection
Somatic mutations compatible with antigen selection were calculated by 2 different methods. First, the ratio of replacement to silent mutations (R/s) in the complementarity-determining region (CDR) 2 and FW3 region was determined. A sequence was considered to be antigen selected when the R/s ratio in the CDR2 was higher than 2.9 and R/s ratio in the FW3 region was lower than 1.5.20 Second, the R/s ratio of the somatic mutations was considered only in the FW3 region. A sequence was regarded as being antigen selected when the R/s ratio was lower than 1.6.21
In situ hybridization
Immunoglobulin gene expression was investigated by radioactive in situ hybridization as previously reported.22 The probes for the Ig light chains kappa and lambda were kindly provided by Dr. Philip Leder (Harvard Medical School, Cambridge, Massachusetts) and probes for the constant regions of the Ig heavy chains (δ, μ, and γ) were prepared from complimentary DNA (cDNA) obtained from peripheral blood lymphocytes or B-cell lines. The nucleotide sequence of all probes was determined and proved to conform to published data.
Cell culture and transfection
Namalwa, BJA-B, DG-75, Raji, and Jok-1 (control B-cell lines), and L428 and KM-H2 (HD cell lines) cells were grown in RPMI 1640 medium (Life Technologies, Karlsruhe, Germany) supplemented with 10% fetal calf serum (FCS), antibiotics, l-glutamate, and 50 μM β-mercaptoethanol at 37°C and 5% CO2. For electroporation 5 × 106 cells were gently pelleted and resuspended in 300 μL RPMI medium with 10% FCS. Fifteen micrograms of the luciferase reporter plasmid (μET1.luc and μET1M.luc, pTATA and pTKL, and 100 ng pRL-TK [Promega, Mannheim, Germany]) was added to the cells.23 The mixture was then transferred to a cuvette with a gap of 0.4 cm and electroporated at 230 V and 960 μF with a Biorad Gene Pulser. Cells were then immediately transferred to a petri dish containing 10 mL RPMI with 10% FCS and incubated for 20 hours. The cells were harvested and the lysates were prepared and measured with the dual-luciferase reporter assay system (Promega). All luciferase values were calculated against the renilla-luciferase values to correct for different transfection efficiencies.
Results
Phenotype
The immunophenotypes of the 25 classical HD cases of nodular sclerosing type are summarized in Table 1. In brief, all HRS cells were CD30+ but negative for T- and B-cell antigens as well as for the ALK protein. An EBV infection of the HRS cells was demonstrated by expression of the virally encoded protein LMP-1 in 12 cases.
Clinical data and immunophenotype of Hodgkin and Reed-Sternberg cells analyzed for immunoglobulin gene rearrangements from 25 patients with classical Hodgkin's disease of nodular sclerosing type
| Patients . | Age/Sex . | Localization . | CD30 . | CD15 . | CD20 . | CD3 . | ALK-1 . | EBV (LMP) . |
|---|---|---|---|---|---|---|---|---|
| 1 | 33/M | SC LN | + | + | − | − | − | − |
| 2 | 30/M | AX LN | + | + | − | − | − | − |
| 3 | 18/F | CV LN | + | + | − | − | − | − |
| 4 | 38/M | LN | + | + | − | − | − | − |
| 5 | 23/M | LN | + | + | − | − | − | − |
| 6 | 60/M | IN LN | + | + | − | − | − | − |
| 7 | 58/M | LN | + | + | − | − | − | − |
| 8 | 23/F | SC LN | + | + | − | − | − | − |
| 9 | 25/F | CV LN | + | + | − | − | − | − |
| 10 | 40/M | SC LN | + | + | − | − | − | − |
| 11 | 39/F | LN | + | + | − | − | − | − |
| 12 | 70/F | CV LN | + | − | − | − | − | − |
| 13 | na | LN | + | + | − | − | − | − |
| 14 | 24/M | SC LN | + | + | − | − | − | + |
| 15 | 35/M | SC LN | + | + | − | − | − | + |
| 16 | 90/M | LN | + | + | − | − | − | + |
| 17 | 27/F | LN | + | + | − | − | − | + |
| 18 | 80/F | SC LN | + | − | − | − | − | + |
| 19 | 26/F | MD | + | − | − | − | − | + |
| 20 | 29/M | PA LN | + | + | − | − | − | + |
| 21 | 65/M | CV LN | + | + | − | − | − | + |
| 22 | 6/M | LN | + | + | − | − | − | + |
| 23 | 22/F | CV LN | + | + | − | − | − | + |
| 24 | 30/M | MD | + | + | − | − | − | + |
| 25 | Na | LN | + | + | − | − | − | + |
| Patients . | Age/Sex . | Localization . | CD30 . | CD15 . | CD20 . | CD3 . | ALK-1 . | EBV (LMP) . |
|---|---|---|---|---|---|---|---|---|
| 1 | 33/M | SC LN | + | + | − | − | − | − |
| 2 | 30/M | AX LN | + | + | − | − | − | − |
| 3 | 18/F | CV LN | + | + | − | − | − | − |
| 4 | 38/M | LN | + | + | − | − | − | − |
| 5 | 23/M | LN | + | + | − | − | − | − |
| 6 | 60/M | IN LN | + | + | − | − | − | − |
| 7 | 58/M | LN | + | + | − | − | − | − |
| 8 | 23/F | SC LN | + | + | − | − | − | − |
| 9 | 25/F | CV LN | + | + | − | − | − | − |
| 10 | 40/M | SC LN | + | + | − | − | − | − |
| 11 | 39/F | LN | + | + | − | − | − | − |
| 12 | 70/F | CV LN | + | − | − | − | − | − |
| 13 | na | LN | + | + | − | − | − | − |
| 14 | 24/M | SC LN | + | + | − | − | − | + |
| 15 | 35/M | SC LN | + | + | − | − | − | + |
| 16 | 90/M | LN | + | + | − | − | − | + |
| 17 | 27/F | LN | + | + | − | − | − | + |
| 18 | 80/F | SC LN | + | − | − | − | − | + |
| 19 | 26/F | MD | + | − | − | − | − | + |
| 20 | 29/M | PA LN | + | + | − | − | − | + |
| 21 | 65/M | CV LN | + | + | − | − | − | + |
| 22 | 6/M | LN | + | + | − | − | − | + |
| 23 | 22/F | CV LN | + | + | − | − | − | + |
| 24 | 30/M | MD | + | + | − | − | − | + |
| 25 | Na | LN | + | + | − | − | − | + |
AX indicates axillar; CV, cervical; IN, inguinal; LN, lymph node; MD, mediastinum; PA, paraaortic; SC, supraclavicular; na, not available.
Clonality of the HRS cells
A total of 1078 single CD30+ HRS cells were isolated from frozen tissue sections of 25 cases, 24 of which disclosed VH or VLκ (or both) rearrangements (96%). Of the 24 classical HD cases, 21 (88%) contained VH rearrangements and 18 of 23 cases (78%; case 5 was not analyzed for VLκ rearrangement due to a lack of material) displayed rearranged VLκ genes as demonstrated by single copy PCR. The number of PCR-positive HRS cells per case ranged from 3 to 28 (195 in total) for VH rearrangements and from 2 to 12 (96 in total) for VLκ rearrangements (Table2).
Analysis of single Hodgkin and Reed-Sternberg cells for immunoglobulin heavy and light chain gene rearrangements
| Patient . | Rearrangements of the Ig Heavy Chain Gene . | Rearrangements of the Ig Kappa Light Chain Gene . | ||||||
|---|---|---|---|---|---|---|---|---|
| Isolated HRS Cells . | HRS Cells with IgH-R . | Sequences Identical/Unrelated . | VH Family . | Isolated HRS Cells . | HRS Cells with IgL-R . | Sequences Identical/Unrelated . | VK Family . | |
| 1 | 54 | 28 | 28/0 | VH3 | 27 | 12 | 12/0 | VK2 |
| 2 | 53 | 12 | 12/0 | VH3 | 15 | 3 | 3/0 | VK2 |
| 3 | 15 | 6 | 6/0 | VH3 | 24 | 0 | 0 | — |
| 4 | 15 | 10 | 10/0 | VH3 | 17 | 5 | 5/0 | VK2 |
| 5 | 37 | 6 | 6/0 | VH1 | na | na | na | na |
| 6 | 33 | 9 | 9/0 | VH3 | 24 | 0 | 0 | — |
| 7 | 29 | 11 | 11/0 | VH3 | 16 | 6 | 6/0 | VK3 |
| 8 | 30 | 5 | 5/0 | VH3 | 14 | 3 | 3/0 | VK1 |
| 9 | 25 | 13 | 13/0 | VH3 | 10 | 2 | 2/0 | VK3 |
| 10 | 54 | 4 | 4/0 | VH3 | 16 | 4 | 4/0 | VK1 |
| 11 | 16 | 0 | 0 | — | 12 | 5 | 5/0 | VK3 |
| 12 | 50 | 0 | 0 | — | 13 | 5 | 5/0 | VK2 |
| 13 | 15 | 8 | 8/0 | VH3 | 12 | 0 | 0 | — |
| 14 | 24 | 0 | 0 | — | 18 | 0 | 0 | — |
| 15 | 48 | 12 | 12/0 | VH3 | 10 | 8 | 6(2)/0* | VK3/VK1 |
| 16 | 32 | 16 | 16/0 | † | 32 | 0 | 0 | — |
| 17 | 35 | 17 | 17/0 | VH1 | 12 | 3 | 3/0 | VK2 |
| 18 | 18 | 7 | 7/0 | VH3 | 11 | 2 | 2/0 | VK3 |
| 19 | 15 | 5 | 5/0 | VH4 | 12 | 5 | 5/0 | VK3 |
| 20 | 9 | 3 | 3/0 | VH4 | 20 | 4 | 4/0 | VK4 |
| 21 | 17 | 6 | 6/0 | VH3 | 15 | 6 | 6/0 | VK2 |
| 22 | 16 | 6 | 6/0 | VH3 | 15 | 10 | 10/0 | VK3 |
| 23 | 12 | 4 | 4/0 | VH2 | 17 | 4 | 4/0 | VK4 |
| 24 | 12 | 7 | 7/0 | VH3 | 10 | 0 | 0 | — |
| 25 | 32 | 0 | 0 | — | 10 | 9 | 9/0 | VK2 |
| Total | 696 | 195 | 195/0 | 382 | 96 | 96/0 | ||
| Patient . | Rearrangements of the Ig Heavy Chain Gene . | Rearrangements of the Ig Kappa Light Chain Gene . | ||||||
|---|---|---|---|---|---|---|---|---|
| Isolated HRS Cells . | HRS Cells with IgH-R . | Sequences Identical/Unrelated . | VH Family . | Isolated HRS Cells . | HRS Cells with IgL-R . | Sequences Identical/Unrelated . | VK Family . | |
| 1 | 54 | 28 | 28/0 | VH3 | 27 | 12 | 12/0 | VK2 |
| 2 | 53 | 12 | 12/0 | VH3 | 15 | 3 | 3/0 | VK2 |
| 3 | 15 | 6 | 6/0 | VH3 | 24 | 0 | 0 | — |
| 4 | 15 | 10 | 10/0 | VH3 | 17 | 5 | 5/0 | VK2 |
| 5 | 37 | 6 | 6/0 | VH1 | na | na | na | na |
| 6 | 33 | 9 | 9/0 | VH3 | 24 | 0 | 0 | — |
| 7 | 29 | 11 | 11/0 | VH3 | 16 | 6 | 6/0 | VK3 |
| 8 | 30 | 5 | 5/0 | VH3 | 14 | 3 | 3/0 | VK1 |
| 9 | 25 | 13 | 13/0 | VH3 | 10 | 2 | 2/0 | VK3 |
| 10 | 54 | 4 | 4/0 | VH3 | 16 | 4 | 4/0 | VK1 |
| 11 | 16 | 0 | 0 | — | 12 | 5 | 5/0 | VK3 |
| 12 | 50 | 0 | 0 | — | 13 | 5 | 5/0 | VK2 |
| 13 | 15 | 8 | 8/0 | VH3 | 12 | 0 | 0 | — |
| 14 | 24 | 0 | 0 | — | 18 | 0 | 0 | — |
| 15 | 48 | 12 | 12/0 | VH3 | 10 | 8 | 6(2)/0* | VK3/VK1 |
| 16 | 32 | 16 | 16/0 | † | 32 | 0 | 0 | — |
| 17 | 35 | 17 | 17/0 | VH1 | 12 | 3 | 3/0 | VK2 |
| 18 | 18 | 7 | 7/0 | VH3 | 11 | 2 | 2/0 | VK3 |
| 19 | 15 | 5 | 5/0 | VH4 | 12 | 5 | 5/0 | VK3 |
| 20 | 9 | 3 | 3/0 | VH4 | 20 | 4 | 4/0 | VK4 |
| 21 | 17 | 6 | 6/0 | VH3 | 15 | 6 | 6/0 | VK2 |
| 22 | 16 | 6 | 6/0 | VH3 | 15 | 10 | 10/0 | VK3 |
| 23 | 12 | 4 | 4/0 | VH2 | 17 | 4 | 4/0 | VK4 |
| 24 | 12 | 7 | 7/0 | VH3 | 10 | 0 | 0 | — |
| 25 | 32 | 0 | 0 | — | 10 | 9 | 9/0 | VK2 |
| Total | 696 | 195 | 195/0 | 382 | 96 | 96/0 | ||
Presence of two clonal IgLκ rearrangements.
A determination of the rearranged VH segment was not possible due to a 160 base pair deletion.
−No Ig rearrangement detectable; na, not analyzed.
IgH-R indicates immunoglobulin heavy chain gene rearrangement; IgLκ-R, immunoglobulin kappa light chain gene rearrangement.
All 291 VH and VLκ PCR products were sequenced and compared to each other and to our own, as well as to published database sequences. The rearranged VH and VLκ revealed a usage of V segments compatible with normal B cells (see Table 2). All rearrangements were clonal in each of the 24 PCR-positive classical HD cases without any exception. No homology was found in the B-cell characteristic CDR3 to other data bank sequences. In 1 patient (case 15) 2 clonal VLκ rearrangements were detectable, indicating a bi-allelic VLκ rearrangement (see Table 2).
VH and VLκ rearrangements and somatic mutations
The sequence alignment with the most homologous VH germline segment disclosed an average mutation frequency of 14.01% in the CDR2 and FW3 region ranging from 5 to 38 base substitutions. In contrast, the rearranged VLκ genes were less often mutated as revealed by comparison to the corresponding germline VLκ segments. Five of the 17 classical HD cases with analyzable VLκrearrangements showed no somatic mutations and in the remaining 12 cases mutations were found with an average frequency of 5% (range of base substitutions, 1-17; Table 3).
Sequence analysis of clonal immunoglobulin chain gene rearrangements of single Hodgkin and Reed-Sternberg cells
| Patient . | Ig Heavy Chain Gene . | Ig Light Chain Gene . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of Sequences Studied . | Total No. of Somatic Mutations . | CDRII R/S . | FW3 R/S . | % of Somatic Mutations CDR2 + FW3 . | No. of Sequences Studied . | Total No. of Somatic Mutations . | CDRII R/S . | FW3 R/S . | % of Somatic Mutations CDR2 + FW3 . | |
| 1 | 283-a | 31 | 15/1 | 10/5 | 21 | 12 | 3 | 1/0 | 2/0 | 2.5 |
| 2 | 12 | 17 | 5/2 | 7/3 | 11.6 | 3 | 1 | 1/0 | 0 | 0 |
| 3 | 6 | 17 | 6/2 | 7/2 | 11.1 | — | — | — | — | — |
| 4 | 10 | 7 | 4/0 | 2/1 | 5 | 5 | 8 | 1/0 | 4/3 | 6.8 |
| 5 | 6 | 32 | 6/3 | 16/7 | 21.7 | na | na | na | na | na |
| 6 | 9 | 28 | 9/3 | 11/5 | 19.4 | — | — | — | — | — |
| 7 | 113-b | 28 | 12/1 | 8/7 | 19.4 | 6 | 15 | 0/2 | 9/4 | 12.8 |
| 83-c | 5 | 27 | 9/2 | 8/8 | 18.8 | 3 | 0 | 0 | 0 | 0 |
| 9 | 133-d | 38 | 12/2 | 17/7 | 26 | 2 | 0 | 0 | 0 | 0 |
| 10 | 4 | 35 | 9/6 | 14/6 | 23.8 | 43-i | nc | nc | 19/1 | ? |
| 11 | — | — | — | — | — | 5 | 0 | 0 | 0 | 0 |
| 12 | — | — | — | — | — | 5 | 0 | 0 | 0 | 0 |
| 13 | 8 | 24 | 9/1 | 6/8 | 16.3 | — | — | — | — | — |
| 14 | — | — | — | — | — | — | — | — | — | — |
| 15 | 12 | 14 | 4/2 | 5/3 | 9.7 | 83-j | 6 | 2/0 | 2/1 | 4.2 |
| 16 | 16 | 3-e | nc | nc | nc | — | — | — | — | — |
| 17 | 17 | 16 | 2/2 | 7/5 | 11.1 | 33-k | 8 | 1/3 | 4/0 | 6.6 |
| 18 | 7 | 14 | 5/2 | 2/5 | 9.5 | 2 | 6 | 3/1 | 2/0 | 5.1 |
| 19 | 53-f | 28 | 9/2 | 13/4 | 20.2 | 5 | 17 | 4/0 | 8/5 | 14.5 |
| 20 | 3 | 11 | 5/1 | 3/2 | 7.6 | 4 | 1 | 0 | 1/0 | 0 |
| 21 | 6 | 5 | 0/1 | 3/1 | 3.4 | 6 | 3 | 0 | 1/2 | 2.5 |
| 22 | 6 | 11 | 3/2 | 4/2 | 7.5 | 10 | 3 | 1/0 | 1/1 | 2.5 |
| 23 | 43-g | 15 | 2/1 | 7/5 | 10.2 | 4 | 3 | 1/0 | 2/0 | 2.5 |
| 24 | 73-h | 10 | 4/0 | 6/0 | 6.9 | — | — | — | — | — |
| 25 | — | — | — | — | — | 91 | 0 | 0 | 0 | 0 |
| Total/Average | 195 | 20.4 | 6.5/1.8 | 7.8/4.3 | 14.01 | 95 | 4.4 | 0.9/0.4 | 3.2/1.0 | 5 |
| Patient . | Ig Heavy Chain Gene . | Ig Light Chain Gene . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of Sequences Studied . | Total No. of Somatic Mutations . | CDRII R/S . | FW3 R/S . | % of Somatic Mutations CDR2 + FW3 . | No. of Sequences Studied . | Total No. of Somatic Mutations . | CDRII R/S . | FW3 R/S . | % of Somatic Mutations CDR2 + FW3 . | |
| 1 | 283-a | 31 | 15/1 | 10/5 | 21 | 12 | 3 | 1/0 | 2/0 | 2.5 |
| 2 | 12 | 17 | 5/2 | 7/3 | 11.6 | 3 | 1 | 1/0 | 0 | 0 |
| 3 | 6 | 17 | 6/2 | 7/2 | 11.1 | — | — | — | — | — |
| 4 | 10 | 7 | 4/0 | 2/1 | 5 | 5 | 8 | 1/0 | 4/3 | 6.8 |
| 5 | 6 | 32 | 6/3 | 16/7 | 21.7 | na | na | na | na | na |
| 6 | 9 | 28 | 9/3 | 11/5 | 19.4 | — | — | — | — | — |
| 7 | 113-b | 28 | 12/1 | 8/7 | 19.4 | 6 | 15 | 0/2 | 9/4 | 12.8 |
| 83-c | 5 | 27 | 9/2 | 8/8 | 18.8 | 3 | 0 | 0 | 0 | 0 |
| 9 | 133-d | 38 | 12/2 | 17/7 | 26 | 2 | 0 | 0 | 0 | 0 |
| 10 | 4 | 35 | 9/6 | 14/6 | 23.8 | 43-i | nc | nc | 19/1 | ? |
| 11 | — | — | — | — | — | 5 | 0 | 0 | 0 | 0 |
| 12 | — | — | — | — | — | 5 | 0 | 0 | 0 | 0 |
| 13 | 8 | 24 | 9/1 | 6/8 | 16.3 | — | — | — | — | — |
| 14 | — | — | — | — | — | — | — | — | — | — |
| 15 | 12 | 14 | 4/2 | 5/3 | 9.7 | 83-j | 6 | 2/0 | 2/1 | 4.2 |
| 16 | 16 | 3-e | nc | nc | nc | — | — | — | — | — |
| 17 | 17 | 16 | 2/2 | 7/5 | 11.1 | 33-k | 8 | 1/3 | 4/0 | 6.6 |
| 18 | 7 | 14 | 5/2 | 2/5 | 9.5 | 2 | 6 | 3/1 | 2/0 | 5.1 |
| 19 | 53-f | 28 | 9/2 | 13/4 | 20.2 | 5 | 17 | 4/0 | 8/5 | 14.5 |
| 20 | 3 | 11 | 5/1 | 3/2 | 7.6 | 4 | 1 | 0 | 1/0 | 0 |
| 21 | 6 | 5 | 0/1 | 3/1 | 3.4 | 6 | 3 | 0 | 1/2 | 2.5 |
| 22 | 6 | 11 | 3/2 | 4/2 | 7.5 | 10 | 3 | 1/0 | 1/1 | 2.5 |
| 23 | 43-g | 15 | 2/1 | 7/5 | 10.2 | 4 | 3 | 1/0 | 2/0 | 2.5 |
| 24 | 73-h | 10 | 4/0 | 6/0 | 6.9 | — | — | — | — | — |
| 25 | — | — | — | — | — | 91 | 0 | 0 | 0 | 0 |
| Total/Average | 195 | 20.4 | 6.5/1.8 | 7.8/4.3 | 14.01 | 95 | 4.4 | 0.9/0.4 | 3.2/1.0 | 5 |
na indicates not analyzed; nc, not calculable; −, no immunoglobulin gene rearrangement found; 0, absence of somatic mutations.
Two HRS cells had an insertion of 22 base pair, causing a frame shift and a stop codon.
All HRS cell sequences harbor a 27 base pair insertion that contains a 19 base pair duplication of the neighboring CDR2.
VH: 3 base pair deletion in CDR2; VLκ: stop codon and frame shift in CDR3.
Deletion (2 base pairs) and insertion (1 base pair) in CDR2; deletion (1 base pair) in FW2 region; stop codon in CDR3.
The same 160 base pair deletion was detectable in all 16 HRS cells.
In 2 of 5 HRS cells a duplication of 98 base pairs was observed.
Two base pair insertion in the FW3 region.
Two base pair deletion in the FW3 region and two stop codons in the CDR2.
Deletion of 12 base pairs in CDR2.
Two of the eight VLκ gene rearrangements were unrelated and showed no somatic mutations.
Three base pair insertion and a stop codon in the FW3 region.
A stop codon in the FW3 region.
Functionality of the rearranged Ig genes
In 6 of the 24 patients (25%), the Ig coding capacity was disrupted in the amplified proportion of the Ig gene by stop codons or frame shifts or both. The VH rearrangement was affected in 3 cases and the VLκ rearrangement in another 3 cases. The Ig coding capacity was not altered in the remaining 18 cases (Table4). In 10 of these 18 cases the PCR was extended to amplify the CDR1 and FW2 region. However, in none of these additionally analyzed sequences could a disruption of the VH coding capacity be detected (data not shown).
Analysis of the coding capacity of the rearranged immunoglobulin heavy and light chain genes from single isolated Hodgkin and Reed-Sternberg cells
| Patient . | Alterations in the Ig Heavy Chain Genes . | Alterations in the Ig Light Chain Genes . | Ig Coding Capacity . | Ig mRNA Expression . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % of Somatic Mutations . | Stop Codons . | Deletion . | Insertion . | Duplication . | In Frame . | % of Somatic Mutations . | Stop Codons . | Deletion . | Insertion . | Duplication . | In Frame . | |||
| 1 | 214-150 | + (−) | − | (−) | − | + (−) | 2.5 | − | − | − | − | + | + (−) | − |
| 2 | 11.6 | − | − | − | − | + | 0 | − | − | − | − | + | + | − |
| 3 | 11.1 | − | − | − | − | + | nc | + | + | − | ||||
| 4 | 5 | − | − | − | − | + | 6.8 | − | − | − | − | + | 4-151κ+ | |
| 5 | 21.7 | − | − | − | − | + | nc | + | − | |||||
| 6 | 19.4 | − | − | − | − | + | nc | + | − | |||||
| 7 | (19.4) | − | − | + | + | + | 12.8 | − | − | − | − | + | + | − |
| 10 | 23.8 | − | − | − | − | + | nc | − | + | − | − | + | + | − |
| 11 | nc | 0 | − | − | − | − | + | + | − | |||||
| 12 | nc | 0 | − | − | − | − | + | + | − | |||||
| 13 | 16.3 | − | − | − | − | + | nc | + | − | |||||
| 15 | 9.7 | − | − | − | − | + | 4.2 | − | − | − | − | + | + | − |
| 18 | 9.5 | − | − | − | − | + | 5.1 | − | − | − | − | + | + | − |
| 19 | 20.24-150 | − | − | − | − | + | 14.5 | − | − | − | − | + | + | − |
| 20 | 7.6 | − | − | − | − | + | 0 | − | − | − | − | + | + | − |
| 21 | 3.4 | − | − | − | − | + | 2.5 | − | − | − | − | + | + | − |
| 22 | 7.5 | − | − | − | − | + | 2.5 | − | − | − | − | + | + | − |
| 23 | 10.2 | − | − | + | − | + | 2.5 | − | − | − | − | + | + | − |
| 8 | 18.8 | − | + | − | − | + | 0 | + | − | − | − | − | − | − |
| 9 | 26 | + | + | + | − | + | 0 | − | − | − | − | + | − | − |
| 16 | nc | − | + | − | − | − | nc | − | − | |||||
| 17 | 11.1 | − | − | − | − | + | 6.6 | + | − | + | − | nc | − | − |
| 24 | 6.9 | + | − | − | − | − | nc | − | − | |||||
| 25 | nc | 0 | + | + | − | − | + | − | − | |||||
| Patient . | Alterations in the Ig Heavy Chain Genes . | Alterations in the Ig Light Chain Genes . | Ig Coding Capacity . | Ig mRNA Expression . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % of Somatic Mutations . | Stop Codons . | Deletion . | Insertion . | Duplication . | In Frame . | % of Somatic Mutations . | Stop Codons . | Deletion . | Insertion . | Duplication . | In Frame . | |||
| 1 | 214-150 | + (−) | − | (−) | − | + (−) | 2.5 | − | − | − | − | + | + (−) | − |
| 2 | 11.6 | − | − | − | − | + | 0 | − | − | − | − | + | + | − |
| 3 | 11.1 | − | − | − | − | + | nc | + | + | − | ||||
| 4 | 5 | − | − | − | − | + | 6.8 | − | − | − | − | + | 4-151κ+ | |
| 5 | 21.7 | − | − | − | − | + | nc | + | − | |||||
| 6 | 19.4 | − | − | − | − | + | nc | + | − | |||||
| 7 | (19.4) | − | − | + | + | + | 12.8 | − | − | − | − | + | + | − |
| 10 | 23.8 | − | − | − | − | + | nc | − | + | − | − | + | + | − |
| 11 | nc | 0 | − | − | − | − | + | + | − | |||||
| 12 | nc | 0 | − | − | − | − | + | + | − | |||||
| 13 | 16.3 | − | − | − | − | + | nc | + | − | |||||
| 15 | 9.7 | − | − | − | − | + | 4.2 | − | − | − | − | + | + | − |
| 18 | 9.5 | − | − | − | − | + | 5.1 | − | − | − | − | + | + | − |
| 19 | 20.24-150 | − | − | − | − | + | 14.5 | − | − | − | − | + | + | − |
| 20 | 7.6 | − | − | − | − | + | 0 | − | − | − | − | + | + | − |
| 21 | 3.4 | − | − | − | − | + | 2.5 | − | − | − | − | + | + | − |
| 22 | 7.5 | − | − | − | − | + | 2.5 | − | − | − | − | + | + | − |
| 23 | 10.2 | − | − | + | − | + | 2.5 | − | − | − | − | + | + | − |
| 8 | 18.8 | − | + | − | − | + | 0 | + | − | − | − | − | − | − |
| 9 | 26 | + | + | + | − | + | 0 | − | − | − | − | + | − | − |
| 16 | nc | − | + | − | − | − | nc | − | − | |||||
| 17 | 11.1 | − | − | − | − | + | 6.6 | + | − | + | − | nc | − | − |
| 24 | 6.9 | + | − | − | − | − | nc | − | − | |||||
| 25 | nc | 0 | + | + | − | − | + | − | − | |||||
Intraclonal diversity.
Weak expression of IgLκ in some HRS cells.
Blank lines indicate cases without detectable rearrangement; nc, not calculable.
Antigen selection of the rearranged immunoglobulin genes
To determine the VH mutation pattern compatible with antigen selection, we applied different methods. When the ratio of replacement to silent mutations (< 1.6) was calculated on the basis of their occurrence in the FW region, 9 of the 20 cases (45%) fulfilled the criteria of antigen selection. However, when both, the CDR (R/s > 2.9) and FW region (R/s < 1.5) were included, only 5 of 19 (26%) displayed signs of an antigen selection (Table5).
Determination of antigen selection in single isolated Hodgkin and Reed-Sternberg cells
| Case No. . | Antigen Selection (R/S for FW3 ≤1.6) . | Antigen Selection (R/S for CDR2 ≥2.9 and for FW3 ≤1.5) . |
|---|---|---|
| 1 | − | − |
| 2 | − | − |
| 3 | − | − |
| 4 | + | − |
| 5 | − | − |
| 6 | − | − |
| 7 | + | nc |
| 8 | + | + |
| 9 | − | − |
| 10 | − | − |
| 13 | + | + |
| 15 | + | + |
| 17 | + | − |
| 18 | + | − |
| 19 | − | − |
| 20 | + | + |
| 21 | − | − |
| 22 | − | − |
| 23 | + | + |
| 24 | − | − |
| Total | 9/20 (45%) | 5/19 (26%) |
| Case No. . | Antigen Selection (R/S for FW3 ≤1.6) . | Antigen Selection (R/S for CDR2 ≥2.9 and for FW3 ≤1.5) . |
|---|---|---|
| 1 | − | − |
| 2 | − | − |
| 3 | − | − |
| 4 | + | − |
| 5 | − | − |
| 6 | − | − |
| 7 | + | nc |
| 8 | + | + |
| 9 | − | − |
| 10 | − | − |
| 13 | + | + |
| 15 | + | + |
| 17 | + | − |
| 18 | + | − |
| 19 | − | − |
| 20 | + | + |
| 21 | − | − |
| 22 | − | − |
| 23 | + | + |
| 24 | − | − |
| Total | 9/20 (45%) | 5/19 (26%) |
nc indicates not calculable.
Intraclonal variation
In 2 of the 25 classical HD cases (cases 1 and 19), differences within the clonal HRS cell populations were observed. In case 1, 25 of the 28 Ig sequences proved to be identical. In 2 sequences the same 22 base pair insertion was observed, resulting in a disruption of the coding capacity by a frame shift and a stop codon in the FW3 region. In the remaining sequence the same 22 base pair insertion was present, however, without the stop codon detectable in 2 other sequences. All 28 sequences harbored the same CDR3 and showed the same mutation pattern. In case 19, 2 of the 5 sequences differed by a 98 base pair insertion in the FW3 region which represented a duplication of a neighboring sequence proportion (see Table 2).
Ig expression in HRS cells
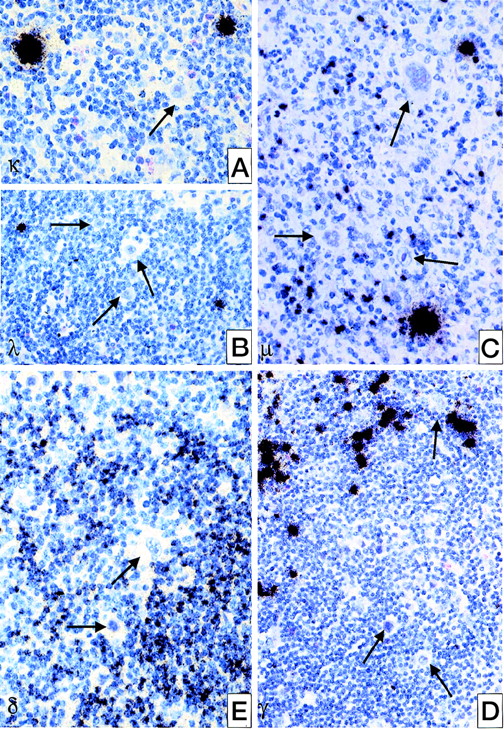
All 25 cases were analyzed for Ig expression by in situ hybridization using radioactively labeled probes for the constant region of kappa and lambda light chains and of δ, μ, and γ heavy chain genes. In none of these cases, could an expression of any of the heavy chain genes be observed. Only in 1 case was a weak expression of the kappa light chain gene found in some of the HRS cells (see Table 4, Figure 1). Normal resting B cells showed moderate amounts of Ig mRNA, whereas plasma B cells revealed extremely strong signals (see Figure 1).
Detection of Ig heavy and light chain transcripts in classical HD by in situ hybridization using radioactively labeled probes for the according constant region.
(A) IgL kappa; (B) IgL lambda; (C) IgH mu; (D) IgH delta; (E) IgH gamma. HRS cells are negative for all types of Ig expression (→) whereas plasma cells are intensively labeled; normal B-cells are weakly or moderately positive.
Detection of Ig heavy and light chain transcripts in classical HD by in situ hybridization using radioactively labeled probes for the according constant region.
(A) IgL kappa; (B) IgL lambda; (C) IgH mu; (D) IgH delta; (E) IgH gamma. HRS cells are negative for all types of Ig expression (→) whereas plasma cells are intensively labeled; normal B-cells are weakly or moderately positive.
Controls
Extensive controls were performed to evaluate the reliability of our single cell approach. Aliquots of the buffers that were taken after each HRS cell isolation (in total 1078) as well as after each of the 50 T cells isolated from areas rich in B cells did not give rise to PCR products in any instances. Normal B cells isolated from reactive lymph nodes or from the classical HD cases demonstrated VH rearrangements in 28% (16 of 58 isolated cells) and VLκ rearrangements in 22% (14 of 65 isolated cells). Sequencing and comparison of these rearrangements disclosed unique (polyclonal) sequences in all instances.
Ig gene transcription activity in HD cell lines
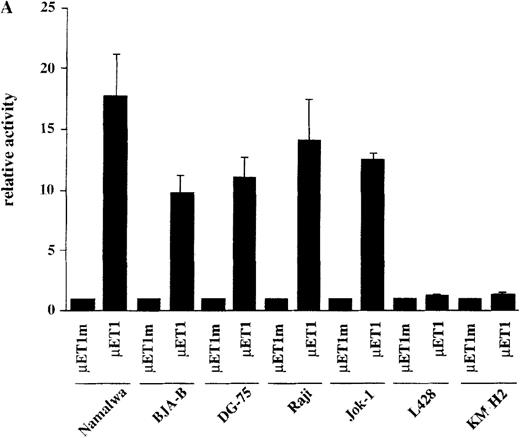
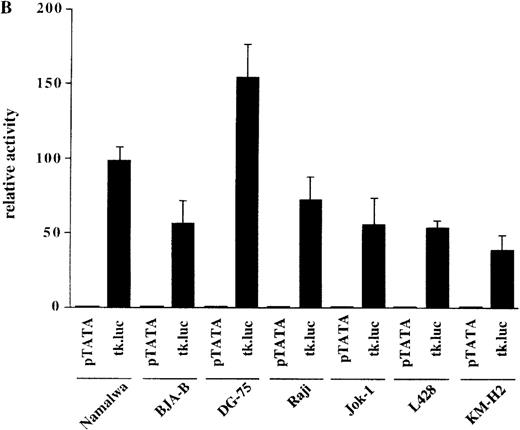
The observation that HRS cells consistently lacked Ig transcripts prompted the analysis of the activity of reporter constructs driven by Ig regulatory elements in the HD cell lines L428 and KM-H2. A luciferase reporter containing the murine kappa T1 promoter and the intronic heavy chain enhancer element was used.24 To assay for specific promoter activity, we included a control where the kappa T1 promoter was inactivated by mutating the octamer motif. When these constructs were tested in 5 control B-cell lines, the activity of the wild-type promoter construct was high, whereas the mutant construct produced very low signals (Figure 2A). In contrast, when transfected into the 2 HD cell lines, the wild-type as well as the mutant construct showed virtually the same low level of activity (see Figure 2A). This lack of Ig-specific transcriptional activity was not due to a general transcriptional incompetence of HD cell line cells, because the activity of the HSV thymidine kinase (tk) promoter was comparable in 5 control B-cell lines and in the 2 HD cell lines (Figure 2B). These findings indicate that the HD cell lines are specifically defective in mediating transcriptional activity of Ig gene regulatory elements.
Transcriptional activity of reporter constructs driven by Ig regulatory elements or the HSV-tk promoter.
(A) Namalwa, BJA-B, DG-75, Raji and Jok-1 cells (control B-cell lines), and L428 and KM-H2 cells (HD cell lines) were transfected with the wild type promoter-driven luciferase reporters (μET1) and the mutant promoter-driven reporters (μET1m). The activity of the mutant promoters was set to 1 for all cell lines and the specific activity of the wild-type promoter-driven reporter is shown as relative activity. All transfections were independently repeated minimally 3 times and in all cases a tk-driven renilla-luciferase reporter was co-transfected to correct for differences in transfection efficiencies. (B) Similar transfection as in A with luciferase reporters containing the intact tk-promoter (tk.luc, −109 to +52) or a truncated version of this promoter (pTATA, −38 to +52). The activity of the truncated promoter was set to 1 and tk.luc activity is shown relative to pTATA. (C) Schematic representation of the reporter constructs used in A and B. O = octamer motif (the mutated octamer motif is indicated by the crossed out symbol); T = TATA box. The enhancer fragment is not drawn to scale (the 1000 bp XbaI fragment of the enhancer was used).40
Transcriptional activity of reporter constructs driven by Ig regulatory elements or the HSV-tk promoter.
(A) Namalwa, BJA-B, DG-75, Raji and Jok-1 cells (control B-cell lines), and L428 and KM-H2 cells (HD cell lines) were transfected with the wild type promoter-driven luciferase reporters (μET1) and the mutant promoter-driven reporters (μET1m). The activity of the mutant promoters was set to 1 for all cell lines and the specific activity of the wild-type promoter-driven reporter is shown as relative activity. All transfections were independently repeated minimally 3 times and in all cases a tk-driven renilla-luciferase reporter was co-transfected to correct for differences in transfection efficiencies. (B) Similar transfection as in A with luciferase reporters containing the intact tk-promoter (tk.luc, −109 to +52) or a truncated version of this promoter (pTATA, −38 to +52). The activity of the truncated promoter was set to 1 and tk.luc activity is shown relative to pTATA. (C) Schematic representation of the reporter constructs used in A and B. O = octamer motif (the mutated octamer motif is indicated by the crossed out symbol); T = TATA box. The enhancer fragment is not drawn to scale (the 1000 bp XbaI fragment of the enhancer was used).40
Discussion
The aim of the present study was to throw light on the conflicting results concerning the frequency of Ig rearrangements and their clonality12-17 in HRS cells. We investigated the HRS cell populations of 25 cases of nodular sclerosis, the main histotype of classical HD, for the presence of Ig rearrangements and analyzed their sequence to clarify whether polyclonal or mixed polyclonal/clonal HRS cell populations exist. In contrast to previous single cell studies, an optimized single cell isolation technique, whose contamination risk was close to zero, was used.3
Ig rearrangements were found in 24 of 25 (96%) of classical HD cases investigated. Rearranged VH genes were present in 21 (87.5%) and rearranged VLκ genes in 18 (75%) of the PCR-positive cases. An occurrence of both types of rearrangements was detectable in 15 of these cases. All 195 VH and 96 VLκ rearrangements, in total 291, amplified from 1078 HRS cells proved to be monoclonal. These data indicate that HD is in nearly all cases, irrespective of phenotype and EBV infection, a B-cell-derived lymphoproliferative disorder and that the HRS cells with rearranged Ig genes represent without any exception a clonal outgrowth of a single transformed B cell. This shows that the absence of Ig rearrangements in HRS cells reported by Roth et al12 is due to technical pitfalls, most likely in the cell isolation. Our present findings further demonstrate that polyclonal rearrangements described in our previous study15 and in the publications of Delabie et al16 as well as Kanzler et al14 were not derived from HRS cells but from contaminating DNA released into the covering buffer during the cell isolation procedure from B lymphocytes. The occurrence of these polyclonal rearrangements could be eliminated by reducing the buffer volume used to transfer the isolated cells into the PCR tube. This was already shown in our recent study of 11 cases of LPHD in which we disclosed exclusively clonal VH rearrangements in the L&H cells.3
The sequence analysis of the Ig rearrangements allows us not only to assign cells to the B-cell lineage and to determine their clonal relationship but also to specify their stage of differentiation. Naive B cells harbor unmutated VH genes, whereas antigen-activated B cells acquire somatic mutations within their VH genes in the course of the germinal center reaction. Thus, somatically mutated VH genes are specific markers for germinal center B cells and their descendants.25 Previous studies have revealed a high load of somatic VH mutations in the clonal HRS cell populations.13-15 In the present study, we found VH mutations in all cases ranging from 5 to 38 base substitutions (average 20.4). This confirms the previous findings and substantiates the conclusion that HRS cells correspond in their developmental stage to germinal or postgerminal center B cells.
The origin of HRS cells from germinal or postgerminal center B cells raises the question whether the pattern of their VH mutations is compatible with antigen selection. Of the 21 cases with VH rearrangements only 5 (26%) or 9 (45%), respectively, showed signs of antigen selection, depending on whether the CDR2 and FW3 or only the FW3 were considered. This coincides with the high load of somatic mutations (average 14%) against a selection of HRS cells by antigen.
Although overwhelming evidence now indicates a B-cell derivation of HRS cells, previous studies failed to detect Ig protein and Ig light chain mRNA expression by HRS cells in nearly all instances. Kanzler et al14 explained this nonexpression with the presence of crippling mutations in the V genes, disrupting their coding capacity. In contrast, our study revealed a disruption of the VH rearrangements in only 3 of 21 (14%) and of the VLκ rearrangements in only 3 of 18 (17%), preventing Ig production in 6 of 23 (26%) of the cases. This prompted us to examine the expression of Ig mRNA in HRS cells by in situ hybridization. In keeping with previous investigations, Ig light chain expression was not demonstrable in all but 1 case in which exclusively IgLκ mRNA was present in only a small proportion of HRS cells.26 27 To exclude that the absence of Ig expression was restricted to Ig light chains we extended our investigation to the analysis of Ig heavy chain mRNA. This new data revealed a complete absence of Igμ, Igδ, and Igγ mRNA in all HRS cells of the cases studied. This nondetection of Ig heavy and light chain mRNA was not due to a lack of sensitivity of the method used because normal resting B cells showed a moderately strong labeling and plasma cells an extremely strong labeling. The correlation of the RNA expression with the coding capacity of the Ig genes disclosed that the absence of Ig RNA in the HRS cells is independent from the presence of crippling mutations in the rearranged Ig genes. Furthermore, the absence of Ig mRNA expression as revealed by in situ hybridization rules out the possibility that additional functional and transcriptionally active Ig rearrangements might be present in the HRS cells, which escaped detection by single cell PCR.
As a reason for the nonexpression of Ig, 3 possibilities are conceivable. The first possibility is that the promoter or enhancer region is mutated with the effect that the transcription factors are incapable of binding and initiating the RNA synthesis. This possibility is supported by the detection of a transcription-inhibiting mutation in the octamer binding site of the Ig promoter in the HD-derived cell line L1236 and in the in situ HRS cells of the corresponding case.28 Own studies showed, however, that in other HD cell lines mutations in the octamer binding site are not detectable, suggesting that mutations in the octamer motif cannot account for the non-Ig transcription in all cases (unpublished observation). The second possibility is that the HRS cells originate by fusion of a B cell with another cell type, which causes a rapid suppression of Ig expression, a phenomenon termed extinction.29,30 This possibility appears to be less likely in view of our findings and those of others showing that the expression of the B-cell-specific transcription factors PU .1, Pip/IRF4 (manuscript in preparation), Pax-531 is not suppressed and NF-κB is constitutively activated32 in primary or in cultured HRS cells. The third possibility is that the transforming event leading to the generation of HRS cells deregulates the Ig transcription machinery, preventing the transcription of Ig. In support of this possibility is our finding of a highly reduced Ig gene transcription activity in the HD cell lines L428 and KM-H2 as revealed by transfection experiments with Ig promoter reporter constructs. Further studies performed not only on the HD cell lines but also on primary HRS cells are needed to more precisely determine the mechanism responsible for the deficient Ig transcription activity in HRS cells.
The absence of Ig expression in classical HD contrasts with the findings in LPHD where the tumor cells nearly constantly express Ig at the transcriptional and protein level as well as B-cell-associated molecules.33-38 A further difference concerns the presence of ongoing mutations. In HRS cells of classical HD intraclonal diversities are rare, but they are frequent in the L&H cells of LPHD.3,4 In classical HD the intraclonal diversities mainly comprise insertions or deletions occurring in the CDR2 or FW3 region, whereas in LPHD they resemble ongoing mutations typical for germinal center B cells. These observations add to the differences between LPHD and classical HD. Despite these differences both lesions have a high rate of VH gene mutations in common. The high load of VH gene mutations suggests that the tumor cells of both the LPHD and the classical HD are derived from B cells that have resided in a germinal center for an abnormally long period of time. The origin of the classical HRS cells from germinal center B cells is substantiated by the observation that one of the HD patients developed follicular lymphoma 2 years after a successfully treated HD. The analysis of the amplified Ig genes of both tumors showed the same rearrangement but different numbers of somatic mutations being 7 in the HRS cells and 22 in the follicular lymphoma tumor cells. Six of these mutations were identical in both tumor cell populations.39 This identifies a germinal center B cell as a common progenitor of both lesions. Interestingly, the Ig rearrangements in both the HRS cells and in the follicular lymphoma cells were functional despite the mutations, but expression of Ig molecules (IgM/Igκ) was only detectable in the follicular lymphoma cells. This finding was confirmed at the transcriptional level by demonstrating Igμ and Igκ mRNA in the non-Hodgkin lymphoma cells and total absence of Igμ mRNA and nearly complete down-regulation of Igκ mRNA in the HRS cells. These findings highlight that the final neoplastic event leading to classical HD can be associated with a complete change of morphology, immunophenotype, and clinical behavior and underscore the conclusion that crippling mutations are not the cause for the absent expression of Ig by the HRS cells but instead a disturbance of the transcription mechanism.
In summary, classical HD is a B-cell neoplasm in nearly all instances. The HRS cells represent a clonal expansion of cells that originate from a single germinal center B cell. The B-cell origin of the HRS cells is not related to the presence of B cell antigens and their clonality is not associated with an EBV infection. The Ig gene coding capacity is preserved in the majority of cases, demonstrating that the absence of Ig expression is not due to crippling mutations but to defects in the transcription regulation. This down-regulation of Ig expression appears to be closely linked with the final neoplastic event as revealed by the derivation of a HD and a FL from the same germinal center B cell. The fact that the same precursor B cell can lead to completely different tumor lesions including differences in Ig expression indicates that the transforming event(s) can be more important than the cell of origin in determining a disease entity.
Acknowledgments
We thank H.-H. Müller, D. Jahnke, H. Protz, and E. Berg for their excellent technical assistance and L. Udvarhelyi for his editorial assistance.
Sponsored by grants of the Deutsche Krebshilfe (70-2202-Mü3) by the Berliner Krebsgesellschaft and Deutsche Forschungsgemeinschaft DFG (Ste 318/5-2, SFB 366 B4), (SFB 465 B7 to T.W.).
Reprints:Harald Stein, Institute of Pathology, Benjamin Franklin University Hospital, Free University Berlin, Hindenburgdamm 30, 12200 Berlin, Germany.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal