CD99, the product of the MIC2 gene, exhibits an erythroid-specific quantitative polymorphism coregulated with the polymorphism of the XG blood group gene. As a preliminary study of this phenomenon, human XG and CD99 recombinant proteins were expressed in murine RAG cells and analyzed by flow cytometry. Both proteins were expressed independently and at a similar level in single and double transfectants. Immunoprecipitation and Western blot analysis, using the murine monoclonal antibodies NBL-1 and 12E7, revealed species of 26 kd (XG) and 32 kd (CD99), respectively. A putative 28-kd intracellular precursor of CD99 was also detected, as was a 26-kd species after neuraminidase treatment of CD99-expressing cells. No evidence of association or complex formation between XG and CD99 proteins could be proven, either on transfected RAG cells or on human erythrocytes. These results were confirmed using somatic hybrids between single transfectants. These findings suggest that the phenotypic relationship between XG and CD99 is mostly regulated at the transcriptional level, but they do not formally exclude some posttranscriptional effect. Studies on the tissue specificity of XG expression showed that surface expression of the XG protein could not be restored in somatic hybrids between B-lymphoblastoid cell lines from Xg(a+) persons and fibroblasts (RAG) or erythroid (MEL) cells. RT-PCR analysis of the transcripts revealed the existence of an XG mRNA in each cell line, suggesting that the tissue-specific regulation of cell surface XG expression occurs either at a quantitative transcriptional level or is a posttranscriptional event. By Northern blot analysis,XG transcripts were detected in erythroid tissues and several nonerythroid tissues.
The human erythrocyte antigen Xga, a 26-kd glycoprotein, was identified with an antibody found in a patient who underwent multiple transfusions.1,2 It is inherited as a sex-linked dominant character,2 and its phenotypic frequencies are different in males and females. The polymorphism is defined by the Xg(a+) and Xg(a−) phenotypes because no antigen antithetical to Xga has been found. Approximately 89% of females and 66% of males are Xg(a+).1 TheXG polymorphism could result from different amounts of the XG protein on Xg(a+) and Xg(a−) erythrocytes,3 but this has yet to be proven. One of the 2 X chromosomes in a female is inactivated early in embryonic development.4 However, unlike most X-linked genes, the XG locus escapes this inactivation.1 The Xga antigen has also been found on human fibroblasts in culture.5
The murine monoclonal antibody 12E7, raised against lymphocytes from a patient with T-cell acute lymphocytic leukemia,6 recognizes a cell surface glycoprotein of 32 kd, called CD99, which is encoded by the MIC2 gene.7,8 The MIC2 gene is borne by the X and Y chromosomes9 and is localized in the pseudoautosomal regions of the short arms Xp and Yp,8 which pair and exchange in male meiosis. The MIC2 locus on the X chromosome is not subject to inactivation.10
The MIC2 gene is expressed in many tissues and shows an erythrocyte-specific quantitative polymorphism that is coregulated with the polymorphism of the XG gene.11 All Xg(a+) persons are CD99-high expressers, and all Xg(a−) females are CD99-low expressers. Xg(a−) males can be CD99-high or CD99-low expressers. To explain this phenotypic relation, the existence of a regulatory locus XGR, present on both X and Y chromosomes, has been postulated.12 In this model, XGR controls the expression of MIC2 and XG and is polymorphic with 2 alleles, A and B. The A allele induces Xga antigen expression and high-level CD99 expression; theB allele fails to express Xga and induces low-level CD99 production. The coexpression of the 2 genes, XG andMIC2, would lead to the cell surface production of the 2 proteins and would be controlled at the transcriptional level.3
MIC213,14 and XG15 genes have been cloned, and their organization on the short arm of the X chromosome has been (Xp) determined. The 2 genes encompass 150 kb of DNA located on both sides of the pseudoautosomal boundary of Xp, and both are oriented toward the centromere. The first 3 exons ofXG are situated in the pseudoautosomal region, approximately 10 kb downstream of MIC2 (10 exons), whereas the other 7 exons are in the Xspecific region.
The CD99 and XG proteins are sequence related (48% homology) and share some unusual motifs in a similar order, suggesting that the 2 genes originated from the duplication of an ancestral gene. The predicted structure of CD9916 and, by analogy, of XG defines an integral membrane protein with a single transmembrane region, an extracellular amino terminus, and a cytosolic carboxy terminus. Immunochemical studies on Xg(a+) and Xg(a−) erythrocytes demonstrated the quantitative polymorphism of CD99 at the cell surface and indicated that the XG and CD99 proteins from Xg(a+) red blood cells (RBCs) can be copurified with the 12E7 antibody17 or coprecipitated with anti-Xga antibodies.18These data suggested that the 2 proteins may be closely associated in the erythrocyte membrane, possibly as a heterodimer.
To understand the molecular basis of the phenotypic relationship between XG and CD99, we analyzed the coexpression of the XG andMIC2 cDNAs in transfected mammalian cells, either in double transfectants or in somatic hybrids from single transfectants. We also studied the tissue specificity of XG expression at the RNA and protein levels.
Materials and methods
Subcloning of the XG and MIC2 cDNAs in the expression vector pcDNA3 and its derivative pcDNA-Hyg
Full-length cDNAs were cloned by polymerase chain reaction (PCR) from a human fetal liver cDNA library using published information13-15 and sequenced on both strands by the dideoxy chain termination method.19 The XG andMIC2 cDNAs were subcloned as BamHI–EcoRI fragments in the pcDNA3 expression vector (Invitrogen, San Diego, CA) to give pcDNA3-XG and pcDNA3-MIC2, respectively. For cotransfection experiments, pcDNA3 was modified by replacement of the selection marker as follows: the 1.9-kb NruI-HindIII fragment from p220.2 (gift from Dr B. Sugden, University of Wisconsin, Madison, WI), containing the hygromycin resistance gene(hygR) under the control of promoter and processing signals from the tk gene of HSV1, was filled in with the Klenow fragment of Escherichia coli DNA polymerase I and inserted between the blunt-ended SmaI and BsmI sites (removal of the neomycin resistance cassette,neoR) of pcDNA3 to obtain pcDNA-hyg. The 0.6-kb, filled-in BamHI-EcoRI XG orMIC2 fragments were introduced into the EcoRV site of pcDNA-hyg to give the plasmids pcDNA-hyg-XG and pcDNA-hyg-MIC2, respectively. All plasmids used were propagated in E. coli XL1 blue.
Cell lines and cell culture
RAG cells (mouse adenocarcinoma cell line) were maintained in IMDM (Life Technologies, Paisley, UK) supplemented with 10% (vol/vol) fetal calf serum (FCS). MEL cells (murine erythroleukemia) were obtained from the American Type Culture Collection (Rockville, MD) and were cultured in Dulbecco's modified Eagle's medium (DMEM; Life Technologies) supplemented with 20% FCS. Epstein-Barr-derived B-lymphoblastoid cell lines (B-LCLs) obtained from 2 healthy male donors CT and CL, typed as Xg(a+) and Xg(a−)/CD99-high expressor, respectively, were established in our laboratory and maintained in IMDM supplemented with 20% FCS. NBL-1,3 a mouse hybridoma cell line producing a monoclonal anti-Xga antibody, was cultured in RPMI 1640 supplemented with 10% FCS and HAT (Sigma, St. Louis, MO). Red blood cells from Xg(a+) and Xg(a−) donors typed as CD99-high or CD99-low expressers were from the Centre National de Référence pour les Groupes Sanguins (Paris, France).
Antibodies
The murine monoclonal anti-Xga was purified from NBL-13 hybridoma supernatants by protein A–Sepharose chromatography, as described.20 The 12E7 murine monoclonal antibody6 was kindly provided as an ascites fluid by Dr Peter Goodfellow (University of Cambridge, Cambridge, UK) and was purified by the same procedure. Binding constants for 12E7 and NBL-1 antibodies were determined by Scatchard analysis as described before.17A polyclonal antibody directed against residues 1 to 13 of the mature XG protein was produced in rabbits according to standard protocol. The human anti-Xga (serum Alo.) was from the Centre National de Référence pour les Groupes Sanguins.
Stable transfection of RAG cells
RAG cells were transfected with the different expression vectors by calcium phosphate coprecipitation.21 NeoR and hygR transfectants were selected in IMDM plus 0.35 mg/mL G418 (Geneticin; Life Technologies) or 1 mg/mL hygromycin (Life Technologies), respectively. When less than 50% of the resistant cells expressed the protein of interest, they were sorted as follows: the cells were incubated with the appropriate monoclonal antibody (NBL-1 or 12E7) and then with magnetic beads covered with sheep antimouse IgG (Dynal A.S., Oslo, Norway) according to the instructions of the manufacturer. The sorted cells and resistant pools containing more than 50% expressing cells were then subjected to limiting dilution cloning in 96-well microtiter plates.
Cell fusions (somatic cell hybrids)
All fusion experiments were performed with polyethylene glycol 400 (Merck, Darmstadt, Germany).22 Adherent hybrids generated by the fusion of RAG cells (hypoxanthine phosphoribosyltransferase-deficient or HPRT−) with B-LCLs (CT and CL) were selected in DMEM plus HAT because B-LCLs are HPRT+ and nonadherent. Hybrids between RAG transfectants were selected in IMDM plus 0.35 mg/mL G418 and 1 mg/mL hygromycin. Hybrids between the clone RAG-XG13 (neoR) and B-LCLs were selected in IMDM plus HAT and 0.35 mg/mL G418. Before the fusion of MEL cells with somatic hybrids RAG × B-LCLs (CT or CL), MEL cells and RAG × B-LCLs were stably transfected with pcDNA3 and pcDNA-hyg, respectively. The MEL/neoR × RAG B-LCL/hygR hybrids were subsequently selected in DMEM plus HAT, 0.55 mg/mL G418, and 0.5 mg/mL hygromycin.
Flow cytometry analysis
Surface expression of the XG and CD99 antigens was determined after incubation with NBL-1 or 12E7 monoclonal antibodies. The antibody-binding capacity was determined using calibration mouse IgG-coated beads (Qifikit; DAKO, Glostrup, Denmark) as standards. Before analysis, adherent cells were detached from plastic with 50 mmol/L HEPES (pH 7.3), 125 mmol/L NaCl, 5 mmol/L KCl, and 1 mmol/L EDTA because the XG and CD99 proteins are sensitive to trypsin treatment.17 23 Cell lines (5 × 105), in a phosphate-buffered saline (PBS)/0.2% (wt/vol) bovine serum albumin suspension, were incubated for 1 hour at 4°C with antibodies used at saturation. After they were washed with PBS, cells were incubated for 30 minutes at 4°C with R-phycoerythrin (RPE)-conjugated F(ab′)2 fragment of goat antimouse immunoglobulins (DAKO). TO-PRO-1 iodide (Interchim, Montluçon, France)-positive cells (dead cells) were excluded from analysis. Fluorescence was measured on a FACScan flow cytometer (Becton Dickinson, San Jose, CA).
Northern blot analysis
Total RNAs from cell lines were extracted by the TRIZOL reagent (Life Technologies), and 20 μg was resolved by electrophoresis on a 6% (wt/vol) formaldehyde, 1% (wt/vol) agarose gel and transferred to nylon filters Zeta-probe GT (Bio-Rad, Hercules, CA). Hybridization with the 32P-labeled XG or MIC2 cDNA probes and stringent washes were performed as described.24 Human multiple-tissue Northern blots (Clontech, Palo Alto, CA) containing polyA+ RNA from different human tissues were hybridized with the XGor MIC2 cDNA probes in the ExpressHyb solution (Clontech), according to the manufacturer's instructions.
Western blot analysis
Membrane proteins from 107 cells (transfectants, somatic hybrids) were solubilized with 1% (wt/vol) Triton X-100 in ice-cold 10 μmol/L Na-phosphate (pH 7.4), 150 mmol/L NaCl (PBS) containing the protease inhibitors 1 mmol/L 4-(2 aminoethyl)-benzenesulfonylfluoride (Pefabloc-SC; Boehringer Mannheim, Germany), 10 μg/mL leupeptin (Sigma); and 5 μg/mL pepstatin (Sigma) for 30 minutes at 4°C with gentle shaking. After centrifugation at 37,000g for 45 minutes, one-fortieth supernatant (cell lysate) was subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by immunoblotting with NBL-1 or 12E7 monoclonal antibodies, essentially as described,17 except that incubation with antibodies and washings were performed in PBS, 5% (wt/vol) skimmed milk and PBS, and 0.1% (wt/vol) Tween, respectively. Specifically bound antibodies were detected by chemiluminescence with the ECL reagent (Amersham, Bucks, UK). For neuraminidase treatment, 107 transfected cells or 5 × 109 RBCs were resuspended in 1 vol receptor-destroying enzyme (RDE; or neuraminidase from Vibrio cholerae; Boehringer Mannheim) diluted (0.1 or 0.25 U) in 50 mmol/L Na-acetate (pH 5.5), 150 mmol/L NaCl, 9 mmol/L CaCl2, and 100 μg/mL bovine serum albumin and were incubated for 30 minutes at 37°C. After 3 washes in PBS, transfected cells were lysed and proteins were analyzed as above. Membranes from treated or untreated RBCs were prepared,25 and protein content was determined.26 Fifty micrograms was analyzed by SDS-PAGE and immunoblotting with NBL-1 or 12E7. Specifically bound antibodies were detected by chemiluminescence with the ECL reagent (Amersham).
Immunoprecipitation analysis
Intact erythrocytes (109 cells) or cell lines (107 cells) were sodium iodide 125-labeled by IODO-GEN (1,3,4,6-tetrachloro-3α-6α-diphenylglycoluril; Pierce, Rockford, IL) as described before.20 RBC membranes were prepared and incubated with 10 μg NBL-1 (or the anti-Xga serum Alo.) or 12E7 overnight at 4°C, whereas labeled cells were incubated for 1 hour with the antibodies. After membrane solubilization (RBCs and cell lines), a preformed complex of protein A–Sepharose (10% wt/vol) and of rabbit antimouse IgG (10 μg) in PBS containing 1% (wt/vol) Triton X-100 was then added to the mixture (100 μL) for at least 1 hour at 4°C. The immune complexes were washed on sucrose gradients,20 and specifically bound proteins were eluted from the final pellet by boiling for 5 minutes in 10 mmol/L Tris-HCl (pH 6.8), 1 mmol/L EDTA, and 5% (wt/vol) SDS separated by SDS-PAGE and detected by autoradiography.
Reverse transcription-PCR analysis of the XG transcripts
Ten micrograms total RNAs from cell lines was reverse transcribed using the first-strand cDNA synthesis kit (Pharmacia, Uppsala, Sweden). The XG fragment (551 bp) was amplified by a nested PCR between primers XG1 (sense primer, positions −36 to −16 from the translation initiation codon) and XG4 (antisense primer, positions +571 to +548), then between primers XG2(sense primer, positions −6 to +15) and XG3 (antisense primer, positions +545 to +522). All reactions were performed in the Expand High Fidelity PCR System (Boehringer Mannheim) under the following conditions: 30 cycles of 1 minute at 94°C, 1 minute at 60°C, 1 minute at 72°C, and a final elongation for 7 minutes at 72°C. Specific products were analyzed by agarose gel electrophoresis and hybridization with an internal [32P]-labeled oligonucleotide probe.
Results and discussion
Cell surface expression of the XG and MIC2 cDNAs in RAG cells and somatic cell hybrids
Full-length cDNAs corresponding to the known coding sequences ofXG15 and MIC227 were cloned by PCR from a human fetal liver cDNA library. The sequence of theXG cDNA was identical to the published sequence, whereas sequence analysis of the MIC2 cDNA revealed 1 base substitution G493A changing Asp to Asn at position 165 of the CD99 protein (cytosolic tail). Whether this change reflected a polymorphism of the population was not investigated.
Murine fibroblastic RAG cells were first stably transfected with eitherXG or MIC2 cDNA in either pcDNA3 or pcDNA-hyg expression vectors. NeoR and hygR cells expressing either XG or CD99 proteins at their surfaces were selected and cloned as described in “Materials and methods.” Flow cytometry analysis revealed positive staining with either NBL-1 or 12E7 antibodies on transfected cells, indicating that the XG and CD99 proteins, respectively, are readily transported to the plasma membrane of RAG cells. This is the first report showing the expression of recombinant XG protein at the surface of transfected cell lines, whereas CD99 has already been produced in mouse L cells.28
The relative amount of XG and CD99 expressed at the surface of the transfectants was estimated through the antibody-binding capacity of the monoclonal antibodies NBL-1 and 12E7, respectively. Preliminary experiments by Scatchard analysis indicated that the binding constant (Kd) for 12E7 was 2.5 × 10−8mol/L,17 whereas the Kd for NBL-1 was 0.5 × 10−8 mol/L (data not shown). Antigen-site determination by flow cytometry analysis was performed using saturation conditions for each antibody (antibody concentration equal to 10 times the Kd values), using calibrated mouse IgG-coated beads. Accordingly, the estimated number of XG and CD99 copies on these cells is an approximate extrapolation from the maximum number of antibody molecules bound to the cells (assuming an ideal molar 1:1 ratio of antibody bound per antigen). Although absolute values of XG and CD99 on each transfectant is difficult to compare (see below), it is clear that in all cases more molecules of CD99 than XG were expressed on the transfectants (and red cells), but the exact ratio between the 2 values is uncertain. These estimates of the number of XG and CD99 molecules at the cell surface also assumed that the antigenic determinants were fully accessible to mAbs on intact cells. The clones described in Table 1 represent the best XG and CD99 expressers among a total of 24 clones tested for each transfection. All clones apparently express a higher level of CD99 than XG at the cell surface. When cDNAs were expressed from the pcDNA3 vector (selection with G418), the difference was approximately 10-fold (XG13, 3 × 104 copies/cell; MIC22, 3.1 × 105 copies/cell), whereas with the pcDNA-hyg vectors (hygromycin selection), there was a 30-fold difference (h.XG66, 7.8 × 104 copies/cell; h.MIC11, 2.5 × 106 copies/cell). Such a difference was also seen with erythrocytes from Xg(a+) persons, in whom the average numbers of XG and CD99 molecules per cell are approximately 102 and 103, respectively (unpublished results). More molecules were expressed on transfectants when hygromycin rather than neomycin selection was used, and this increase was greater for the MIC2cDNA (XG copies/cell increase only 2-fold between XG13 and h.XG66; CD99 copies/cell increase 10-fold between MIC22 and h.MIC11). The higher level of CD99 expression, compared with XG, was apparently not caused by the toxicity of XG affecting cell growth because we found that the average doubling rate of clones that expressed XG (XG13 and h.XG66) or did not express XG (MIC22 and h.MIC11) was similar (5 doublings per 24 hours).
Relative levels of the XG and CD99 antigens at the surface of transfected RAG clones and somatic hybrid derivatives
| Clones . | Selection Marker . | XG Copies/cell (×10−3) . | CD99 Copies/cell (×10−3) . |
|---|---|---|---|
| Single transfectants | |||
| XG13 | neoR | 30 | 0 |
| h.XG66 | hygR | 78 | 0 |
| MIC22 | neoR | 0 | 310 |
| h.MIC11 | hygR | 0 | 2 500 |
| Double transfectants | |||
| XG13-h.MIC15 | neoR + hygR | 28 | 2 200 |
| XG13-h.MIC19 | neoR + hygR | 26 | 2 000 |
| MIC22-h.XG12 | neoR + hygR | 34 | 420 |
| MIC22-h.XG22 | neoR + hygR | 40 | 370 |
| Somatic hybrids | |||
| XG13 × h.MIC11 | neoR + hygR | 15 | 1 800 |
| MIC22 × h.XG66 | neoR + hygR | 49 | 705 |
| XG13 × hygR | neoR + hygR | 21 | 0 |
| MIC22 × hygR | neoR + hygR | 0 | 750 |
| neoR × h.XG66 | neoR + hygR | 45 | 0 |
| neoR × h.MIC11 | neoR + hygR | 0 | 1 900 |
| Clones . | Selection Marker . | XG Copies/cell (×10−3) . | CD99 Copies/cell (×10−3) . |
|---|---|---|---|
| Single transfectants | |||
| XG13 | neoR | 30 | 0 |
| h.XG66 | hygR | 78 | 0 |
| MIC22 | neoR | 0 | 310 |
| h.MIC11 | hygR | 0 | 2 500 |
| Double transfectants | |||
| XG13-h.MIC15 | neoR + hygR | 28 | 2 200 |
| XG13-h.MIC19 | neoR + hygR | 26 | 2 000 |
| MIC22-h.XG12 | neoR + hygR | 34 | 420 |
| MIC22-h.XG22 | neoR + hygR | 40 | 370 |
| Somatic hybrids | |||
| XG13 × h.MIC11 | neoR + hygR | 15 | 1 800 |
| MIC22 × h.XG66 | neoR + hygR | 49 | 705 |
| XG13 × hygR | neoR + hygR | 21 | 0 |
| MIC22 × hygR | neoR + hygR | 0 | 750 |
| neoR × h.XG66 | neoR + hygR | 45 | 0 |
| neoR × h.MIC11 | neoR + hygR | 0 | 1 900 |
The different clones were obtained as described in “Materials and methods” and “Results.” The somatic hybrids neoR × h.XG66 and neoR × h.MIC11 were generated by fusion of single transfectants expressing XG or CD99 and RAG cells transfected with the empty pcDNA3 vector (selected for the neoR marker). The somatic hybrids XG13 × hygRand MIC22 × hygR were generated by fusion of single transfectants expressing XG or CD99 and RAG cells transfected with the empty pcDNA-hyg vector (selected for the hygR marker). Results represent the mean of three measurements, each at a different cell passage.
The XG13 and MIC22 clones were next cotransfected with the plasmids pcDNA-hyg-MIC2 and pcDNA-hyg-XG, respectively, and new clones were obtained under G418 plus hygromycin selection. Again the same characteristics of expression were observed as with the single transfectants (see Table 1)—a higher cell surface level of CD99 than XG in every clone and a higher expression of the MIC2 gene when expressed from the pcDNA-hyg vector than with the pcDNA3 (neoR) vector. The number of XG or CD99 antigen copies/cell was equivalent between double and single transfectants for the same vector, pcDNA3 or pcDNA-hyg (Table 1), suggesting that the level of cell surface expression of these proteins is not interdependent because the coexpression of 1 protein (XG or CD99) has apparently no effect on the cell surface expression of the other. The same result was obtained with double transfectants of the clone h.XG66 with pcDNA3-MIC2or of the clone h.MIC11 with pcDNA3-XG (not shown).
To further confirm these data, we generated somatic cell hybrids by fusion of the individual clones and analyzed the expression of the 2 proteins (Table 1). Within experimental limits, there was no significant variation in the number of XG or CD99 molecules between single transfectants (eg, XG13, 3 × 104 XG copies/cell; h.MIC11, 2.5 × 106 CD99 copies/cell) and double hybrids (eg, XG13 × h.MIC11, 1.5 × 104XG copies/cell and 1.8 × 106 CD99 copies/cell). Single hybrids used as controls also showed equivalent amounts of surface proteins (XG13 × hygR, 2.1 × 104 XG copies/cell; neoR × h.MIC11, 1.9 × 106 CD99 copies/cell).
Transcripts and genomic analysis of transfectants
The XG and MIC2 cDNAs are under the control of the cytomegalovirus promoter in the expression plasmid pcDNA3. To explain the difference in the cell surface expression of XG and CD99, we estimated the transcription efficiency of each plasmid in the RAG transfectants by measuring the number of integrated plasmid copies and the approximative level of each transcript. Southern blot analysis of genomic DNA indicated that the expression plasmids were integrated at approximately 1 copy per cell in each transfectant (not shown), excluding gene dosage as the principal cause of the apparent difference in levels of cell surface expression of CD99 and XG. Next, RNA from the different clones and somatic hybrids was extracted and analyzed by Northern blot. A major transcript of the expected size (1 kb XGplus vector sequences) and a minor transcript (3 kb) were detected with the XG cDNA probe (Figure 1A), whereas a single band of 1 kb (expected size) was revealed with theMIC2 probe (Figure 1B). The minor 3-kb XG species might be a product generated by an alternative termination of transcription downstream of the specific sequences of the bovine growth hormone gene (BGH) present in the pcDNA3 and pcDNA-hyg vectors. Semiquantitative analysis by densitometry (not shown) indicate that the hybrids XG13 × hygR and XG13 × hMIC11 expressed fewer XG transcripts (Figure 1A) and fewer XG copies (Table 1) than the other clones. Similarly, the transfectants MIC22 and the hybrids MIC22 × hygR and MIC22 × h.XG66 expressed fewer MIC2 transcripts (Figure 1B) and fewer CD99 copies (Table 1). In all cases, therefore, the relative amounts ofXG and MIC2 mRNA correlated well with the number of XG and CD99 molecules found at the cell surface, as determined by flow cytometry.
Northern blot analysis of the XG and MIC2transcripts from transfected RAG clones and somatic hybrid derivatives.
Twenty micrograms total RNA from the different cell lines were hybridized under high-stringency conditions with either the XG(A) or MIC2 (B) cDNA probes. Identical amounts of RNA were deposited in each lane, as determined by densitometry analysis of ribosomal RNA under ultraviolet light illumination (not shown).
Northern blot analysis of the XG and MIC2transcripts from transfected RAG clones and somatic hybrid derivatives.
Twenty micrograms total RNA from the different cell lines were hybridized under high-stringency conditions with either the XG(A) or MIC2 (B) cDNA probes. Identical amounts of RNA were deposited in each lane, as determined by densitometry analysis of ribosomal RNA under ultraviolet light illumination (not shown).
The differences in cell surface expression of XG and CD99 seen with all transfectants and hybrids might have resulted from some difference in the transcription efficiency (plasmid integration-site dependency), the stability of the mRNA or posttranscriptional events, or a combination of these factors. This may also be the case for the 10-fold difference between XG and CD99 molecules at the surface of Xg(a+) erythrocytes, though the genes are under the control of different promoters in human erythroblasts. Altogether, these data did not reveal any obvious mechanism of coregulation of XG and CD99 at the protein level because there was no change in the steady state levels of XG and CD99 expression when they were expressed together. This is in accordance with a transcriptional mechanism of control of the coexpression of the 2 genes,3 but it does not exclude some form of translational or posttranslational regulation of cell surface expression of the 2 proteins.
Characterization of the XG and CD99 proteins in recombinant cells and in erythrocytes
Western blot analysis.
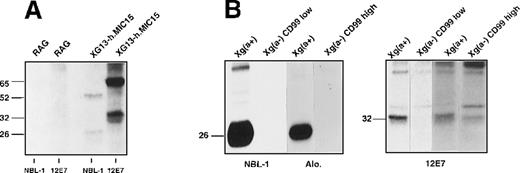
Solubilized proteins from the different clones and somatic cell hybrids were separated on SDS-PAGE and immunoblotted using the NBL-1 or 12E7 monoclonal antibodies. NBL-1 reacted with a single band corresponding to a protein of 26 kd in XG13, XG13-h.MIC15, and XG13-h.MIC19 extracts, whereas 12E7 recognized a major species of 32 kd in MIC22 and the double transfectants (Figure 2A). On the blots, which were loaded with the same amount of total membrane proteins, the staining intensity of the bands was rather similar (Figure 2A, left and middle panels). However, the exposure times were much longer for NBL-1 blots (5 minutes) than for 12E7 blots (3 seconds), suggesting that more CD99 than XG molecules were present in the transfectants. This result correlates well with those obtained by flow cytometry and Northern blot analysis (see above). The results from Figure 2A clearly indicate that the 26- and 32-kd components are related to XG and CD99, respectively, because no signal could be detected with lysates from nontransfected (murine) RAG cells that did not express either of these proteins. Indeed, examination of the evolutionary conservation of the 2 genes on zoo blots showed thatXG (unpublished results) and MIC214 are undetectable in nonprimate species. The point mutation in ourMIC2 cDNA (see above) did not affect the reactivity of recombinant CD99 with 12E7 because the Asp165Asn substitution occurs in the cytosolic tail of the protein and the epitope recognized by 12E7 has been mapped to the luminal domain of the protein.16 The 26-kd protein detected by NBL-1 is present in Xg(a+) RBCs (female, CD99 high-expressor) but not in Xg(a−) RBCs (male, CD99-high expressor), whereas the 32-kd species detected by 12E7 is present in Xg(a−) RBCs and Xg(a+) RBCs, which are CD99-high expressers (Figure 2B). However, this protein is undetectable in RBCs from an Xg(a−), CD99-low expressor (see Figure3B). These results strongly suggest that the presence or absence of the 26-kd component (XG) determines the basis of the Xg(a+)/Xg(a−) polymorphism and that the quantitative polymorphism of CD99 is coregulated with XG.
Western blot analysis of the XG and CD99 proteins from transfected RAG clones, somatic hybrid derivatives, and erythrocytes.
Proteins from cell lysates (RAG transfectants or somatic hybrids) or erythrocyte membranes were subjected to SDS-PAGE on a 12% (wt/vol) polyacrylamide gel, transferred to nitrocellulose sheets (Schleicher and Schuell, Dassel, Germany), probed with either NBL-1 or 12E7 monoclonal antibodies (as indicated below the blots), and revealed by chemiluminescence (ECL reagent, Amersham). (A) Western blot analysis, using indicated antibodies, of lysates from single or double transfectants (names above blots; details of transfectants given in Table 1) or nontransfected RAG cells. Right-hand panel represents Western blot analysis with monoclonal antibody 12E7 after treatment of the clone XG13-h.MIC15 (CD99-expressing, RAG transfectant) with 0, 0.1 (RDE1), or 0.25 (RDE2) units of neuraminidase (as described in “Materials and methods”). Exposure times to radiographic films were 5 minutes for NBL-1 blots, 3 and 6 seconds for 12E7 blots on middle and right hand panels, respectively. (B) Western blot analysis, using indicated antibodies, of membrane lysates from erythrocytes from Xg(a−) (male, CD99-high expressor) or Xg(a+) (female) (exposure time, 1 minute). Right-hand panel represents Western blot analysis with monoclonal antibody 12E7 after treatment of Xg(a+) (female) erythrocytes with 0, 0.1 (RDE1), or 0.25 (RDE2) units of neuraminidase (as described in “Materials and methods”) (exposure time, 10 minutes). (C) Western blot analysis, using indicated antibodies, of lysates from somatic hybrids of the single transfectants h.XG66 (XG-expressing) and MIC22 (CD99-expressing) (details of transfectants given in Table 1) (exposure time, 6 minutes). The size (kd) of the detected bands was determined using rainbow-colored protein molecular weight markers.
Western blot analysis of the XG and CD99 proteins from transfected RAG clones, somatic hybrid derivatives, and erythrocytes.
Proteins from cell lysates (RAG transfectants or somatic hybrids) or erythrocyte membranes were subjected to SDS-PAGE on a 12% (wt/vol) polyacrylamide gel, transferred to nitrocellulose sheets (Schleicher and Schuell, Dassel, Germany), probed with either NBL-1 or 12E7 monoclonal antibodies (as indicated below the blots), and revealed by chemiluminescence (ECL reagent, Amersham). (A) Western blot analysis, using indicated antibodies, of lysates from single or double transfectants (names above blots; details of transfectants given in Table 1) or nontransfected RAG cells. Right-hand panel represents Western blot analysis with monoclonal antibody 12E7 after treatment of the clone XG13-h.MIC15 (CD99-expressing, RAG transfectant) with 0, 0.1 (RDE1), or 0.25 (RDE2) units of neuraminidase (as described in “Materials and methods”). Exposure times to radiographic films were 5 minutes for NBL-1 blots, 3 and 6 seconds for 12E7 blots on middle and right hand panels, respectively. (B) Western blot analysis, using indicated antibodies, of membrane lysates from erythrocytes from Xg(a−) (male, CD99-high expressor) or Xg(a+) (female) (exposure time, 1 minute). Right-hand panel represents Western blot analysis with monoclonal antibody 12E7 after treatment of Xg(a+) (female) erythrocytes with 0, 0.1 (RDE1), or 0.25 (RDE2) units of neuraminidase (as described in “Materials and methods”) (exposure time, 10 minutes). (C) Western blot analysis, using indicated antibodies, of lysates from somatic hybrids of the single transfectants h.XG66 (XG-expressing) and MIC22 (CD99-expressing) (details of transfectants given in Table 1) (exposure time, 6 minutes). The size (kd) of the detected bands was determined using rainbow-colored protein molecular weight markers.
Immune precipitation of the Xga and CD99 antigens from transfected RAG cells and erythrocytes.
(A) Intact nontransfected RAG cells or the clone XG13-h.MIC15 were125I-labeled and incubated with NBL-1 or 12E7 antibodies, as noted below the autoradiograph. Solubilized complexes were precipitated, separated on SDS-PAGE, and autoradiographed. (B) Erythrocyte surface proteins from Xg(a+) (female), Xg(a−) (male, CD99-high expressor) or Xg(a−) (female, CD99-low expressor) donors were 125I-labeled. Cell membranes were prepared, incubated with NBL-1 (or the anti-Xga serum Alo.) or 12E7 antibodies, and solubilized complexes were treated as in A. The size (kd) of the detected bands was determined using rainbow-colored protein molecular weight markers (Amersham).
Immune precipitation of the Xga and CD99 antigens from transfected RAG cells and erythrocytes.
(A) Intact nontransfected RAG cells or the clone XG13-h.MIC15 were125I-labeled and incubated with NBL-1 or 12E7 antibodies, as noted below the autoradiograph. Solubilized complexes were precipitated, separated on SDS-PAGE, and autoradiographed. (B) Erythrocyte surface proteins from Xg(a+) (female), Xg(a−) (male, CD99-high expressor) or Xg(a−) (female, CD99-low expressor) donors were 125I-labeled. Cell membranes were prepared, incubated with NBL-1 (or the anti-Xga serum Alo.) or 12E7 antibodies, and solubilized complexes were treated as in A. The size (kd) of the detected bands was determined using rainbow-colored protein molecular weight markers (Amersham).
In previous immunoblot studies with human RBC, diffuse bands ranging from 24 to 29 kd, probably resulting from heterogeneity of glycosylation of the XG protein, were detected in membranes prepared from Xg(a+) erythrocytes using either human anti-Xgaantisera or rabbit polyclonal antibodies,3,18,29whereas a faint band at approximately 26 kd was revealed with NBL-1.3 The CD99 glycoprotein was detected as a single 32-kd band with the 12E7 antibody in such studies.18,27,28Examination of the immunoblot with 12E7 (Figure 2A, middle panel) revealed a minor 28-kd band in clones expressing high levels of CD99 (double transfectants XG13-h.MIC15 and XG13-h.MIC19) but not in clone MIC22 (except after long exposures). A 28-kd molecule has also been reported after neuraminidase (RDE) treatment of CD99 immunoprecipitates from thymocyte surface proteins30 or in lysates from untreated or RDE-treated RBCs.27 This protein was assumed to be the intracellular unsialylated precursor of the membrane-associated polypeptide. The double transfectant XG13-h.MIC15 was treated with neuraminidase (RDE) before solubilization of proteins and immunoblotting with 12E7 (Figure 2A, right panel; exposure time 6 seconds). With 0.1 U neuraminidase (RDE 1), the 28-kd band was present in greater amounts than it was in untreated cells, and a new band of approximately 26 kd (not identical to the XG band) became detectable. When the cells were treated with 0.25 U neuraminidase (RDE 2), a partial shift from the 28- to the 26-kd species was observed. Whether this 26-kd component is a more completely desialylated species or a degradation product was not investigated. Treatments with higher amounts of neuraminidase (0.5 U) or for a longer period of time (16 hours) did not allow a complete shift of the 32-kd component to molecules of lower molecular mass (not shown), perhaps because of the high number of CD99 molecules (2 × 106) at the cell surface and incomplete removal of sialic acid residues. It is also possible that there is heterogeneous terminal sialylation of CD99, with only a subset of molecules susceptible to cleavage by the specific neuraminidase used in these studies.
A Western blot of RDE-treated RBCs with 12E7 revealed 2 faster migrating bands of 30 and 28 kd, the latter accumulating in higher amounts with 0.25 U than with 0.1 U enzyme (Figure 2B, right panel). Similar results were obtained by Latron et al,17 except that the reported apparent molecular mass of the different species was 28 kd for untreated RBCs and 25 and 22 kd for RDE-treated RBCs. The 26-kd species obtained after neuraminidase treatment of the RAG clone XG13-h.MIC15 expressing CD99 (Figure 2A) probably is the unsialylated precursor equivalent of the intracellular 28-kd molecule in RBCs27 (Figure 2B). The difference in size might correspond to differences of glycosylation between molecules synthesized in human (RBC) and murine (RAG) cells. A 29-kd intracellular protein recognized by the 12E7 antibody and probably not related to the 32-kd CD99 has been found in human and mouse cells, including RAG cells.28The 29-kd protein was weakly detected in all RAG clones and nontransfected cells by longer exposures of the Western blots (not shown).
NBL-1 and 12E7 also recognized XG and CD99 proteins in somatic cell hybrids of RAG transfectants (Figure 2C). Interestingly, though only the 32-kd band was revealed by 12E7 in the clone MIC22, the 28-kd species was also detected in lysates from both MIC22 × hygR (Figure 2C, right panel) and MIC22 × h.XG66 hybrids (seen only on long exposure), where CD99 expression was higher (Table 1). These data strongly suggest that the 28-kd molecule is the intracellular, partially sialylated precursor of the 32-kd CD99 antigen.
Immunoprecipitation studies.
To determine whether the XG and CD99 proteins are associated in the plasma membrane and could be detected by coprecipitation analysis, the double transfectant XG13-h.MIC15 (expressing high levels of these proteins; Table 1) was 125I-labeled and incubated with NBL-1 or 12E7, and the immunoprecipitates recovered in the cell lysates were analyzed by SDS-PAGE (Figure 3A). Each antibody immunoprecipitated the corresponding protein (26 and 32 kd, respectively) but neither NBL-1 nor 12E7 coprecipitated CD99 or XG, respectively. Additional proteins of 52 and 65 kd were also precipitated with NBL-1 and 12E7, respectively. These higher molecular weight proteins could correspond to SDS-resistant homodimers of XG and CD99 because there is some evidence that CD99 exists as a dimer at the erythrocyte surface.17 Immuno-cross-blot analysis failed to detect CD99 or XG proteins in the immunoprecipitates from NBL-1 or 12E7, respectively (not shown).
The absence of coprecipitation of the XG and CD99 molecules from the double transfectant cell line could result from differences of protein conformation or from another unidentified factor, in nonerythroid cells, that would prevent a close association between these molecules. Accordingly, immunoprecipitation studies were carried out using NBL-1 (or the human anti-Xga serum Alo.) or 12E7 and membrane proteins prepared from Xg(a+) and Xg(a−) erythrocytes125I-labeled to a similar extent (3 × 108 cpm/ packed RBCs). The results (Figure 3B) show that XG and CD99 were clearly precipitated by their respective antibodies from the Xg(a+) RBCs (female), but again no coprecipitation of the 2 proteins occurred. Neither XG nor CD99, however, could be precipitated from the Xg(a−) RBCs (female, CD99-low expressor), and only CD99 was precipitated from the Xg(a−) RBCs (male, CD99-high expressor), thus confirming the absence of detectable XG and the quantitative polymorphism of CD99 in Xg(a−) RBCs. Higher molecular weight bands on the gels are probably nonspecific because they were detected in all precipitates. The XG protein from Xg(a+) RBCs was also immunoprecipitated with a rabbit polyclonal antibody (raised in our laboratory) directed against the same peptide as for NBL-1,3 but again CD99 was not present in the precipitate (not shown).
Immunoblotting Xga immunoprecipitates (obtained using NBL-1 or serum Alo.) obtained from nonlabeled RBCs with 12E7 did not reveal any bands corresponding to CD99 (32 kd), as found by Petty and Tippett.18 Moreover, immunoblotting 12E7 immunoprecipitates with NBL-1 did not reveal any band corresponding to XG (26 kd) (data not shown). Using biotin-labeled RBCs, Petty and Tippett18found that 1 human anti-Xga serum could coprecipitate a 32-kd component (tentatively assigned to, but not formerly identified as, CD99), from Xg(a+) RBCs. Our studies indicate, however, that although the NBL-1 antibody could efficiently immunoprecipitate the XG protein, there was no detectable 32-kd component in the precipitate (Figure 3B).
We draw the following conclusions from these experiments: (1) XG and CD99 can be expressed independently in cell transfectants; (2) there is no physical association between XG and CD99 detectable by coprecipitation of the 2 proteins from Xg(a+) membrane preparations or from transfected RAG cells, using either NBL-1 (and the anti-Xga serum Alo.) or 12E7; and (3) the molecular basis of the Xg(a−) phenotype is most likely caused by the lack of the XG protein on Xg(a−) erythrocytes either from a CD99-low expressor (Figure 3B) or a CD99-high expressor (Figures 2B, 3B).
Tissue specificity of XG expression
To elucidate the mechanisms of tissue-specific regulation ofXG expression, we constructed somatic cell hybrids between B-LCLs from Xg(a+) (CT) and Xg(a−) (CL) male donors (CD99-high expressers) and murine fibroblastic RAG cells. We then examined, by flow cytometry, whether a reactivation of XG protein expression at the cell surface would occur. The CD99 protein, but not the XG protein, is present at the surface of B-LCLs of persons of both phenotypes (Table2). RAG cells express neither XG nor CD99 proteins (not shown). Our results indicate that the CD99 protein, but not the XG protein, is present on the surface of somatic hybrids RAG × CL or RAG × CT (Table 2). We then fused these hybrids with MEL cells to test whether an erythroid environment would restore a cell surface expression of XG, but again no Xgaantigen appeared at the plasma membrane. Finally, somatic hybrids between B-LCLs and the XG13 clone were constructed to investigate the influence of a nonerythroid context on XG protein expression. No effect was detected because the numbers of XG molecules/cell (2 × 104 and 1.6 × 104) were similar to those observed on the different somatic hybrids of RAG transfectants, including the XG13 clone as 1 fusion partner (see Table 1).
Relative levels of the XG and CD99 antigens at the surface of B-LCLs and somatic hybrid derivatives
| Cell Lines . | Xga Status of Donor . | XG Copies/cell (×10−3) . | CD99 Copies/cell (×10−3) . |
|---|---|---|---|
| B-LCLs | |||
| CL | Xg (a−) | 0 | 125 |
| CT | Xg (a+) | 0 | 164 |
| Somatic hybrids | |||
| RAG × CL | Xg (a−) | 0 | 77 |
| RAG × CT | Xg (a+) | 0 | 117 |
| MEL × (RAG × CL) | Xg (a−) | 0 | 70 |
| MEL × (RAG × CT) | Xg (a+) | 0 | 92 |
| XG13 × CL | Xg (a−) | 20 | 124 |
| XG13 × CT | Xg (a+) | 16 | 177 |
| Cell Lines . | Xga Status of Donor . | XG Copies/cell (×10−3) . | CD99 Copies/cell (×10−3) . |
|---|---|---|---|
| B-LCLs | |||
| CL | Xg (a−) | 0 | 125 |
| CT | Xg (a+) | 0 | 164 |
| Somatic hybrids | |||
| RAG × CL | Xg (a−) | 0 | 77 |
| RAG × CT | Xg (a+) | 0 | 117 |
| MEL × (RAG × CL) | Xg (a−) | 0 | 70 |
| MEL × (RAG × CT) | Xg (a+) | 0 | 92 |
| XG13 × CL | Xg (a−) | 20 | 124 |
| XG13 × CT | Xg (a+) | 16 | 177 |
The generation of the successive somatic cell hybrids was described in “Materials and methods.” Results represent the mean of three measurements, each at a different cell passage.
To determine whether the tissue-specific regulation of XGexpression occurs at the transcriptional level, we analyzed (by RT-PCR) the XG transcripts from the B-LCLs and the different somatic hybrids, using primers bordering the coding sequence of the XGcDNA (Figure 4). The transcripts from each cell line generated a specific band of the expected size (551 bp) and of normal sequence compared to the transcripts from erythroid cells (not shown). Thus, the absence of XG molecules at the surface of these nonerythroid cells might be explained by a low transcription rate of the gene or by some posttranscriptional event altering the stability or translation of the messenger RNA, glycosylation of the protein, or its translocation to the cell surface. We reasoned that as a hybrid partner, MEL cells should have brought the necessary erythroid factors for cell surface expression of XG, but this may not be true because the hybrid cells may have eliminated some MEL chromosomes (not determined) encoding these factors. An alternative possibility is that human B-LCLs do not possess the program (sequence of events including the putative erythroid factors) needed for cell surface expression of XG; therefore, the supply of erythroid factor(s) by the MEL cells is ineffective. Indeed, only adult hemoglobin was produced in fetal nonerythroid (fibroblasts or lymphoblasts) × MEL cell hybrids.31
RT-PCR analysis of the XG transcripts from B-LCLs and somatic hybrid derivatives.
Total-cell RNA from the indicated cell lines was reverse transcribed and PCR-amplified to obtain a fragment of 551 bp corresponding to the coding sequence of the XG cDNA. The PCR products were electrophoresed on a 1.2% (wt/vol) agarose gel, transferred to a nylon membrane (Hybond N+; Amersham), and hybridized with a32P-labeled internal oligonucleotide probe. C, control PCR without template cDNA. The size (bp) of the PCR products was checked with EcoRI–HindIII-digested λ DNA markers.
RT-PCR analysis of the XG transcripts from B-LCLs and somatic hybrid derivatives.
Total-cell RNA from the indicated cell lines was reverse transcribed and PCR-amplified to obtain a fragment of 551 bp corresponding to the coding sequence of the XG cDNA. The PCR products were electrophoresed on a 1.2% (wt/vol) agarose gel, transferred to a nylon membrane (Hybond N+; Amersham), and hybridized with a32P-labeled internal oligonucleotide probe. C, control PCR without template cDNA. The size (bp) of the PCR products was checked with EcoRI–HindIII-digested λ DNA markers.
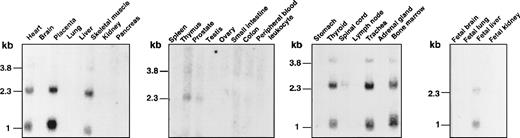
Finally, we studied the tissue distribution of XG gene expression by Northern blot analysis (Figure5). Two major transcripts of 2.3 and 1.0 kb and 1 minor species of 3.8 kb were detected in erythroid tissues (thymus, bone marrow, and fetal liver) and several nonerythroid tissues (heart, placenta, skeletal muscle, prostate, thyroid, spinal cord, and trachea). No XG mRNA was revealed in peripheral blood leukocytes, whereas we found a specific band by RT-PCR analysis on transcripts from B-LCLs (Figure 4), indicating that the XG gene is expressed at a low level in these cells. XG mRNA was also found by Northern analysis in human skin fibroblasts15 and by RT-PCR analysis in other adult (lung, kidney, testis) and fetal (spleen, adrenal glands, brain, pancreas, small intestine) tissues.3 Only a few lymphoid cell lines gave a weak positive signal.3MIC2 transcripts of 1.2 kb and 2 minor species of 3.6 and 6.2 kb were detected in all the tissues tested for XG mRNA expression (not shown), as expected, because the CD99 antigen is present on most human cells.32 The 1.2-kb transcript was also found in several T-lymphocytic and monocytic cell lines and in peripheral blood lymphocytes.27
Tissue distribution of the XG transcripts.
Human multiple-tissue Northern blots from Clontech were hybridized under high-stringency conditions with the 32P-labeledXG cDNA probe and autoradiographed.
Tissue distribution of the XG transcripts.
Human multiple-tissue Northern blots from Clontech were hybridized under high-stringency conditions with the 32P-labeledXG cDNA probe and autoradiographed.
Conclusion
The expression data obtained with the transfected RAG cells and somatic hybrid derivatives are consistent with a transcriptional regulation of XG and MIC2 gene expression because no influence of either protein on the surface production of the other was found. Cloning, sequencing, and functional tests of promoter and upstream regulatory regions of the XG gene, contained within the 10-kb stretch downstream of the MIC2 gene, may help to elucidate the molecular basis of the coexpression of these 2 genes and by extension of the XG polymorphism and the quantitative polymorphism of MIC2. Indeed, according to the model proposed by Goodfellow et al,12 the putative XGR polymorphic regulatory locus might be located between the 3′ end ofMIC2 and the 5′ end of XG. However, as long as formal proof of this model is not given, this regulatory element can be anywhere else within the 100-kb region, from the 5′ end ofMIC2 to the pseudoautosomal boundary. We could not confirm a physical association between the XG and CD99 proteins at the cell surface of RBCs or transfected RAG cells. If such an association exists, it may be of low affinity and may be disrupted during analysis. Future investigation along this line will be carried out by the 2-hybrid system.33 In addition, further analysis on the tissue distribution of the XG protein and precise identification of which cells express this protein may help to clarify its biologic role.
Acknowledgments
We thank Anne-Marie D'Ambrosio (INTS, Paris) for the production of Epstein-Barr virus-positive B-lymphoblastoid cell lines, Peter Goodfellow (University of Cambridge, UK) for the gift of mAb 12E7, and Dr PierreYves Le Pennec (CNRGS, Paris) for the gift of serum Alo. We also thank Nicole Souleyreau (Institut Pasteur, Paris, France) for the transmission of the cell fusion method and Patrick Lambin and Martine Debbia (INTS, Paris) for immunochemistry analysis. We thank Patricia Hermand and Pascal Bailly (INTS, Paris) for helpful discussions.
C.F. was supported in part by Ortho-Clinical Diagnostics.
Reprints:Jean-Pierre Cartron, Inserm U76, Institut National de la Transfusion Sanguine, 6 rue Alexandre Cabanel, 75015, Paris, France; e-mail: cartron@infobiogen.fr.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal