Mutations of coding repeats within the E2F4, TGF-βRII, BAX, IGFIIR, and hMSH3 are critical targets of microsatellite instability (MSI) in many kinds of cancers. We analyzed 9 childhood acute lymphoblastic leukemia (ALL) samples, 5 acute myelocytic leukemia (AML) samples, and 10 adult T-cell leukemia (ATL) samples having MSI to determine whether they had mutations of the E2F4, TGF-βRII, BAX, IGFIIR, and hMSH3 genes. Frameshift mutations were found at trinucleotide repeats within a coding exon of the E2F4 gene in 2 of 10 (20%) ATL samples and 1 of 9 (11%) childhood ALL samples. No mutations were found in the TGF-βRII, BAX, IGFIIR, andhMSH3 genes. E2F4 is a transcription factor that influences the cell-cycle progression. These results suggest that mutations of the E2F4 gene, presumably caused by an abnormality of one of the DNA repair genes, may play an important role in development of ATL and childhood ALL.
Microsatellite instability (MSI), representing either an expansion or a reduction of (C-A)n repeats, has been reported in many kinds of human malignancies.1-3 MSI appears to reflect multiple replication errors because of a defective mismatch repair gene, including hMSH2, hMLH1,hPMS1, and hPMS2.4,5 Cancers with MSI show exaggerated genomic instability at simple repeat sequences that generate somatic frameshift mutations in genes containing these repeat sequences. Somatic frameshift mutations of coding repeats within the E2F4, TGF-βRII, BAX, IGFIIR, and hMSH3 are recognized as critical targets of MSI in many kinds of cancers.6-10 We previously found that MSI is present in childhood acute lymphoblastic leukemia (ALL), acute myelocytic leukemia (AML), and adult T-cell leukemia (ATL).11-13However, the genes that are the target for mutation in hematological malignancies with MSI are not known. In this study, we analyzed 24 hematological malignancies having MSI to determine whether mutations occurred at coding repeats of 5 target genes: E2F4, TGF-βRII, BAX, IGFIIR, and hMSH3.
Study design
We previously analyzed for MSI using 48 childhood ALL, 17 AML, and 22 ATL samples.11-13 MSI was present in 9 (19%) childhood ALL specimens, 5 (29%) AML specimens, and 10 (45%) ATL specimens. These 24 samples with MSI were used in this study. Informed consent was obtained from the patients, their parents, or both, as appropriate. Mutation analysis of the E2F4, TGF-βRII, BAX, IGFIIR, andhMSH3 genes were performed as reported previously.6-10 A trinucleotide (AGC) repeat spanning codons 306-321, which normally encodes 13 serine residues of theE2F4 gene, was studied.6 For the TGF-βRIIgene, the poly (A) sequence (nucleotides 709-718) was analyzed.7 For the BAX gene, the poly (G) sequence in the third coding exon was analyzed.8 A fragment containing the deoxyguanine repeat (nucleotides 4030-4140) was investigated in the IGFIIR gene.9 For the hMSH3gene, the track of 8 deoxyadenine in exon 7 was assessed.10
Results and discussion
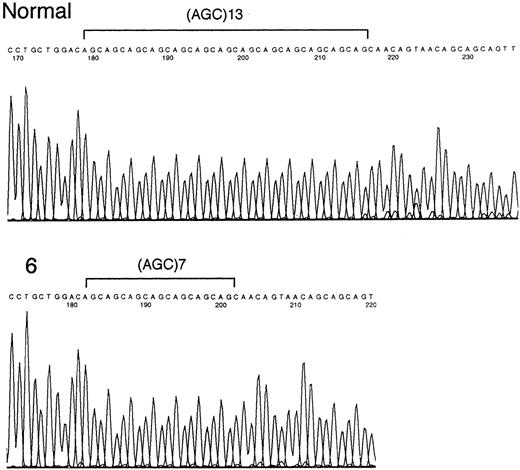
Three (13%) samples contained mutations of the poly (AGC) tract within E2F4 (Table 1). One sample contained an E2F4 allele that was 6 codons (18 nucleotides) shorter than the normal sequence. The 2 other samples containedE2F4 alleles that were 3 codons (9 nucleotides) longer than the normal sequence (Figure 1). By leukemia subtype, 1 of 9 (11%) childhood ALL samples and 2 of 10 (20%) ATL samples contained E2F4 mutations. The ALL patient with anE2F4 mutation had a T-cell phenotype; 2 ATL patients withE2F4 mutations had acute-type ATL. All 3 samples withE2F4 mutations had MSI at 1 or 2 loci. None of the 5 AML samples had mutations within E2F4. No mutations were found within the repeat sequence of TGF-βRII, BAX, IGFIIR, andhMSH3 genes (data not shown).
Summary of clinicopathologic data, MSI status of patients with E2F4 mutations
| Patient No. . | Diagnosis . | MSI Status . | Mutational Analysis . | ||||
|---|---|---|---|---|---|---|---|
| E2F4 (AGC) 13* . | TGF-βRII (A) 10 . | BAX (G) 8 . | IGFIIR (G) 8 . | hMSH3 (A) 8 . | |||
| 54 | T-ALL | + | Deletion (7) | N | N | N | N |
| 6 | Acute ATL | + | Insertion (16) | N | N | N | N |
| 18 | Acute ATL | ++ | Insertion (16) | N | N | N | N |
| Patient No. . | Diagnosis . | MSI Status . | Mutational Analysis . | ||||
|---|---|---|---|---|---|---|---|
| E2F4 (AGC) 13* . | TGF-βRII (A) 10 . | BAX (G) 8 . | IGFIIR (G) 8 . | hMSH3 (A) 8 . | |||
| 54 | T-ALL | + | Deletion (7) | N | N | N | N |
| 6 | Acute ATL | + | Insertion (16) | N | N | N | N |
| 18 | Acute ATL | ++ | Insertion (16) | N | N | N | N |
MSI indicates microsatellite instability; +, MSI at 1 locus; ++, MSI at 2 loci; N, negative for mutation.
The number in parentheses on the E2F4 gene indicates total number of AGC repeats observed in the E2F4 gene in each of the samples.
Sequencing of the mutated E2F4 gene.
Only the (AGC)13 tract and flanking regions of the E2F4 gene are indicated. Sample 6 (acute ATL) shows a mutant nucleotide sequence.
Sequencing of the mutated E2F4 gene.
Only the (AGC)13 tract and flanking regions of the E2F4 gene are indicated. Sample 6 (acute ATL) shows a mutant nucleotide sequence.
The present study has found for the first time that mutations within the important cell cycle gene E2F4 were present in hematological malignancies with MSI. E2F is a family of transcription factors, the activity of which influences the progression through the G1-S transition of the cell cycle.14 E2F consensus binding sites have been identified in the promoters of several growth-regulatory genes, including c-myc, cyclin-dependent kinases, and cyclin D1.15-17 E2F-mediated transcriptional activation is repressed through a physical association between E2F and proteins of the retinoblastoma family (pRB, p107, p130). Five different E2Fs have been identified: E2F1, E2F2, and E2F3 interact preferentially with pRB18; E2F5 with p13019; and E2F4 associates with all 3 of these proteins, especially p107 and p130 in a cell cycle–dependent manner.14,18,20,21 The p107 and p130 proteins inhibit the ability of E2F4 to transactivate genes whose promoter contains E2F binding sites, whereas the capacity of E2F4 to bind DNA is retained.22 This regulation of E2F4 is important in the normal control of cell growth. For example, an E2F4 mutation that prevented the protein from binding to p107 resulted in transformation of NIH3T3 cells, allowing the cells to form tumors.14 The poly (AGC) tract of E2F4 lies between the “marked box” region and the binding region of the retinoblastoma family of proteins. This serine repeat domain may induce transactivation of target genes.6 Even though a functional effect of a mutation of this region has not been studied, changes in this region might potentiate transcriptions of growth-stimulatory E2F4 target genes.
In this study, mutations were found only at the trinucleotide repeat sequence within the E2F4 gene, and no mutations were found at the single nucleotide repeat within the TGF-βRII, BAX, IGFIIR,and hMSH3 genes. This finding is congruent with the previous findings that no mutations of the TGF-βRII andBAX genes were found in childhood ALL.23,24Moreover, Kaneko et al25 reported that mutations of theTGF-βRII gene were not found in MDS patients with MSI. Taken together, these findings suggest that the mechanisms underlying the occurrence of such trinucleotide replication errors could differ from those of the single nucleotide frameshifts in hematological malignancies.
Acknowledgment
We thank Naoko Hayase for excellent secretarial help.
Supported in part by grant-in-aid from the Ministry of Education, Science, Sports, and Culture of Japan; the Parker Hughes Trust; the C. and H. Koeffler FUND; and the Gladys Lichtenstein Grant.
Reprints:Seisho Takeuchi, 3rd Department of Medicine, Kochi Medical School, Okohcho, Nankoku, Kochi 783-8505, Japan; e-mail: takeuti@kochi-ms.ac.jp.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal