Elevated leukotriene (LT)C4 synthase activity was observed in peripheral blood granulocyte suspensions from patients with chronic myeloid leukemia (CML). Magnetic cell sorting (MACS) with CD16 monoclonal antibodies (mAbs), which were used to fractionate granulocytes from CML patients and healthy individuals, yielded highly purified suspensions of CD16+ neutrophils. The purity of these cell fractions was verified by extensive morphologic examination. Reverse transcriptase–polymerase chain reaction (RT-PCR) analyses, demonstrating the absence of interleukin-4 messenger RNA (IL-4 mRNA), further confirmed the negligible contamination of eosinophils in these fractions. Notably, purified CML CD16+ neutrophils from all tested patients transformed exogenous LTA4 to LTC4. These cells also produced LTC4 after activation with ionophore A23187 or the chemotactic peptide fMet-LeuPhe (N-formylmethionyl-leucyl-phenylalanine). Subcellular fractionation revealed that the enzyme activity was exclusively distributed to the microsomal fraction. Expression of LTC4 synthase mRNA in CML CD16+neutrophils was confirmed by RT-PCR. Furthermore, Western blot analyses consistently demonstrated expression of LTC4 synthase at the protein level in CML CD16+ neutrophils, whereas expression of microsomal glutathione S-transferase 2 occurred occasionally. Expectedly, LTC4 synthase activity or expression of the protein could not be demonstrated in CD16+ neutrophil suspensions from any of the healthy individuals. Instead, these cells, as well as CML CD16+neutrophils, transformed LTA4 to LTB4. The results indicate that aberrant expression of LTC4 synthase is a regular feature of morphologically mature CML CD16+neutrophils. This abnormality, possibly associated with malignant transformation, can lead to increased LTC4 synthesis in vivo. Such overproduction may be of pathophysiological relevance because LTC4 has been demonstrated to stimulate proliferation of human bone marrow–derived myeloid progenitor cells.
Chronic myeloid leukemia (CML) is characterized by markedly accelerated myelopoiesis, with increased numbers of immature and mature myeloid cells in peripheral blood and bone marrow. A characteristic feature of the disease is a specific chromosome abnormality, the t9;22 reciprocal translocation, which creates the Philadelphia (Ph) chromosome. The Ph chromosome is the product of a molecular rearrangement between the c-ABLproto-oncogene situated on chromosome 9 and the breakpoint cluster region (BCR) gene on chromosome 22. The BCR-ABL fusion gene encodes a 210-kd protein, which is an aberrant highly active tyrosine kinase. It is currently believed that the initiation, maintenance, and progression of CML are consequences ofBCR-ABL–induced activation of multiple signal transduction pathways. This leads to, for example, cytokine-independent uncontrolled proliferation of myeloid cells in the bone marrow and to resistance to apoptosis.1
We have previously reported that suspensions of peripheral white blood cells from patients with CML possess a markedly elevated capacity to produce leukotriene (LT)C4 from endogenous arachidonic acid.2 Furthermore, overproduction of LTC4 was observed also in granulocyte suspensions2 and unfractioned bone marrow cells3 from CML patients. These findings are of potential interest, since LTs, which are endogenously formed mediators of inflammation and asthma,4,5 may also play a role in the regulation of human myelopoiesis. Thus LTC4 and, to a lesser extent, LTB4 have been demonstrated to potentiate granulocyte-macrophage colony-stimulating factor–induced (GM-CSF–induced) myeloid progenitor cell (CFU-GM) proliferation.6,7 In addition, inhibition of LT formation attenuated CFU-GM growth, an effect that could be counteracted by the addition of exogenous LTC4 or LTB4.6 Accordingly, 5-lipoxygenase (5-LO) inhibitors have been observed to suppress the proliferation and DNA synthesis of normal CFU-GM and myeloid leukemic cell lines.8-11 Furthermore, inhibition of LT synthesis has been reported to induce programmed cell death in U937 cells and in CML blast cells.12 13
Cells of myeloid origin are the main producers of LTs in humans. Upon cell activation, the LT biosynthesis is initiated by phospholipase A2–dependent release of arachidonic acid from membrane phospholipids.14 Subsequently, 5-LO translocates to the nuclear membrane, where it cooperates with the 5-LO activating protein (FLAP) in the conversion of arachidonic acid to the unstable epoxide LTA4.15 This intermediate can be metabolized either by a cytosolic LTA4 hydrolase to the leukocyte-activating dihydroxy acid LTB416 or by a membrane-bound LTC4 synthase,17 which specifically catalyzes conjugation of LTA4 with the tripeptide glutathione, thereby yielding LTC4. Once formed, LTC4 is actively exported to the extracellular space, a process involving the multidrug resistance-associated protein (MRP).18-20 Extracellularly, LTC4 is converted by γ-glutamyl transpeptidase to the biologically active LTD4 and is further metabolized by dipeptidase to LTE4 via successive elimination of a γ-glutamyl residue and glycine, respectively.4
Among human blood cells, neutrophils, eosinophils, and basophils, as well as monocytes, possess the enzymatic machinery needed for LTA4production from endogenous substrate and are thus capable to produce LTs after cell stimulation. However, the subsequent metabolism of LTA4 differs among these cell types. Thus, neutrophils, equipped with LTA4 hydrolase but lacking LTC4synthase, produce LTB4 but not LTC4, whereas the reverse applies for eosinophils and basophils.21,22Monocytes express both these enzymes, consequently producing both LTB4 and LTC4.23 In addition, platelets24-26 and erythrocytes27 lack 5-LO/FLAP but express LTC4 synthase and LTA4hydrolase, respectively. Recent findings demonstrate that activated neutrophils release the main part of produced LTA4 extracellularly upon stimulation with ionophore A23187 or physiological stimuli.28,29 These results clearly indicate that platelets and erythrocytes may participate in LT biosynthesis in vivo via transcellular mechanisms.30
LTC4 synthase is a highly specific glutathione S-transferase that comprises 2 subunits (18 kd each) and is active as a homodimer.17 This enzyme has been purified from several sources,31,26 and cloning of the gene, as well as expression of the protein, has been reported.32,33LTC4 synthase is a member of the membrane-associated proteins involved in the eicosanoid and glutathione metabolism (MAPEG) protein family, which also includes FLAP, microsomal glutathione S-transferase (MGST) 1-334 and prostaglandin (PG) E2 synthase.66 Among these proteins, MGST2 and MGST3 have also been shown to possess LTC4 synthase activity.35,36 The activity of LTC4 synthase has been demonstrated to be regulated by phosphorylation-dependent mechanisms.37-41 A recent report describing LTC4 synthase deficiency in relation to a fatal developmental syndrome further suggests that the enzyme may be of importance also in areas not related to allergy and inflammation.42
The gene encoding human LTC4 synthase has been mapped to chromosome 5, long arm, band 35 (5q35) and contains several transcription factor–binding motifs in the 5′ flanking region, suggesting a regulated mode of expression.43,44 In agreement, the transcription of the LTC4synthase gene was recently demonstrated to be up-regulated by the transforming growth factor–β (TGF-β) family of cytokines in the monocyte-like cell line THP-1.45Interestingly, in close proximity to the LTC4synthase gene on chromosome 5, there is a cluster of genes for cytokines and receptors (eg, GM-CSF, interleukin-3 [IL-3], IL-4, macrophage-CSF receptor [M-CSFR], and platelet-derived growth factor receptor [PDGFR]). These cytokines and receptors are involved in the regulation of hematopoiesis, as well as in hypersensitivity and inflammation. Notably, various chromosomal aberrations on chromosome 5q are well recognized in myeloid leukemias.46
In this report we address the abnormal LTC4 synthase activity in CML granulocytes. The results, obtained after magnetic cell sorting (MACS) with monoclonal antibodies (mAbs), demonstrate a regularly occurring aberrant expression of active LTC4synthase in highly purified CD16+ neutrophils from CML patients.
Materials and methods
Materials
The following materials were used in this study: blood collection tubes (Vacutainer; Becton Dickinson, Rutherford, NJ); sodium metrizoate (Lymphoprep; Nyegaard, Oslo, Norway); paramagnetic microbeads conjugated to mouse antihuman CD16 mAbs, magnetic stands, and LS+ MACS columns (Miltenyi Biotec, Bergisch Gladbach, Germany); ionophore A23187 (Calbiochem-Boehring, La Jolla, CA); arachidonic acid (Nu-Check Prep, Elysian, MN); fMet-Leu-Phe (N-formylmethionyl-leucyl-phenylalanine [fMLP]) and agarose (Sigma, St. Louis, MO); LTA4 methyl ester, saponified before use as previously described39 (gift from Dr Robert Zipkin, Biomol Research Laboratories, Plymouth Meeting, PA); LTB4, LTC4, and PGB2(Biomol Research Laboratories); nitrocellulose membranes, precast polyacrylamide gels, and protein standards (Novex, San Diego, CA); LTC4 synthase and MGST2 antibodies, developed as previously described47 48 (gift from Merck Frosst, Montreal, Quebec, Canada); horseradish peroxidase–coupled antirabbit secondary antibodies as well as a chemoluminescence detection kit (ECL plus; Amersham, Buckinghamshire, England); film (Biomax MR; Kodak, Rochester, NY); RNA extraction kit (Ultraspec II; Biotecx, Houston, TX); first strand cDNA synthesis kit, protease inhibitor cocktail, and amplification mixture (Complete, PCR Master; Boehringer Mannheim, Mannheim, Germany); LTC4 synthase PCR primers (Scandinavian Gene Synthesis, Köping, Sweden); IL-4 PCR primers, β-actin PCR primers, and IL-4 complementary DNA (cDNA) (Clontech, Palo Alto, CA); and a PCR cloning kit (pCR-Script Amp SK+; Stratagene, La Jolla, CA).
Patients and healthy control subjects
Fresh blood samples were obtained from a total of 17 patients in the chronic phase of Ph chromosome-positive (Ph+) CML and 15 healthy medication-free volunteers. All blood samples were collected with the informed consent of involved individuals in accordance with the approval of the project from the Ethics Committee of Karolinska Institutet (Stockholm, Sweden). The peripheral white blood cell differential counts of the patients were routinely analyzed at the time of blood collection.
Granulocyte isolation
Peripheral venous blood was drawn into blood collection tubes containing ethylenediaminetetraacetic acid (EDTA). After centrifugation at 200g for 15 minutes, the platelet-rich plasma was removed, and granulocytes were isolated from the remaining lower phase by dextran sedimentation, hypotonic ammonium chloride lysis, and sodium metrizoate centrifugation, as previously described.49
Magnetic cell sorting
The granulocyte fraction obtained after centrifugation was resuspended in phosphate-buffered saline (PBS, pH 7.4) supplemented with 0.1% bovine serum albumin (BSA) and 2 mmol/L EDTA (buffer A) to a cell concentration of 10 × 107 cells/mL and incubated with anti-CD16 microbeads at 8°C for 30 minutes. The cells were applied to an LS+ MACS column, and the CD16− eosinophil-enriched cell fraction was eluted with 9 mL buffer A. Thereafter, the column was washed with 3 mL buffer A, removed from the magnetic field, and eluted with 5 mL buffer A in order to collect CD16+ neutrophils.
Morphologic examination
Duplicate aliquots of the cell fractions were subjected to cytocentrifugation and May-Grünwald-Giemsa staining. Thereafter, the slides were numbered by code and evaluated blindly by an experienced hematology morphologist (S. Widell). Two hundred cells were counted on each slide.
Preparation of subcellular fractions
CD16+ neutrophils were centrifuged at 300g for 10 minutes, resuspended in 0.125 mol/L potassium phosphate buffer (2.5 × 107 cells/mL, pH 7.4), and sonicated at 0°C for 5 × 15 seconds prior to centrifugation at 1400g for 15 minutes. The supernatant was collected and further centrifuged at 100 000g for 60 minutes. After collection of the supernatant (cytosolic fraction), the pellet (microsomal fraction) was resuspended in 0.125 mol/L potassium phosphate buffer (equal volume as cytosolic fraction) prior to assay of enzyme activity in the fractions, as described below.
Determination of LTC4 synthase activity
Intact cell preparations were centrifuged at 300g for 10 minutes and resuspended in PBS (containing 0.9 mmol/L calcium chloride and 0.03% human serum albumin [HSA], pH 7.4) to a final concentration of 1.5 × 107 or 0.3 × 107 (CD16−eosinophil-enriched fraction) cells/mL. The metabolism of LTA4 in intact cells was determined by incubation with 10 μmol/L LTA4 at 37°C for 5 minutes. Subcellular fractions of CD16+ cells were incubated with 60 μmol/L LTA4 and 5 mmol/L reduced glutathione at 20°C for 10 minutes in the presence of 0.05% BSA. The capacity of normal and CML CD16+ neutrophils to produce LTC4 from endogenous LTA4 was determined by incubation with 1 μmol/L A23187 or 1 μmol/L fMLP in combination with 8 μmol/L arachidonic acid at 37°C for 5 minutes. All incubations were stopped by the addition of 5 volumes of ethanol containing prostaglandin B2 as an internal standard. Prior to analysis, the samples were centrifuged, evaporated, dissolved in 250 μL high-pressure liquid chromatography (HPLC) mobile phase, and recentrifuged. Thereafter, LTs were analyzed by reversed-phase HPLC as described,39 using a 3.9 × 150–mm column (Nova-Pak C18; Waters Associates, Milford, MA), and eluted with a mixture of acetonitrile, methanol, water, and acetic acid at a ratio of 27:18:54:0.8, vol/vol (apparent pH 5.6). Eluted compounds were quantified using a variable wavelength ultraviolet (UV) detector (LDC Spectromonitor III, Stone, England) connected to an integrator (LDC/Milton Roy CI-4000, Laboratory Data Control). The compounds were identified by cochromatography with authentic standards and on-line UV spectroscopy.
RNA isolation, cDNA synthesis, and polymerase chain reaction
Total RNA was extracted from isolated cells by the guanidinium-phenol-chloroform extraction technique50(Ultraspec II, Biotecx). The yield and purity of RNA were examined by spectrophotometrical measurements at 260 and 280 nm. For the preparation of cDNA, 1 μg RNA was used in a 20 μL cDNA synthesis reaction with 20 units avian myeloblastosis virus RT, 3.2 μg random hexamer primers, 10 mmol/L Tris (tris[hydroxymethyl aminomethane]), 50 mmol/L potassium chloride (KCl), 5 mmol/L magnesium dichloride (MgCl2), deoxynucleotide (dNTP) mix (1 mmol/L each), and 50 units ribonuclease (RNase) inhibitor. The reaction mixture was incubated for 10 minutes at 25°C (primer annealing), 60 minutes at 42°C (transcription), and 5 minutes at 99°C (inactivation of RT). Thereafter, 1 μL of the cDNA synthesis reaction was added to a PCR reaction mixture (total volume 50 μL) containing 1.25 units Thermus aquaticus (Taq) polymerase, 10 mmol/L Tris-hydrochloride, 50 mmol/L KCl, 1.5 mmol/L MgCl2, dNTP mix (0.2 mmol/L each) and 0.3 μmol/L primers. The following primers were used: LTC4 synthase: 5′-ACCTGGGCTCGGTAGAC-3′ and 5′-GAGTCCTGCTGCAAGCCTACTTC-3′; β-actin: 5′-GAGGAGCACCCC GTGCTGCTGA-3′ and 5′-CTAGAAGCATTTGCGGTGG 3′; IL-4: 5′-CGGCAACTTTGACCACGGACACAAGTGCGATA-3′ and 5′-ACGTA CTCTGGTTGGCTTCCTTCACAGGACAG-3′. The expected sizes of the DNA fragments amplified with these primers were 119 base pairs (bp) for LTC4 synthase, 784 bp for β-actin, and 344 bp for IL-4.
Temperature cycling (Mastercycler 5330; Eppendorf, Hamburg, Germany) was performed as follows: First cycle, denaturation at 94°C for 60 seconds, annealing at 55°C for 15 seconds, and extension at 72°C for 15 seconds; subsequent cycles: denaturation at 94°C for 15 seconds, annealing at 55°C for 15 seconds, and extension at 72°C for 15 seconds. In total, 30 cycles were performed for β-actin, 35 cycles for LTC4 synthase, and 40 cycles for IL-4. PCR products were analyzed by agarose gel electrophoresis. To verify specific LTC4 synthase amplification, the PCR product was ligated into a plasmid vector (pCR-Script Amp SK+, Stratgene) following manufacturer instructions. Bacteria were transformed, and double-stranded plasmid DNA was isolated from recombinant clones and sequenced according to standard protocols.
Immunoblotting
Normal and CML CD16+ neutrophils, as well as normal CD16− eosinophil-enriched cell preparations, were isolated as described above and suspended to a cell concentration of 15 × 106 cells/mL in lysis buffer (PBS without Ca2+ and Mg2+, supplemented with 1 × protease inhibitor cocktail) (Complete, Boehringer Mannheim). Thereafter, cells were sonicated at 0°C for 3 × 10 seconds prior to centrifugation at 1000g for 15 minutes. The supernatant was removed and further centrifuged at 100 000gfor 60 minutes. The pellet (microsomal fraction) was resuspended in lysis buffer, mixed with 1 volume 2 × loading buffer (containing 125 mmol/L Tris, 20% glycerol, 4% SDS [sodium dodecyl sulfate], 10% 2-mercaptoethanol, and 0.002% bromophenol blue; pH 6.8), and boiled for 5 minutes. Protein levels were determined, and 20 μg of protein was separated on 14% SDS-PAGE (polyacrylamide gel electrophoresis). Proteins were electroblotted onto a nitrocellulose membrane followed by blocking of the membrane with 5% milk powder in 100 mmol/L Tris (containing 0.9% NaCl and 0.1% Tween 20, pH 7.5) for 60 minutes. After blocking, the membranes were washed and incubated for 60 minutes with antibodies against LTC4synthase or MGST2 (both 1:1000 dilution) in 100 mmol/L Tris (containing 0.9% NaCl, 0.05% Tween 20, and 2% milk powder; pH 7.5) prior to being washed and incubated with secondary antirabbit antibody1:1000 in 100 mmol/L Tris (containing 0.9% NaCl, 0.05% Tween 20, and 2% milk powder; pH 7.5) for 60 minutes. Enhanced chemiluminescence (ECL plus, Amersham) was used for detection per manufacturer instructions.
Statistics
The 2-sided Student t test for unpaired samples was employed to compare LT production in leukemic and normal cells.
Results
Morphological and RT-PCR analyses of granulocyte preparations
Morphological analyses demonstrated that normal and CML granulocyte suspensions obtained after density gradient centrifugation contained approximately 90% neutrophils and 6%-7% eosinophils together with minute amounts of lymphocytes (Table 1). Since eosinophils are potent producers of LTC4, MACS was employed in order to eliminate eosinophil contamination. After immunomagnetic separation of the granulocyte suspensions with CD16 mAbs, CD16+ cell suspensions from CML patients and controls contained more than 99% neutrophils (Table 1). The mean eosinophil content in these fractions was estimated to be less than 0.04%. The presence of platelets was evaluated in these cell fractions, and platelet contamination in the normal and CML CD16+neutrophil preparations was always less than 1 platelet per 20 neutrophils. The CD16− cell preparations contained predominantly eosinophils together with other cell types, mostly neutrophils and lymphocytes. In addition, the CML CD16− cell preparations contained immature myeloid cells, particularly myelocytes (Table 1). Messenger RNA (mRNA) encoding the cytokine IL-4 was present in normal and CML CD16−fractions, as demonstrated by RT-PCR amplification of a DNA fragment of the expected size (344 bp) (Table2). In contrast, IL-4 expression could not be observed in normal or CML CD16+ neutrophil fractions. The integrity of the RNA was verified by amplification with β-actin–specific primers, and an IL-4 cDNA was used as a positive control to verify specific amplification with the IL-4 primers (results not shown). The findings are in accordance with the negligible contamination of eosinophils in the CD16+ fractions, because IL-4 mRNA has been demonstrated to be expressed in eosinophils but not in neutrophils.51-53
Percent of cell-type distribution before and after CD16 MACS of human granulocyte suspensions
| . | Before MACS . | CD16+ . | CD16− . | |||
|---|---|---|---|---|---|---|
| Control (n = 7) . | CML (n = 7) . | Control (n = 8) . | CML (n = 11) . | Control (n = 7) . | CML (n = 6) . | |
| Neutrophils | 92.1 ± 2.1 | 90.1 ± 4.3 | 99.1 ± 0.4 | 99.5 ± 0.3 | 10.7 ± 8.2 | 17.0 ± 5.2 |
| Eosinophils | 6.1 ± 1.6 | 7.0 ± 3.8 | 0 | 0 | 78.4 ± 9.7 | 63.2 ± 13.9 |
| Basophils | 0 | 0.7 ± 0.5 | 0.1 ± 0.1 | 0.1 ± 0.1 | 0 | 2.7 ± 1.8 |
| Lymphocytes | 1.6 ± 0.8 | 1.1 ± 0.7 | 0.5 ± 0.3 | 0.2 ± 0.1 | 10.0 ± 6.8 | 4.0 ± 1.4 |
| Monocytes | 0.1 ± 0.2 | 0.4 ± 0.2 | 0 | 0.1 ± 0.1 | 0.7 ± 0.5 | 0.8 ± 0.5 |
| Metamyelocytes | 0 | 0.1 ± 0.2 | 0.1 ± 0.1 | 0 | 0.1 ± 0.2 | 2.0 ± 1.1 |
| Myelocytes | 0 | 0.4 ± 0.3 | 0.1 ± 0.1 | 0.1 ± 0.1 | 0 | 7.9 ± 4.5 |
| Promyelocytes | 0 | 0 | 0 | 0 | 0 | 1.8 ± 1.1 |
| Blasts | 0 | 0 | 0 | 0.1 ± 0.1 | 0 | 0.7 ± 0.7 |
| . | Before MACS . | CD16+ . | CD16− . | |||
|---|---|---|---|---|---|---|
| Control (n = 7) . | CML (n = 7) . | Control (n = 8) . | CML (n = 11) . | Control (n = 7) . | CML (n = 6) . | |
| Neutrophils | 92.1 ± 2.1 | 90.1 ± 4.3 | 99.1 ± 0.4 | 99.5 ± 0.3 | 10.7 ± 8.2 | 17.0 ± 5.2 |
| Eosinophils | 6.1 ± 1.6 | 7.0 ± 3.8 | 0 | 0 | 78.4 ± 9.7 | 63.2 ± 13.9 |
| Basophils | 0 | 0.7 ± 0.5 | 0.1 ± 0.1 | 0.1 ± 0.1 | 0 | 2.7 ± 1.8 |
| Lymphocytes | 1.6 ± 0.8 | 1.1 ± 0.7 | 0.5 ± 0.3 | 0.2 ± 0.1 | 10.0 ± 6.8 | 4.0 ± 1.4 |
| Monocytes | 0.1 ± 0.2 | 0.4 ± 0.2 | 0 | 0.1 ± 0.1 | 0.7 ± 0.5 | 0.8 ± 0.5 |
| Metamyelocytes | 0 | 0.1 ± 0.2 | 0.1 ± 0.1 | 0 | 0.1 ± 0.2 | 2.0 ± 1.1 |
| Myelocytes | 0 | 0.4 ± 0.3 | 0.1 ± 0.1 | 0.1 ± 0.1 | 0 | 7.9 ± 4.5 |
| Promyelocytes | 0 | 0 | 0 | 0 | 0 | 1.8 ± 1.1 |
| Blasts | 0 | 0 | 0 | 0.1 ± 0.1 | 0 | 0.7 ± 0.7 |
CD16+ neutrophils and eosinophil-enriched CD16− cell suspensions were prepared by MACS of the granulocyte fractions from CML patients and healthy controls. Duplicate cytocentrifugation slides were prepared from each cell fraction and stained with May-Grünwald-Giemsa. Thereafter, differential counting of a total of 200 cells per slide (ie, 400 cells per cell fraction from each individual) was performed. The numbers indicate percentage ± SEM of nucleated cells.
RT-PCR analysis of IL-4 mRNA in CD16+ and CD16− cell fractions
| Fraction . | IL-4 . | β-Actin . |
|---|---|---|
| CML CD16+ neutrophils | − | ++ |
| CML CD16− eosinophil-enriched | ++ | +++ |
| Normal CD16+ neutrophils | − | ++ |
| Normal CD16− eosinophil-enriched | + | ++ |
| Fraction . | IL-4 . | β-Actin . |
|---|---|---|
| CML CD16+ neutrophils | − | ++ |
| CML CD16− eosinophil-enriched | ++ | +++ |
| Normal CD16+ neutrophils | − | ++ |
| Normal CD16− eosinophil-enriched | + | ++ |
CD16+ neutrophils and eosinophil-enriched CD16− cell suspensions were prepared by MACS of the granulocyte fractions from CML patients and healthy controls. Total RNA was extracted and used as a template in an RT-PCR reaction using IL-4 or β-actin specific primers. The levels of specific amplification products were noted as −, +, ++, and +++, which correlate to absent, low, intermediate, and high expression, respectively. One representative experiment out of 3 is shown.
Leukotriene C4 synthase activity in granulocyte suspensions
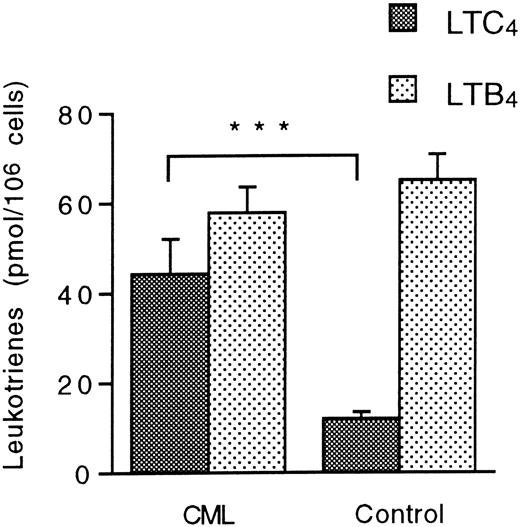
The metabolism of exogenous LTA4 was investigated in unfractionated granulocyte suspensions that were not subjected to MACS. Granulocytes from CML patients and normal controls transformed LTA4 to LTC4 and LTB4. Notably, the levels of LTC4 were almost 4 times higher in CML granulocyte suspensions compared with normal granulocytes, as measured by the mean ± SEM (standard error of the mean) (44.4 ± 7.7 pmol/106 cells [n = 13] versus 12.0 ± 1.5 pmol/106 cells [n = 11]; P = .0007), although the eosinophil content was similar in these suspensions (Table 1). In contrast, the production of LTB4 was unaltered in the CML suspensions (Figure 1). These data indicate increased LTC4 synthase activity in CML granulocytes.
Leukotriene A4 metabolism in CML and normal granulocytes.
Unfractionated granulocyte suspensions (15 × 106cells/mL) from CML patients and healthy controls were incubated with LTA4 (10 μmol/L) at 37°C for 5 minutes. Levels of LTC4 and LTB4 were determined by HPLC. Each value represents the mean obtained from duplicate experiments with cells from 13 CML patients and 11 healthy controls. Error bars indicate SEM. ***Indicates P < .001.
Leukotriene A4 metabolism in CML and normal granulocytes.
Unfractionated granulocyte suspensions (15 × 106cells/mL) from CML patients and healthy controls were incubated with LTA4 (10 μmol/L) at 37°C for 5 minutes. Levels of LTC4 and LTB4 were determined by HPLC. Each value represents the mean obtained from duplicate experiments with cells from 13 CML patients and 11 healthy controls. Error bars indicate SEM. ***Indicates P < .001.
Leukotriene C4 synthase activity in CD16+neutrophils and CD16− eosinophil-enriched cell preparations
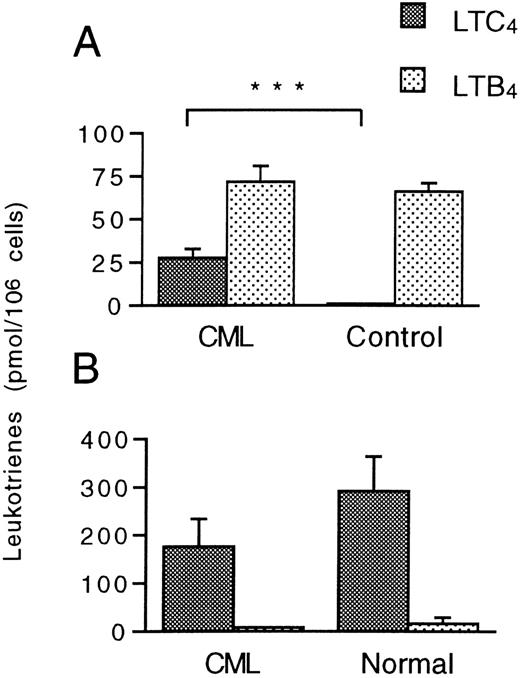
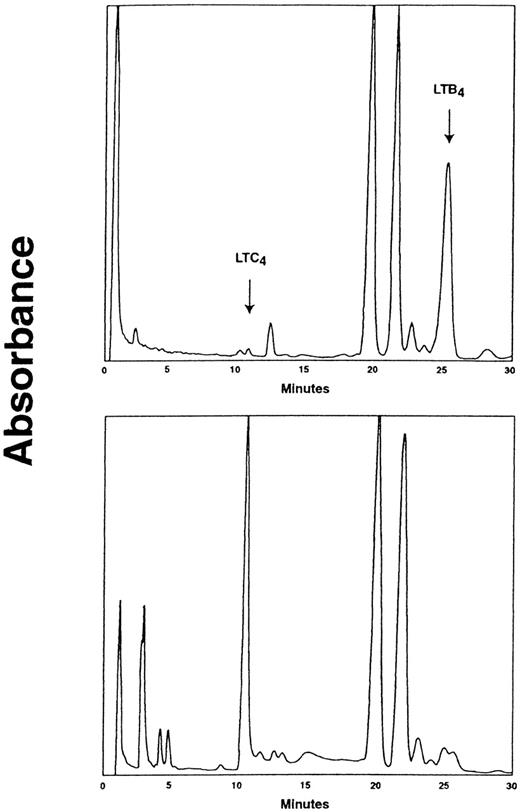
The metabolism of LTA4 in CD16+ neutrophils and CD16− eosinophil-enriched cell preparations isolated by MACS was investigated. Notably, CML CD16+neutrophils from all patients tested transformed LTA4 to LTC4, (range, 8.1-44.2 pmol/106 cells; mean ± SEM 27.7 ± 5.1 pmol/106 cells [n = 9]) (Figure 2A). In contrast and as anticipated, normal CD16+ neutrophils failed to produce significant amounts of LTC4 from LTA4 (range, 0-2.9 pmol/106 cells; 1.3 ± 0.3 pmol/106cells [n = 11]; P < .0001). The capacity to convert LTA4 to LTB4 was similar in CML and normal CD16+ neutrophils (Figure 2A). Expectedly, the CD16− eosinophil-enriched cell preparations from leukemic patients and normal individuals efficiently transformed LTA4 to LTC4 (176.7 ± 56.9 and 291.7 ± 72.8 pmol/106 cells, respectively) and minor amounts of LTB4 (Figure 2B). Determination of LTA4 metabolism in subcellular fractions obtained from CML CD16+ neutrophil suspensions revealed that the LTC4 synthase activity was almost exclusively confined to the microsomal fraction (Figure 3), in accordance with the known subcellular distribution of LTC4synthase.17 In contrast and as expected,54 the LTA4 hydrolase activity was essentially located to the cytosolic fraction. Formation of LTD4 or LTE4was not observed in any of the cell fractions.
Leukotriene A4 metabolism in CML and normal CD16+ neutrophils and CD16−eosinophil-enriched cell suspensions.
(A) CD16+ neutrophils (15 × 106cells/mL) and (B) CD16− eosinophil-enriched cell suspensions (3 × 106 cells/mL) from CML patients and healthy controls were incubated with LTA4 (10 μmol/L) at 37°C for 5 minutes. Levels of LTC4 and LTB4 were determined by HPLC. Each value represents the mean obtained from duplicate experiments with cells from 9 (neutrophil) or 6 (eosinophil-enriched) CML patients and 11 (neutrophil) or 4 (eosinophil-enriched) healthy controls. Error bars indicate SEM. ***Indicates P < .001.
Leukotriene A4 metabolism in CML and normal CD16+ neutrophils and CD16−eosinophil-enriched cell suspensions.
(A) CD16+ neutrophils (15 × 106cells/mL) and (B) CD16− eosinophil-enriched cell suspensions (3 × 106 cells/mL) from CML patients and healthy controls were incubated with LTA4 (10 μmol/L) at 37°C for 5 minutes. Levels of LTC4 and LTB4 were determined by HPLC. Each value represents the mean obtained from duplicate experiments with cells from 9 (neutrophil) or 6 (eosinophil-enriched) CML patients and 11 (neutrophil) or 4 (eosinophil-enriched) healthy controls. Error bars indicate SEM. ***Indicates P < .001.
HPLC chromatograms demonstrating the subcellular distribution of LTC4 synthase activity in CML CD16+ neutrophils.
CD16+ neutrophils from CML patients were subjected to subcellular fractionation as described in Materials and Methods. Cytosolic (upper panel) and microsomal (lower panel) fractions were incubated with LTA4 (60 μmol/L) and glutathione (5 mmol/L) in the presence of 0.05% BSA at 20°C for 10 minutes. Absorbance was measured at 280 nm. The retention times of LT standards are indicated. One representative experiment out of 4 is shown.
HPLC chromatograms demonstrating the subcellular distribution of LTC4 synthase activity in CML CD16+ neutrophils.
CD16+ neutrophils from CML patients were subjected to subcellular fractionation as described in Materials and Methods. Cytosolic (upper panel) and microsomal (lower panel) fractions were incubated with LTA4 (60 μmol/L) and glutathione (5 mmol/L) in the presence of 0.05% BSA at 20°C for 10 minutes. Absorbance was measured at 280 nm. The retention times of LT standards are indicated. One representative experiment out of 4 is shown.
Endogenous formation of LTC4 in CML CD16+neutrophils
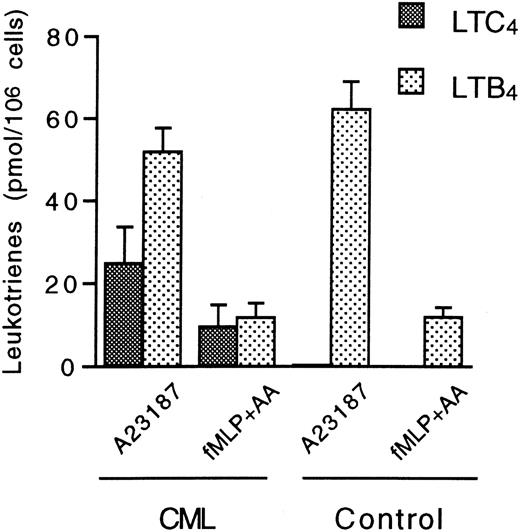
The capacity of CML CD16+ neutrophils to produce LTC4 from endogenous arachidonic acid was investigated by incubation with 1 μmol/L ionophore A23187 for 5 minutes. Again, these cells readily produced LTC4 (24.8 ± 8.9 pmol/106 cells [n = 4]) (Figure 4). Furthermore, CML CD16+ neutrophils also produced LTC4 (9.6 ± 5.2 pmol/106 cells [n = 4]) when activated with the chemotactic tripeptide fMLP in the presence of low amounts of exogenous arachidonic acid (8 μmol/L) for 5 minutes. Again, further conversion of LTC4 to LTD4 or LTE4 could not be observed. In addition, these cells produced LTB4 after stimulation with ionophore A23187 or fMLP plus arachidonic acid. In comparison, normal CD16+ neutrophils only produced LTB4 when stimulated with these agents (Figure 4). LT production was not observed after the addition of 8 μmol/L arachidonic acid to normal or CML CD16+ neutrophils (results not shown).
Leukotriene biosynthesis in normal and CML CD16+ neutrophils after stimulation with ionophore A23187 or a combination of fMLP and arachidonic acid.
CD16+ neutrophils (15 × 106 cells/mL) from CML patients and healthy controls were incubated with 1 μmol/L of either A23187 or fMLP at 37°C for 5 minutes. The latter compound was combined with 8 μmol/L arachidonic acid. Levels of LTC4 and LTB4 were determined by HPLC. Each value represents the mean obtained from duplicate experiments with cells from 4 CML patients and 4 healthy controls. Error bars indicate SEM.
Leukotriene biosynthesis in normal and CML CD16+ neutrophils after stimulation with ionophore A23187 or a combination of fMLP and arachidonic acid.
CD16+ neutrophils (15 × 106 cells/mL) from CML patients and healthy controls were incubated with 1 μmol/L of either A23187 or fMLP at 37°C for 5 minutes. The latter compound was combined with 8 μmol/L arachidonic acid. Levels of LTC4 and LTB4 were determined by HPLC. Each value represents the mean obtained from duplicate experiments with cells from 4 CML patients and 4 healthy controls. Error bars indicate SEM.
Expression of LTC4-producing enzymes in CML CD16+ neutrophils
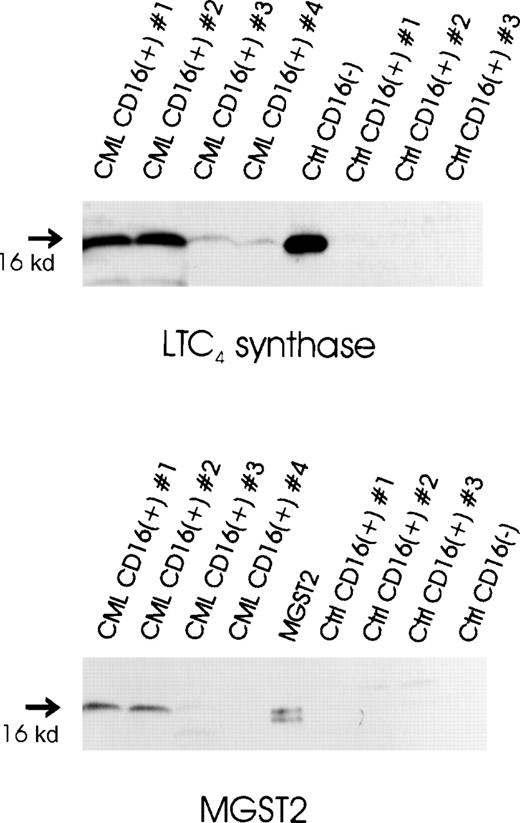
Expression of LTC4 synthase protein in CML and normal CD16+ neutrophils was investigated by immunoblot analysis. Protein samples obtained from normal CD16−eosinophil-enriched cell preparations were used as positive controls. LTC4 synthase was consistently expressed at intermediate to high levels in CML CD16+ neutrophils (Figure5). In contrast, LTC4 synthase immunoreactivity was not observed in normal CD16+neutrophils. The 2 CML CD16+ neutrophil samples displaying the highest levels of LTC4 synthase also contained detectable amounts of MGST2. However, in the 2 additional CML samples, as well as in the samples obtained from normal controls, MGST2 immunoreactivity could not be observed. The positive control for MGST2 was prepared from MGST2-transfected Sf9 cells, as described previously.48
Expression of LTC4-producing enzymes in normal and CML CD16+ neutrophils.
Microsomal fractions of CML CD16+ neutrophils, normal CD16+ neutrophils, normal CD16−eosinophil-enriched cell preparations (20 μg protein per sample), and an MGST2 standard obtained from MGST2-transfected Sf9 cells (1 μg protein) were electrophoresed through 14% polyacrylamide gels and electroblotted onto nitrocellulose. Immunoblot analysis was performed using antibodies against LTC4 synthase and MGST2, respectively.
Expression of LTC4-producing enzymes in normal and CML CD16+ neutrophils.
Microsomal fractions of CML CD16+ neutrophils, normal CD16+ neutrophils, normal CD16−eosinophil-enriched cell preparations (20 μg protein per sample), and an MGST2 standard obtained from MGST2-transfected Sf9 cells (1 μg protein) were electrophoresed through 14% polyacrylamide gels and electroblotted onto nitrocellulose. Immunoblot analysis was performed using antibodies against LTC4 synthase and MGST2, respectively.
Expression of mRNA encoding LTC4 synthase was demonstrated in CML CD16+ neutrophils by RT-PCR using samples from normal CD16− eosinophil-enriched cells as positive controls (Figure 6). To verify RNA integrity, amplifications with β-actin–specific primers were also performed. The identity of the LTC4 synthase amplification product was confirmed by cloning and sequencing (results not shown).
RT-PCR analysis of LTC4 synthase mRNA expression in CML CD16+ neutrophils.
Total RNA was extracted from CML CD16+ neutrophils and normal CD16− eosinophil-enriched cell preparations and used as templates in RT-PCR reactions using LTC4synthase or β-actin specific primers. M represents a 50-2000 bp molecular weight marker. The expected lengths of amplification products are 119 bp (LTC4 synthase) and 784 bp (β-actin). One representative experiment out of 4 is shown.
RT-PCR analysis of LTC4 synthase mRNA expression in CML CD16+ neutrophils.
Total RNA was extracted from CML CD16+ neutrophils and normal CD16− eosinophil-enriched cell preparations and used as templates in RT-PCR reactions using LTC4synthase or β-actin specific primers. M represents a 50-2000 bp molecular weight marker. The expected lengths of amplification products are 119 bp (LTC4 synthase) and 784 bp (β-actin). One representative experiment out of 4 is shown.
Discussion
In previous studies we reported increased production of LTC4 in myeloid cells from patients with CML.2 3 The LTC4 overproduction, consistently observed after ionophore A23187 stimulation of unfractionated leukocytes, bone marrow cells, or granulocyte preparations strongly indicated enhanced LTC4 synthase activity in CML. In the current investigation, elevated LTC4 synthase activity in CML granulocyte suspensions, as compared with the enzyme activity in normal granulocytes, was confirmed by significantly enhanced LTC4 formation after incubation of unstimulated intact cells with exogenous LTA4, the immediate enzyme substrate. Notably, cautious morphological analyses demonstrated that the number of eosinophils was not essentially higher in CML granulocyte suspensions than in control preparations. These findings exclude the possibility that the increased LTC4 synthase activity was related to the number of eosinophils. However, these data could not conclude whether the increased LTC4 synthesis was due to elevated intrinsic LTC4 synthase activity in CML eosinophils or to an aberrant expression of the enzyme in the leukemic neutrophils.
To elucidate this question, MACS methodology based on CD16 antigen immunoselection was applied in order to obtain eosinophil-free, highly purified neutrophil preparations. Expression of the CD16 surface antigen on neutrophils but not on eosinophils has been demonstrated.55 Furthermore, CD16 expression on CML neutrophils has been confirmed.56 Using the MACS technique, we obtained CD16+ neutrophil preparations with more than 99% purity and no detectable eosinophil contamination, as judged by morphological examination. The high purity of these cells was further indicated by negative RT-PCR analysis of mRNA encoding IL-4, which is known to be expressed in lymphocytes and eosinophils but not in neutrophils.51-53 57 CML CD16+cells were as potent as the corresponding control cells in transforming LTA4 to LTB4, indicating unaltered LTA4 hydrolase activity in the cells. Since LTA4 hydrolase expression is a characteristic of neutrophils, these findings further confirm the identity of the leukemic CD16+ neutrophils. Interestingly, however, all CD16+ neutrophil preparations from CML patients also expressed LTC4 synthase activity, as judged by the ability of these cells to convert LTA4 to LTC4.
Experiments with subcellular fractions demonstrated that the enzyme activity was solely distributed to the microsomal fraction, in agreement with the known intracellular localization of LTC4synthase.17 In contrast, the LTC4 synthase activity in normal CD16+ neutrophil suspensions was negligible. The contamination of platelets, a potential source of LTC4 synthase,24-26 was always low in both normal and CML CD16+ neutrophil fractions, with less than 50 000 platelets per 106 neutrophils. In an earlier study using platelet preparations from 44 normal donors, platelet LTC4 formation under conditions identical to those used in the present study was approximately 3 pmol/106platelets.39 Furthermore, we have observed a similar LTC4 synthase activity in CML and normal platelets (Lindgren et al, unpublished observation). This excludes the possibility that the LTC4 synthase activity observed in CML CD16+ neutrophil preparations was the result of platelet contamination.
Further investigation of the LTC4-producing capacity in CML CD16+ neutrophils revealed that these cells also synthesized LTC4 from endogenous substrate upon challenge with calcium ionophore A23187. Similarly, LTC4 synthesis was provoked in these cells by the chemotactic tripeptide fMLP. Again, normal CD16+ neutrophils failed to produce LTC4under these conditions, whereas the synthesis of LTB4 was similar in normal and CML neutrophils. The findings demonstrate that the LTC4 synthase activity in CML neutrophils is efficiently coupled to the enzymatic machinery providing endogenous LTA4 and strongly suggest that these cells are capable of LTC4 synthesis in vivo.
The expression of mRNA encoding LTC4 synthase in CML CD16+ neutrophils was demonstrated by RT-PCR, indicating that the gene is transcriptionally active in these cells. Moreover, expression of LTC4 synthase protein in all CML CD16+ neutrophil preparations tested was verified by immunoblot analysis. In contrast, LTC4 synthase immunoreactivity could not be observed in protein extracts from normal CD16+ neutrophils.
Messenger RNA encoding MGST2 was previously found to be highly expressed in K-562, a myeloid leukemia cell line,35 suggesting expression of this enzyme in myeloid leukemia. It was therefore of interest to investigate the expression of this enzyme on the protein level in freshly isolated CD16+neutrophils from CML patients. Interestingly, CD16+neutrophil preparations from some of the CML patients displayed MGST2 immunoreactivity. However, significant expression of this enzyme could not be detected in CD16+ neutrophils from other CML patients despite the fact that these cells always possessed LTC4 synthase activity. Thus, these data suggest that the LTC4 synthase activity observed in CML neutrophils depends on the aberrant expression of LTC4 synthase and not the expression of MGST2. Whether abnormal expression of MGST2 is a common feature of CML neutrophils or other myeloid cells and whether this can contribute in any degree to the abnormal LTC4synthase activity need to be further investigated. In addition, our data suggest that MGST2 expression is not a regular feature of neutrophils, because MGST2 immunoreactivity could not be observed in any of the normal CD16+ neutrophil preparations.
The present findings provide new insights regarding the mechanisms behind the previously reported elevated LTC4 synthase activity in myeloid cells from CML patients.2,3Furthermore, the observed aberrant LTC4 synthase expression in highly purified mature CML neutrophils strongly indicates correlation between the abnormal LTC4 synthase activity and the malignant transformation. Interestingly, however, the LTC4 overproduction was much less pronounced in the neutrophils than in unfractionated white blood cell suspensions from CML patients.2 Thus, after ionophore A23187 stimulation, the LTC4 production was twice that of LTB4 in CML white blood cells, whereas this proportion was reversed in CML CD16+ neutrophils. This strongly suggests that less mature leukemic myeloid cells contribute to the overproduction of LTC4 in CML. Presently, we are investigating the expression and activity of LTC4 synthase immature myeloid cells from CML patients.
The CD16− cell fractions were highly enriched in eosinophils (63% and 78% in CML and control preparations, respectively) but also contained other cell types. No attempt was made to further purify the eosinophils. These cell suspensions efficiently produced LTC4 when incubated with LTA4, in agreement with a high LTC4 synthase activity in eosinophils. Notably, there was no sign of elevated LTC4synthase activity in CML eosinophils compared with normal cells. The CD16− eosinophil-enriched cell suspensions also transformed LTA4 to LTB4, reflecting the presence of contaminating LTA4 hydrolase-expressing cells such as neutrophils and lymphocytes58 in these cell preparations.
It can be approximated from the present data that the mean LTC4 synthase activity was roughly 10-fold higher in CML eosinophils than in CML CD16+ neutrophils. However, in CML the normal myelopoiesis is gradually replaced by the production of malignant cells. It has been demonstrated that normal myeloid cells coexist with leukemic cells in the bone marrow as well as in the peripheral blood during the chronic phase of the disease.59 60 Since it can be speculated that normal neutrophils within the CML CD16+ cell fractions lack expression of LTC4 synthase, it may be assumed that the determined enzyme activity in these cell fractions is an underestimate of the real LTC4 synthase activity in leukemic neutrophils. Furthermore, since the number of neutrophils in bone marrow and peripheral blood is usually more than 10 times higher than the number of eosinophils, it is probable that leukemic neutrophil LTC4 synthase activity accounts for a substantial part of the in vivo LTC4 synthesis and that this may lead to a significant overproduction of this compound in patients with CML.
Studies of LTC4 synthase expression and activity in differentiating myeloid cells have suggested up-regulation of LTC4 synthase expression during eosinophilic differentiation.47,61 Furthermore, LTC4synthase activity or expression could not be detected in CD34+ cord blood stem cells.61 In contrast, we have recently obtained data demonstrating LTC4 synthase activity in MACS-purified immature progenitor cells from normal human bone marrow (Sjölinder et al, to be published). Accordingly, the LTC4 synthase activity in white blood cell suspensions obtained from patients with acute myeloid leukemia62 was highly correlated to the proportion of circulating blast cells. Thus, it may be speculated that LTC4 synthase is expressed in stem cells and progenitors in the bone marrow and that the expression is down-regulated before differentiation of the cells into mature neutrophils. It may be further speculated that this down-regulation is disturbed in CML neutrophils, leading to aberrant expression of LTC4 synthase.
The present results demonstrate that abnormal expression of LTC4 synthase is a regular feature in CML. Elevated expression of LTC4 synthase has recently been demonstrated in aspirin-sensitive asthma.63 However, in this condition the increase was merely quantitative, because the altered expression was observed in eosinophils, which normally express LTC4synthase. Thus, the present paper is the first report in which aberrant expression of LTC4 synthase can be coupled to a specific disease.
Several cellular abnormalities in CML have been directly associated to the tyrosine kinase activity possessed by the BCR-ABL fusion protein.64 Tyrosine kinase is known to influence the expression of several genes via a wide range of signal transduction pathways. Therefore it will be of interest to elucidate whether this abnormal enzyme is involved in the mechanisms leading to aberrant expression of LTC4 synthase in CML neutrophils. Notably, we have previously reported a normal LT biosynthesis in white blood cell suspensions from patients with polycythemia vera.2 This Ph chromosome-negative chronic myeloproliferative disorder is typically characterized by excessive erythrocytosis in addition to an accumulation of myeloid cells. Furthermore, various neutrophil abnormalities, such as impaired stimulus-induced activation of phospholipase D and oxidative metabolism, have been described.65
It may be speculated that the aberrant LTC4 synthase gene expression in CML neutrophils may lead to overproduction of LTC4 in the bone marrow of these patients. This may be of potential interest because LTs may play a role in myelopoiesis.6 Further studies are in progress to investigate the pathophysiological importance of the increased LTC4 production in CML.
Acknowledgment
We thank Dr Robert Zipkin, Biomol Research Laboratories (Plymouth Meeting, PA), for the generous gift of LTA4 methyl ester.
Supported by grant 2663 from the Swedish Cancer Society (Stockholm, Sweden), grants 03X-6805 and 03X-12573 from the Swedish Medical Research Council (Stockholm, Sweden), and research funds from the Karolinska Institutet (Stockholm, Sweden).
Reprints:Jan Åke Lindgren, Division of Physiological Chemistry II, Department of Medical Biochemistry and Biophysics, Scheele Laboratory, Karolinska Institutet, S-171 77 Stockholm, Sweden; e-mail: jan.ake.lindgren@mbb.ki.se.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal