We recently described a new low-frequency platelet alloantigen on the human platelet glycoprotein (GP) Ib-IX complex, termed Iya, which was implicated in a severe case of neonatal alloimmune thrombocytopenia. Immunoprecipitation studies with trypsin-treated platelets indicated that the Iyaalloantigenic determinants are formed by the membrane-associated remnant moiety of GP Ib (GP Ibr) together with GP Ibβ and GP IX. To elucidate the molecular basis underlying the Iya alloantigen, we amplifiedGPIbr, GPIbβ, andGPIX genes by polymerase chain reaction (PCR). Nucleotide-sequence analysis of these 3 genes showed a G to A transition at position 141 on GPIbβ gene in a subject positive for Iya. This transition resulted in a Gly15Glu dimorphism on the N-terminal domain ofGPIbβ. This finding was confirmed by genotyping analysis of 6 Iya-positive subjects by restriction fragment length polymorphism (RFLP) studies using NarI endonuclease. In 300 randomly selected healthy blood donors, one Iya-positive individual was found. Phenotypes determined by monoclonal antibody-specific immobilization of platelet antigens assay and genotypes determined by RFLP were identical in this population. Analysis of Iya-positive platelets showed that the point mutation affected neither the degree of surface expression nor the function of the GP Ib-GP Ibβ-IX complex on the platelet surface. Transient expression of the GP Ib-IX complex in CHO cells using wild-type GP Ibβ (Gly15) or mutant GP Ibβ (Glu15) allowed us to demonstrate that this single amino acid substitution is sufficient to induce Iya epitope(s).
Human platelet glycoproteins (GP) are carriers of alloantigenic determinants that can elicit an alloimmune response leading to platelet destruction, such as occurs in neonatal alloimmune thrombocytopenia (NAIT), posttransfusion purpura, and platelet transfusion refractoriness.1 Four GP subunits (GP Ia, GP Ibα, GP IIb, and GP IIIa) on the platelet surface are known to be polymorphic and immunogenic in humans.2,3 Two allelic variants have been found for GP Ia and GP Ibα, bearing human platelet alloantigens (HPA) 5a/5b (Brb/Bra) and 2a/2b (Kob/Koa), respectively.4,5 GP IIb exists in 3 allelic variants carrying HPA-3a/3b (Baka/Bakb) and HPA-9bW (Maxa).6,7 GP IIIa is the most polymorphic molecule. Ten allelic variants encoding GP IIIa have been found in the human gene pool so far, 9 of which are immunogenic as carriers of HPA-1a (PlA1) and HPA-4a (Yukb or Pena), HPA-1b (PlA2), HPA-4b (Yukaor Penb), HPA-6bW (Caa), HPA-7bW (Moa), HPA-8bW (Sra), HPA-10bW (Laa), HPA-11bW (Groa), and Oea alloantigenic determinants.8-15 Most of these alloantigens result from point mutations in wild-type DNA that produce single amino acid substitutions and lead to the expression of the offending alloantigenic determinants. The Oea variant is an exception because it results from a deletion of a codon of the mutated GP IIIa (PlA2) isoform.15
We recently described a new low-frequency platelet alloantigen on the GP Ib-IX complex, termed Iya, that was responsible for a case of severe NAIT.16 GP Ib-IX complex is a receptor for both von Willebrand factor and thrombin and plays an essential role in adhesion of platelets to the subendothelium. GP Ib is a heterodimer consisting of a large α chain (molecular weight [MW], 143 kilodaltons [kd]) and a smaller disulfide-linked β chain (MW, 27 kd). In the platelet membrane, GP Ib forms a noncovalent complex with GP IX (MW, 22 kd).17 GP Ib is also weakly associated with GP V (MW, 82 kd) in a noncovalent manner.18 All 4 GP are members of the leucine-rich GP family containing a variable number of leucine repeats.19-22 GP Ibα, GP Ibβ, GP IX, and GP V are known to be derived from distinct genes, with the entire open reading frame of the mature protein located within a single exon.23-26 The GPIbα gene is located on chromosome 17,27 whereas the GPIbβ gene is on chromosome 22.24 The GPIX and GPV genes are on distinct sites of the long arm of chromosome 3, on band q21 and band q29, respectively.28
We here report the first molecular variant of GP Ibβ responsible for the formation of a clinically important alloantibody in NAIT. Because this new alloantigen was found in 1 of 300 healthy blood donors tested, it may be involved in other cases of alloimmune thrombocytopenia.
Materials and methods
Monoclonal antibodies (MAB)
MAB Gi10 and Gi27 against the remnant moiety of GP Ibα and GP Ibβ, respectively, were raised and characterized in our laboratory.12,29 MAB SZ2 and AN51 specific for GP Ibα30,31 were purchased from Dianova and Dako (both Hamburg, Germany). MAB FMC25 against GP IX32 was provided by H. Zola, Adelaide, Australia.
Phenotyping
Immunoprecipitation analysis
Washed platelets from ACD–anticoagulated blood were labeled with biotin hydrazide (Pierce, Munich, Germany) as described by Fabris et al, with minor modifications.34 Briefly, 109platelets in 900 μL of phosphate-buffered saline (PBS)-EDTA (3.72 g/L of EDTA, 1 μmol/L of leupeptin, 1 mmol/L of phenylmethylsulfonyl fluoride (PMSF), 4 mmol/L of N-ethylmaleimide in PBS; pH 7.4) were exposed to 100 μL of 12 mmol/L sodium metaperiodate at 4°C for 10 minutes; 0.6 mol/L of glycerol was then added. After being washed twice with 500 μL of PBS-EDTA, the platelets were incubated with 3 mmol/L of biotin hydrazide at room temperature for 2 hours. Labeled platelets were washed 4 times and lysed in 1 mL solubilization buffer (25 mmol/L of Tris, 10 mmol/L of EDTA, 100 mmol/L of sodium chloride (NaCl) containing 1% Triton X-100, 2 mmol/L of PMSF, 1 mmol/L of leupeptin, and 2 mmol/L ofN-ethylmaleimide) for 30 minutes at 4°C.
Aliquots of 109 labeled platelets were digested with 100 μL of trypsin (1 mg/mL; Sigma, Deisenhofen, Germany) for 5 minutes at 37°C. Digestion was stopped by adding 200 μL of soybean trypsin inhibitor (1 mg/mL; Sigma). Trypsin-treated platelets were washed twice and then solubilized as described above.
After centrifugation (30 minutes at 16 000g at 4°C), immunoprecipitation was performed as described previously.12 Immunoprecipitates were separated by using 7.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose membrane, and visualized with use of streptavidin-peroxidase and chemiluminescence substrate. Colored protein MW markers (Rainbow; Amersham, Braunschweig, Germany) were used as the standard.
Immunoblotting
Aliquots of 109 washed platelets were lysed in 1 mL of solubilization buffer. After centrifugation (30 minutes at 16 000gat 4°C), proteins were separated with SDS-PAGE and transferred to nitrocellulose membrane. Membrane strips were blocked with 1.5% bovine serum albumin (BSA) in PBS and then incubated with an MAB dilution (20 μg/mL) or serum for 30 minutes at room temperature. The strips were washed twice with Tris buffer (pH 7.4) containing 0.05% Tween 20 and then incubated with peroxidase-conjugated rabbit antimouse or antihuman antibodies (1:200 000 dilution; Dianova). After washing, the recognized protein was visualized by using the enhanced chemiluminescence substrate system (Amersham).
Determination of GP Ib-IX binding sites
Aliquots of 2 × 107 washed platelets were incubated with increasing amounts of MAB Gi10 (50-100 μg) at 37°C for 30 minutes. The sensitized platelets were washed 3 times with 0.2% BSA in isotonic saline before resuspension in 80 μL of isotonic saline. Bound MAB were eluted with 40 μL of 100 mmol/L of NaCl (pH 2.2; adjusted with acetic acid) containing 1.5% BSA for 10 minutes at room temperature. After centrifugation, eluates were neutralized with a predetermined volume (about 3.3 μL) of 2.5 mmol/L of Tris buffer. A sandwich enzyme-linked immunoassay (ELISA) using purified normal mouse IgG (m-IgG) as the standard was used to quantify the number of binding sites, as reported previously.12 In brief, microtiter wells were coated overnight with 100 μL of goat antimouse IgG F(ab′)2 (1:1000 dilution in coating buffer; Dianova) at 4°C. After being washed 3 times with 200 μL of 1% BSA in PBS (PBS-BSA), wells were blocked with 200 μL of PBS-BSA for 30 minutes at 4°C and then incubated with either 100 μL of eluates or 100 μL of various dilutions of m-IgG (2500-50 pg) for 1 hour at 37°C. Afterward, the wells were washed 3 times, and 100 μL of alkaline phosphatase-labeled goat antimouse IgG Fc (1:1000 dilution, Dianova) was added. After 1 hour of incubation at 37°C, the wells were washed 5 times. Finally, 100 μL of p-nitrophenylphosphate substrate solution (Sigma) was added and the plate was incubated at room temperature for 30 minutes. The color reaction was stopped by adding 50 μL of 3 mol/L of sodium hydroxide and was read at 405 nm in a Titertek photometer (Helsinki, Finland). All samples were assayed in duplicate.
Platelet function studies
Platelet-rich plasma (PRP) was obtained by centrifugation (200gfor 15 minutes) of ACD–anticoagulated blood collected from Iya-positive and Iya-negative individuals. The platelet count was adjusted to 3 × 105 per μL by dilution with autologous plasma. To aliquots of 180 μL of PRP, 20 μL of various dilutions of ristocetin (2.5, 5, 10, and 15 μg/mL) were added and the change in optical density was monitored by using an aggregometer, with continuous stirring, at 37°C.
To evaluate the functional effect of anti-Iya antibodies, aliquots of 180 μL of PRP derived from Iya-positive individuals were mixed with either 20 μL of isotonic saline, MAB SZ2 (20 μg/mL), or 20 μL eluates of heat-inactivated serum (normal human serum [NHS] or anti-Iya) and incubated at 37°C for 30 minutes in an atmosphere supplemented with 5% carbon dioxide (CO2). After stimulation with ristocetin (15 μg/mL), platelet aggregation was recorded as described above.
Isolation and amplification of genomic DNA
Genomic DNA was isolated from 10 mL of EDTA–anticoagulated blood from Iya-phenotyped donors as described previously.12 Primers used to amplify theGPIbαr, GPIbβ, andGPIX genes (Table 1) were constructed according to published DNA sequences.20,23 25The coding regions of the GPIbα gene encompassing nucleotides 826 to 1965 were amplified in 2 overlapping fragments (bases 826 to 1518 and bases 1192 to 1965) by using primer pairs GP Ibα 1-GP Ibα 2 and GP Ibα 3-GP Ibα 4, respectively. Amplification was performed in a total volume of 50 μL containing 10 μL of genomic DNA (400-600 ng), 0.3 μmol/L of each primer, 200 μmol/L of each dNTP, 1.5 mmol/L of magnesium chloride (MgCl2), and 1.5 U of TaqGOLD polymerase on a GeneAmp 9600 DNA thermal cycler (Perkin Elmer, Weiterstadt, Germany). After heating at 97°C for 5 minutes, polymerase chain reaction (PCR) was performed under the following conditions. For amplification of the first region (bases 826-1518), denaturation was done for 75 seconds at 94°C, annealing for 120 seconds at 52°C, and extension for 180 seconds at 72°C. For the second region (bases 1192-1965), denaturation was done for 60 seconds at 95°C, annealing for 90 seconds at 55°C, and extension for 120 seconds at 72°C. Both amplifications proceeded for 36 cycles. In the final cycle, all samples were kept at 72°C for 10 minutes and then chilled to 4°C.
Sequences and positions of primers used in the polymerase chain reaction amplification of GPIbr, GPIbβ, and GPIX genes
| Primer . | Position . | Sequence (5′ → 3′) . |
|---|---|---|
| GP Ibα 1 | 826-843 | GTGCAGTGTGACAATTCA |
| GP Ibα 2 | 1518-1501 | GTCATTTCTGGAGCTCTC |
| GP Ibα 3 | 1192-1220 | CCAAGCCCGACCACCTCAGAGCCC |
| GP Ibα 4 | 1965-1942 | AGCCCACAGGCTCTTCTCTCAAGG |
| GP Ibβ 1 | 47-72 | CTGAGCTTACTGCTCCTGCTGCTGGC |
| GP Ibβ 2 | 939-920 | GGGTTTATTCAGCACCAGAG |
| GP Ibβ 3 | 378-359 | ACGCAACGCAGGTCGCGGTA |
| GP IX 1 | 271-288 | ACCAAGGACTGCCCCAGC |
| GP IX 2 | 750-733 | ATCCAGGGCCTCTGTGGT |
| Primer . | Position . | Sequence (5′ → 3′) . |
|---|---|---|
| GP Ibα 1 | 826-843 | GTGCAGTGTGACAATTCA |
| GP Ibα 2 | 1518-1501 | GTCATTTCTGGAGCTCTC |
| GP Ibα 3 | 1192-1220 | CCAAGCCCGACCACCTCAGAGCCC |
| GP Ibα 4 | 1965-1942 | AGCCCACAGGCTCTTCTCTCAAGG |
| GP Ibβ 1 | 47-72 | CTGAGCTTACTGCTCCTGCTGCTGGC |
| GP Ibβ 2 | 939-920 | GGGTTTATTCAGCACCAGAG |
| GP Ibβ 3 | 378-359 | ACGCAACGCAGGTCGCGGTA |
| GP IX 1 | 271-288 | ACCAAGGACTGCCCCAGC |
| GP IX 2 | 750-733 | ATCCAGGGCCTCTGTGGT |
The entire coding region of GPIbβ (nucleotides 47-939) was amplified by using 0.5 μmol/L each of GP Ibβ 1 and GP Ibβ 2 primer, 200 μmol/L each of dNTP, 1.5 mmol/L of MgCl2, 10% dimethyl sulfoxide (DMSO), 1.5 U μL of Taq GOLD polymerase, and 5 μL of 10 × PCR buffer. A cycle consisted of denaturation at 94°C for 75 seconds, annealing at 48°C for 90 seconds, and primer extension at 72°C for 120 seconds and was repeated 35 times.
The coding region of GPIX (nucleotides 271-750) was amplified by using 0.5 μmol/L each of GP IX 1 and GP IX 2 primer, 200 μmol/L each of dNTP, 1.2 mmol/L of MgCl2, 0.01% gelatin, 1.5 U ofTaq GOLD polymerase, and 5 μL 10 × PCR buffer under the same PCR conditions used for the GPIbβ gene except that the annealing was done at 52°C.
Subcloning and sequencing
All PCR products were purified on 1.5% SeaKem agarose gel (FMC, Hessisch Oldendorf, Germany) by using Geneclean (Dianova). Purified DNA was flushed with Klenow DNA polymerase (Biolabs, Schwalbach, Germany) for blunt-end ligation into the EcoRV site of the pGEM5 plasmid and then transformed into DH5α high-efficiency competentEscherichia coli (Gibco BRL, Eggenstein, Germany). Before ligation, the GP Ibβ PCR product was shortened by digestion withSmaI endonuclease (Biolabs). Recombinant colonies were selected by blue-white screening on indicator plates. For single-strand nucleotide sequencing, plasmid DNA from 8 positive clones of each region was amplified with use of biotinylated forward primer 5′-CGC CAG GGT TTT CCC AGT CAC GAC G-3′ and nonbiotinylated reverse primer 5′-GCT TCC GGC TCG TAT GTT GTG TGG-3′ or vice versa. Biotinylated single-strand DNA was isolated by magnetic beads (Dynal, Norway), sequenced with SP6 and T7 primers using a fluorescence DNA-sequencing kit (Perkin Elmer), and then analyzed on ABI Prism 373 DNA Sequencers (Applied Biosystems, Weiterstadt, Germany).
Genotyping by restriction fragment length polymorphism (RFLP)
Genomic DNA was amplified by using primer pair GP Ibβ 1-GP Ibβ 3 as described above, except that “hot start” PCR was saved by application of Taq GOLD polymerase (1.5 U, Perkin Elmer). Aliquots of 7 μL of PCR products were subjected to RFLP using 2 U ofNarI endonuclease (Biolabs) and then analyzed on 3.0% NuSieve agarose gel (Gibco).
Construction of allele-specific GP Ibβ expression vectors
A full-length complementary DNA (cDNA) encoding wild-type GP Ibβ in pDX plasmid was removed with EcoRI (Biolabs) and ligated into the mammalian pcDNA3.1 Zeo expression vector (Invitrogen, Leek, Holland). Specific mutation G→A at position 141 was induced in the wild-type GP Ibβ construct by site-directed mutagenesis with use of a QuickChange Mutagenesis Kit (Strategene, Heidelberg, Germany). For PCR amplification, single nucleotide-mismatched sense primer 5′-GGG ACG CTC GTG GAC TGC GAG CGC CGC GGG CTG ACT TGG-3′ and antisense primer 5′-CCA AGT CAG CCC GCG GCG CTC GCA GTC CAC GAG CGT CCC-3′ corresponding to base 122 to 160 of GP Ibβ cDNA were constructed. After 12 cycles of amplification (denaturation for 30 seconds at 95°C, annealing for 60 seconds at 55°C, and extension for 12 minutes at 68°C) in the presence of 10% DMSO, PCR product was digested with DpnI and transformed into DH5α high-efficiency competent E coli. Plasmid DNA from positive clones was amplified by PCR using GP Ibβ 1 and GP Ibβ 3 primers, and subjected to RFLP analysis withNarI as described above. Purified GP Ibβ allele-specific constructs used for subsequent transfection were validated by nucleotide-sequence analysis.
Cell culture and transfection
CHO cells stably expressing GP Ibα and GP IX35 were transiently transfected by liposome-mediated delivery of plasmid DNA with use of a commercially available kit (Lipofectamine, Gibco BRL, Grand Island, NY). Cells were grown to approximately 70% confluence on 50-mm2 tissue culture dishes. Twelve microliters of liposome suspension and 2 μg of plasmid DNA (either pcDNA 3.1 Zeo vector containing the wild-type or mutated cDNA of GP Ibβ, or the vector alone) were separately mixed in 200 μL of serum-free medium. The 2 suspensions were then combined, mixed gently, and allowed to form DNA-liposome complexes for 30 minutes at room temperature. The mixture was diluted in 1.6 mL of serum-free medium and added to the cells, which had been washed twice with the same medium. The cells were exposed to the mixture for 5 hours under standard culture conditions (37°C in 5% CO2), after which 2 mL of medium containing 10% fetal-calf serum was added. Twenty-four hours later, the medium was changed.
Flow cytometry
Forty-eight hours after transfection, cells were detached from the dishes with 0.54 mmol/L of EDTA and washed twice with PBS. Cells were fixed in 1% paraformaldehyde and washed twice. A total of 400 000 cells were counted by hemacytometer, resuspended in 200 μL of PBS, and incubated with 50 μL of serum (either anti-Iya serum or NHS) for 30 minutes at 37°C. The cells were washed twice with PBS and resuspended in 200 μL of PBS. They were incubated with 40 μL of a 1:40-diluted fluorescein isothiocyanate-conjugated rabbit antihuman antibody (Dianova) for 30 minutes at 37°C. To remove unbound antibody, cells were washed twice. A total of 10 000 cells from each transfection were analyzed with an Ortho Cytoron Absolute flow cytometer (Ortho Diagnostic Systems, Raritan, NJ).
Results
Immunochemical characterization
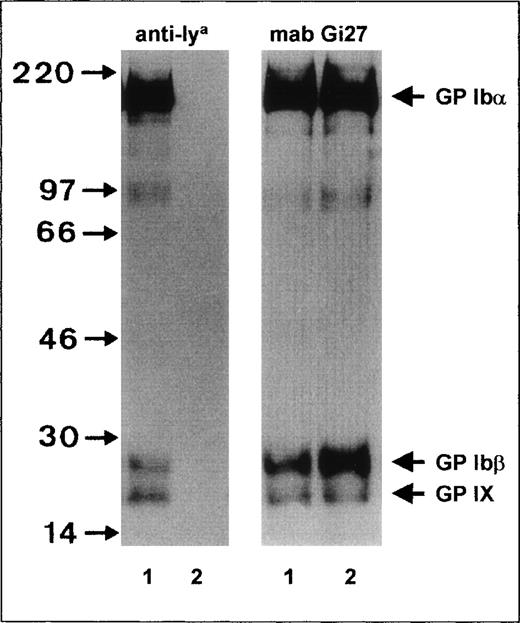
To characterize the Iya antigen, immunoprecipitation studies with surface-labeled biotinylated platelets were performed (Figure 1). When anti-Iyaimmunoprecipitate was electrophoresed under reducing conditions, 3 bands—GPIbα, GPIbβ, and GP IX—could be detected, with apparent MW of 145 kd, 27 kd, and 22 kd, respectively (Figure 1, left panel, lane 1). In the control experiments, these bands could not be precipitated from Iya-negative platelets (Figure 1, lane 2).
Immunoprecipitation analysis of the GPIb-Ibβ-IX complex of Iya-phenotyped platelets.
Platelets from Iya-positive (lane 1) and Iya-negative (lane 2) individuals were surface labeled with biotin, lysed, and immunoprecipitated with anti-Iya (left panel) and monoclonal antibody (MAB) Gi27 (right panel). Immunoprecipitates were analyzed with 7.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under reducing conditions, transferred to nitrocellulose membrane, and visualized by using a streptavidin-horseradish peroxidase–chemiluminescence substrate system.
Immunoprecipitation analysis of the GPIb-Ibβ-IX complex of Iya-phenotyped platelets.
Platelets from Iya-positive (lane 1) and Iya-negative (lane 2) individuals were surface labeled with biotin, lysed, and immunoprecipitated with anti-Iya (left panel) and monoclonal antibody (MAB) Gi27 (right panel). Immunoprecipitates were analyzed with 7.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under reducing conditions, transferred to nitrocellulose membrane, and visualized by using a streptavidin-horseradish peroxidase–chemiluminescence substrate system.
To analyze the surface expression of the GP Ib-IX complex in Iya-positive platelets, we compared the levels of GP Ib-IX precipitates from Iya-positive and Iya-negative platelets by immunoprecipitation (Figure 1, right panel). When MAB Gi27 directed against GP Ibβ was used, similar amounts of GP Ibα, GP Ibβ, and GP IX were precipitated from both platelet phenotypes. In addition, in both platelet types, GP Ibα, GP Ibβ, and GP IX subunits migrated with similar mobility. Similar results were obtained with MAB SZ2 directed against the GP Ibα subunit and with MAB FMC25 directed against the GP IX subunit (data not shown). These observations indicate that normal amounts of GP Ib-IX complex are expressed on the surface of Iya-positive platelets and that the Iya antigen is not associated with an MW polymorphism.
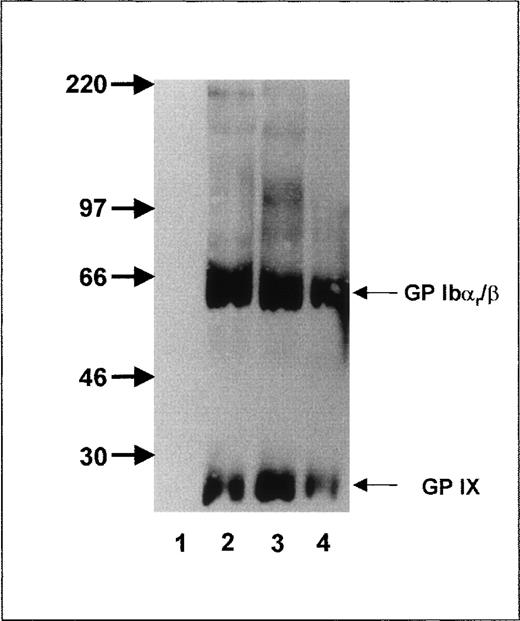
To further localize the epitope recognized by anti-Iyaantibodies, we took advantage of the fact that trypsin cuts off the amino-terminal part of GP Ibα, leaving GP Ibαr as the remnant moiety, which is associated with GP Ibβ and GP IX on the platelet surface.36 After trypsin treatment, anti-Iya antibodies (Figure 2, lane 4) still precipitated the GP Ibαr-Ibβ complex (MW, 65 kd) and GP IX subunit (MW 22, kd) under nonreducing conditions, as did MAB Gi10 and Gi27 (Figure 2, lanes 2 and 3). In contrast, MAB SZ2 (Figure 2, lane 1), directed against the glycocalicin moiety, did not precipitate any platelet proteins.
Immunoprecipitation analysis of the GPIb-Ibβ-IX complex of Iya-phenotyped platelets after trypsin treatment.
Aliquots of 109 biotinylated platelets from an Iya-positive individual were treated with trypsin, washed, lysed, and immunoprecipitated with MAB SZ2 (lane 1), MAB Gi10 (lane 2), MAB Gi27 (lane 3), and anti-Iya antibodies (lane 4). Immunoprecipitates were analyzed with 7.5% SDS-PAGE under nonreducing conditions.
Immunoprecipitation analysis of the GPIb-Ibβ-IX complex of Iya-phenotyped platelets after trypsin treatment.
Aliquots of 109 biotinylated platelets from an Iya-positive individual were treated with trypsin, washed, lysed, and immunoprecipitated with MAB SZ2 (lane 1), MAB Gi10 (lane 2), MAB Gi27 (lane 3), and anti-Iya antibodies (lane 4). Immunoprecipitates were analyzed with 7.5% SDS-PAGE under nonreducing conditions.
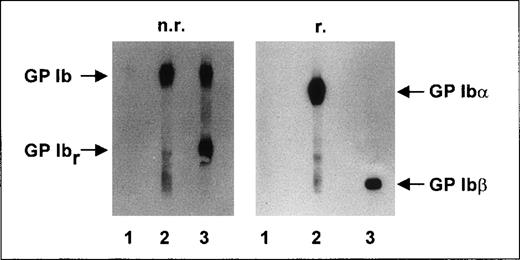
In immunoblotting analysis, no reactivity of anti-Iyaantibodies with any platelet proteins was detectable (Figure3, lane 1) under either nonreducing or reducing conditions. In the control experiments, MAB SZ2 (Figure 3, lane 2) reacted with GP Ib (both α and β subunits) under nonreducing conditions. Under nonreducing conditions, MAB Gi27 (Figure3, lane 3) showed binding to GP Ib and to its proteolytic fragment, GP Ibr. Under reducing conditions, MAB SZ2 recognized the GP Ibα subunit and MAB Gi27 recognized the GP Ibβ subunit.
Immunoblotting studies.
Aliquots of 109 washed platelets from an Iya-positive individual were lysed, and proteins were separated by using 7.5% SDS-PAGE under nonreducing (n.r.) and reducing (r.) conditions. After proteins were transferred to a nitrocellulose membrane, membrane strips were incubated with anti-Iyaantiserum (lane 1), MAB SZ2 (lane 2), and MAB Gi27 (lane 3). Antibody binding was detected by using corresponding peroxidase-conjugated antibodies and chemiluminescence substrate.
Immunoblotting studies.
Aliquots of 109 washed platelets from an Iya-positive individual were lysed, and proteins were separated by using 7.5% SDS-PAGE under nonreducing (n.r.) and reducing (r.) conditions. After proteins were transferred to a nitrocellulose membrane, membrane strips were incubated with anti-Iyaantiserum (lane 1), MAB SZ2 (lane 2), and MAB Gi27 (lane 3). Antibody binding was detected by using corresponding peroxidase-conjugated antibodies and chemiluminescence substrate.
Amplification and analysis of GPIb, GPIbβ, andGPIX genes
Because anti-Iya antibodies bind to trypsin-treated platelets, we predicted that the region formed by amino acid residues 450 to 610 (nucleotides 1440-1920) of GPIbα, together withGPIbβ and GPIX, would carry the Iyaepitope(s). To analyze this region, we amplified the GPIbαgene in 2 overlapping fragments (nucleotides 826-1518 and 1192-1965) and the entire coding regions of the GPIbβ (nucleotides 47-939) and GPIX genes (nucleotides 271-750). PCR products ofGPIbα, GPIbβ, and GPIX genes from an Iya-positive individual migrated with the same electrophoretic mobility as PCR products derived from an Iya-negative individual (data not shown). All PCR products were subcloned, and 6 independent clones from each fragment were subjected to nucleotide-sequence analysis.
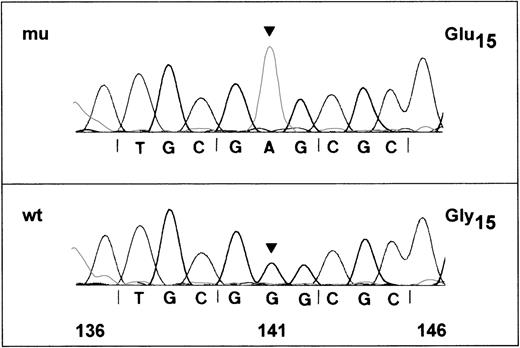
Nucleotide-sequence analysis of GPIbα and GPIXfragments from an Iya-positive individual and an Iya-negative individual did not show any differences in nucleotides (data not shown). However, nucleotide-sequence analysis of the 517-base-pair (bp) fragment of GPIbβ encoding nucleotides 47-563 from an Iya-positive individual revealed a single G to A substitution at base 141 in 3 of 6 subclones examined (Figure 4). In contrast, all clones from an Iya-negative individual encoded a G at this position (data not shown). These results are consistent with the idea that Iya-positive individuals are usually heterozygous for this low-frequency antigen. The G to A substitution changes a GGG codon for glycine to GAG, which encodes for glutamic acid at amino acid 15 of the mature GP Ibβ.
DNA-sequence analysis of amplified GPIbβ gene of an Iya-positive individual.
Polymerase chain reaction (PCR) products of the GPIbβ gene encompassing nucleotides 47-563 were subcloned in the plasmid vector pGEM-5Zf and sequenced on both strands. Nucleotide-sequences of 2 positive clones are shown. (A) The wild-type G in position 141 is changed to an A (arrow), predicting a glycine to glutamic acid (GGG→GAG) polymorphism at position 15 of the mature glycoprotein. (B) The sequence is identical to the published wild-type sequence for GPIbβ.20
DNA-sequence analysis of amplified GPIbβ gene of an Iya-positive individual.
Polymerase chain reaction (PCR) products of the GPIbβ gene encompassing nucleotides 47-563 were subcloned in the plasmid vector pGEM-5Zf and sequenced on both strands. Nucleotide-sequences of 2 positive clones are shown. (A) The wild-type G in position 141 is changed to an A (arrow), predicting a glycine to glutamic acid (GGG→GAG) polymorphism at position 15 of the mature glycoprotein. (B) The sequence is identical to the published wild-type sequence for GPIbβ.20
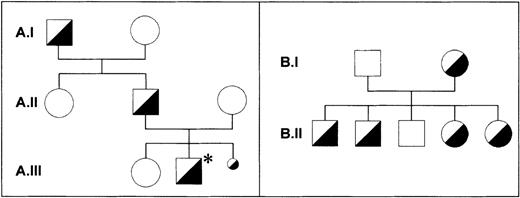
Correlation of the G141A dimorphism with the Iya phenotype
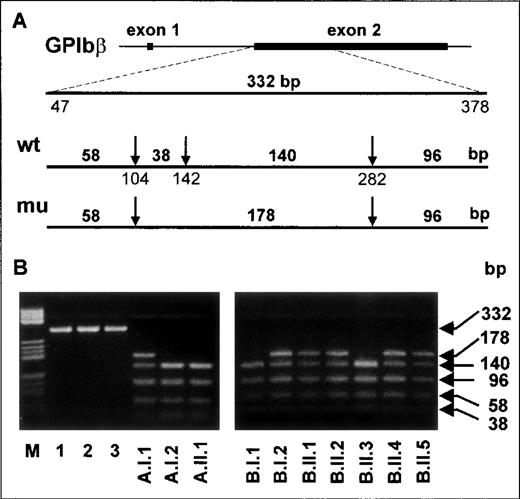
Along with the Iya-positive subjects from the index family,16 one Iya-positive individual (Kr) was identified among 300 German blood donors phenotyped for the Iya antigen with the MAIPA assay. The pedigrees of both families are shown in Figure 5. No case of NAIT was observed in the Kr family. To determine whether the G to A substitution at position 141 of the GPIbβ gene segregates with the Iya phenotype, we established genomic DNA typing with a PCR-RFLP technique. The G to A substitution abolishes a cleavage site for the restriction endonuclease NarI, which cleaves at 5′-GG↓CGCC-3′ but not at 5′-AGCGCC-3′ sequences (Figure 6A). We used this technique to genotype 3 members of the index family (A.I.1, A.I.2, and A.II.1 in Figure 5) and all members of the Kr family (Figure5B and Figure 6B). After amplification of genomic DNA by using primer pair GP Ibβ 1 and GP Ibβ 3, the 332-bp PCR product was digested with NarI. All Iya-negative individuals had 140-bp, 96-bp, 58-bp, and 38-bp restriction fragments. All Iya-heterozygous individuals could be differentiated from the Iya-negative subjects by the presence of an additional 178-bp fragment. The results of the genotyping of 300 unrelated individuals correlated with the phenotyping results.
Pedigrees of the index family and the Kr family.
In the Iy (index) family (A), the family member with the index case of neonatal alloimmune thrombocytopenia (NAIT) is marked with an asterisk (A.III.2). A third pregnancy was interrupted because of massive intraventricular bleeding in the fetus (A.III.3). All individuals were phenotyped with use of the monoclonal antibody-specific immobilization of platelet antigens assay. Material for genotyping was available only from individuals A.I.1, A.I.2, and A.II.1. One member of the Kr family (B) was found in assessing 300 healthy blood donors. All individuals were phenotyped and genotyped. No case of NAIT was observed in the Kr family.
Pedigrees of the index family and the Kr family.
In the Iy (index) family (A), the family member with the index case of neonatal alloimmune thrombocytopenia (NAIT) is marked with an asterisk (A.III.2). A third pregnancy was interrupted because of massive intraventricular bleeding in the fetus (A.III.3). All individuals were phenotyped with use of the monoclonal antibody-specific immobilization of platelet antigens assay. Material for genotyping was available only from individuals A.I.1, A.I.2, and A.II.1. One member of the Kr family (B) was found in assessing 300 healthy blood donors. All individuals were phenotyped and genotyped. No case of NAIT was observed in the Kr family.
PCR-restriction fragment length polymorphism analysis of Iya-phenotyped individuals with use ofNarI.
(A) A 332-base-pair product encompassing nucleotides 47-378 of theGPIbβ gene was obtained from genomic DNA by using primers GPIbβ 1 and GP Ibβ 3. The arrows indicate the cleavage sites recognized by the restriction endonuclease NarI. The length of the expected digested fragments from Iya-positive and Iya-negative alleles is also shown. (B) Analysis ofNarI-digested PCR products from genomic DNA of the members of the Iy family (left panel) and the Kr family (right panel). Lanes are inscribed according to the pedigrees (Figure 4). Undigested products from samples obtained from 3 individuals are shown in lanes 1 to 3 (left panel). In lane M, pBr 322 HaeIII DNA fragments are shown as standards.
PCR-restriction fragment length polymorphism analysis of Iya-phenotyped individuals with use ofNarI.
(A) A 332-base-pair product encompassing nucleotides 47-378 of theGPIbβ gene was obtained from genomic DNA by using primers GPIbβ 1 and GP Ibβ 3. The arrows indicate the cleavage sites recognized by the restriction endonuclease NarI. The length of the expected digested fragments from Iya-positive and Iya-negative alleles is also shown. (B) Analysis ofNarI-digested PCR products from genomic DNA of the members of the Iy family (left panel) and the Kr family (right panel). Lanes are inscribed according to the pedigrees (Figure 4). Undigested products from samples obtained from 3 individuals are shown in lanes 1 to 3 (left panel). In lane M, pBr 322 HaeIII DNA fragments are shown as standards.
Expression of the wild-type and mutated GP Ib-Ibβ-IX complex in CHO cells
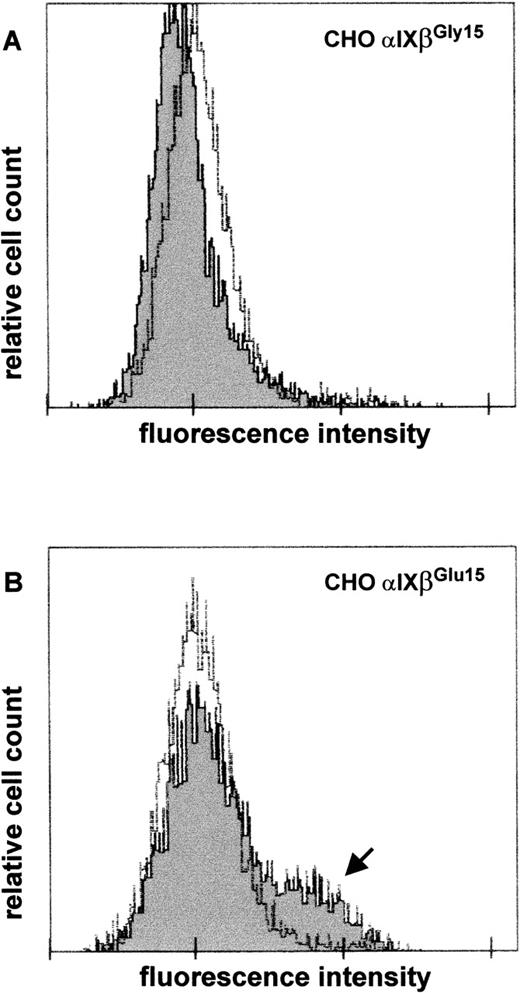
To demonstrate that the G to A substitution at nucleotide 141 of GPIbβ cDNA is sufficient to induce formation of the epitope recognized by anti-Iya serum, we performed transient transfection of CHO cells expressing GP Ibα and GP IX with eucaryotic expression vectors carrying wild-type or mutated GP Ibβ cDNA. Two days after transfection, surface expression of Iyaepitope(s) was examined by flow cytometry after staining with anti-Iya serum. Sham-transfected CHO cells solely expressing GP Ibα and GP IX failed to bind anti-Iyaantibodies (data not shown). CHO cells expressing the wild-type complex also did not bind anti-Iya antibodies (Figure7A, dark curve), whereas those expressing the mutated GP Ib-IX complex showed antibody binding (Figure 7B, arrow). NHS was used as a negative control (Figure 7A and 7B, bright curves). These results were reproduced in independent transfection experiments in 2 different laboratories.
Detection of expression of Iya epitope on recombinant GP Ib-Ibβ-IX complexes by flow cytometry.
CHO cells stably expressing GP Ibα and GP IX were transiently transfected with eucaryotic expression vectors carrying wild-type (A) or mutated (B) complementary DNA for GPIbβ. Forty-eight hours after transfection, expression of the Iya epitope on the cell surface was determined by using anti-Iya antibodies (dark). Normal human serum was used as a negative control (bright). The arrow indicates the subpopulation of cells showing antibody binding.
Detection of expression of Iya epitope on recombinant GP Ib-Ibβ-IX complexes by flow cytometry.
CHO cells stably expressing GP Ibα and GP IX were transiently transfected with eucaryotic expression vectors carrying wild-type (A) or mutated (B) complementary DNA for GPIbβ. Forty-eight hours after transfection, expression of the Iya epitope on the cell surface was determined by using anti-Iya antibodies (dark). Normal human serum was used as a negative control (bright). The arrow indicates the subpopulation of cells showing antibody binding.
Effect of G to A mutation on the expression and function of GP Ib-IX complex
To determine whether the G to A mutation influences the efficiency of expression of the GP Ib-IX complex on the platelet surface, binding isotherms were generated by using MAB Gi10 in a quantitative sandwich ELISA.
In accordance with the findings from our immunoprecipitation analysis, platelets from Iya-positive individuals bound amounts of MAB Gi10 (22 753 ± 200 molecules per platelet; n = 3) similar to the amounts bound by platelets from Iya-negative individuals (23 000 ± 300 molecules per platelet; n = 3).
To determine the possible effect of the point mutation on the ability of the GP Ib-IX complex to bind von Willebrand factor, platelets from an Iya-positive individual were compared with those from 3 Iya-negative individuals in standard platelet aggregation assays. The ristocetin-induced agglutination of Iya-positive platelets was indistinguishable from that of Iya-negative platelets (data not shown). All these results suggest that neither the expression nor the function of the GP Ib-IX complex is affected by the Gly15Glu mutation.
When ristocetin-induced agglutination was performed in the presence of purified anti-Iya antibodies, no inhibition was observed in platelets from either Iya-positive or Iya-negative individuals. In the control experiments, MAB SZ2 (20 μg/mL) completely inhibited the ristocetin-induced agglutination (data not shown).
Discussion
We report the characterization of the new platelet-specific alloantigen, Iya, that was responsible for a severe case of NAIT. Our immunochemical studies demonstrated that anti-Iyaprecipitated GP Ibαr, GP Ibβ, and GP IX from trypsin-treated platelets. Because we were unable to detect reactivity of the Iya antibody with either serum or affinity-purified antibody in immunoblot analysis, we predict that the alloantigenic determinants of Iya are dependent on protein conformation sensitive to denaturation by SDS.
To elucidate the molecular basis underlying the Iyaantigen, we amplified GPIbαr,GPIbβ, and GPIX genes from genomic DNA derived from Iya-positive and Iya-negative individuals. Nucleotide-sequence analysis of the GPIbα, GPIbβ, and GPIX genes showed a single G to A transition at nucleotide 141 in the GPIbβ gene, changing glycine to glutamic acid at residue 15 of the mature GP Ibβ protein. RFLP analysis using the restriction endonuclease NarI, which is capable of discriminating between these 2 alleles, demonstrated that this nucleotide substitution correlated with the serologic phenotypes of 6 Iya-positive individuals from 2 independent families and 300 Iya-negative unrelated blood donors.
The Gly15Glu dimorphism of the GP Ibβ protein represents the only difference between Iya-positive and Iya-negative individuals. Flow cytometry analysis that used CHO cells expressing the wild-type (Gly15) or mutated (Glu15) GP Ibα-Ibβ-IX complex on their surface showed that this single amino acid substitution is directly responsible for formation of the Iya alloantigenic determinant(s).
The actual Iya-antibody binding sites are probably complex-specific, formed by GP Ibβ and other GP Ib subunits. Which GP Ib subunit—GP Ibα, GP IX, or both—is required for the alloantigenic formation remains unclear. However, expression of recombinant mutated GP Ibβ in CHO cells confirmed that this single amino acid substitution is sufficient to induce formation of the epitope(s) recognized by anti-Iya serum.
Bernard-Soulier syndrome (BSS) is an extremely rare autosomal recessive bleeding disorder in which patients have a low platelet count and large platelets that are unable to adhere to subendothelium. The functional defect lies in the inability of the platelets to bind von Willebrand factor.37 In patients with classic BSS, the level of GP Ib-IX complex is greatly reduced because of abnormalities (mutations and deletions) in the GPIb-IX genes that lead to a biosynthetic defect affecting expression, processing, or synthesis of the complex. In contrast, patients with variant BSS have normal amounts of GP Ib-IX complex, but the complex is dysfunctional. Recently, BSS variants due to point mutations in a leucine-rich domain of GP Ibα and GP IX have been identified.38-40 So far, only 2 GP Ibβ mutations have been reported to be associated with BSS. The first involved a giant-platelet syndrome caused by point mutations that led to amino acid substitutions Tyr to Cys at residue 88 and Ala to Pro at residue 108 of the mature glycoprotein.41 The patient was heterozygous for these mutations. It was suspected that the Tyr to Cys transition affected the disulfide linkage between GP Ibα and GP Ibβ. The second mutation was in a patient with velocardiofacial syndrome who had a point mutation at position −133 (C133G) and deletion of the other allele.42
Studies of Iya-positive platelets showed that the Gly15Glu point mutation of the GP Ibβ gene does not impede expression or function of the GP Ib-IX complex. Wright et al39 observed variable band shifts within the GP Ibβ coding region in a few patients with BSS and healthy blood donors by means of single-stranded conformation polymorphism. However, this sequence variation did not show any linkage to the BSS phenotype. Our RFLP analysis of these patients with BSS showed that this polymorphism is not related to the Gly15Glu polymorphism (data not shown).
So far, all the alloantigenic epitopes responsible for the observed cases of NAIT, posttransfusional purpura, and refractoriness to platelet transfusion have been found to be on GP Ia, GP Ibα, GP IIb, or GP IIIa. The elucidation of the Iyaalloantigen represents the first characterization of an immunologically important polymorphism of GP Ibβ.
Acknowledgments
We thank Dr M. Vicariot and Dr Y. Giovangrandi of the Centre Hospitalier et Universitaire de Brest, France, who observed the index case and provided anti-Iya serum; Dr J. Clemetson, Theodor Kocher Institute, University of Bern, Switzerland, for helpful discussions; Dr E. G. D. Tuddenham, Imperial College Medical School, London, United Kingdom, for providing genomic DNA from patients with Bernard-Soulier syndrome; and Ms Dagmar Westrup for technical support in performing nucleotide-sequence analysis at the Institute for Transfusion Medicine and Immunohematology, German Red Cross Blood Donor Service, Frankfurt/ Main, Germany.
Supported by grants from the German Research Foundation (DFG Sa480/2-1). This work is part of a PhD thesis (UJHS).
Reprints:S. Santoso, Institute for Clinical Immunology and Transfusion Medicine, Langhansstr 7, D-35392 Giessen, Germany; e-mail:sentot.santoso@immunologie.med.uni-giessen.de.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal