Abstract
We prospectively analyzed p15 and p16 promoter methylation patterns using methylation-specific polymerase chain reaction (PCR) in patients with adult and childhood acute leukemias and studied the association of methylation patterns with chromosomal abnormalities and prognostic variables. In nearly all French-American-British leukemia subtypes, we found p15methylation in bone marrow or peripheral blood cells from 58% (46/79) of patients with acute myeloid leukemia (AML), acute lymphoblastic leukemia (ALL), or acute biphenotypic leukemia (ABL). An identical alteration was detected in blood plasma from 11 of 12 of these patients (92%). We also demonstrated for the first time concomitant p16and p15 methylation in 22% (8/37) of adults with AML or ALL, exclusively in those with M2, M4, or L2 subtypes. According to cytogenetic data from 35 patients with ALL, AML, or ABL, 82% (14/17) of those with unmethylated p15 alleles had normal karyotypes or hyperdiploidies associated with a favorable prognosis. Conversely, 44% (8/18) of patients with p15 methylation had chromosomal translocations, inversions, or deletions, suggesting an interplay of these abnormalities with p15 methylation. As a prognostic marker for disease monitoring, p15 methylation appears to be more widely applicable than BCR-ABL, AF4-MLL, andAML1-ETO transcripts, which were detectable in only 8% (4/48) of patients by reverse transcriptase-PCR. Thirty-nine of 43 blood samples (91%) sequentially collected from 12 patients with AML, ALL, or ABL showed p15 methylation status in excellent concordance with morphologic disease stage. Early detection of p15methylation at apparent remission or its acquisition during follow-up may prove valuable for predicting relapse. Overall survival of patients with p15 methylation was notably shortened among 38 adults with AML and 12 adults with ALL. Aberrant p15 methylation may have important prognostic implications for clinical monitoring and risk assessment.
Loss of cell-cycle regulation through changes in the cyclin D-retinoblastoma (Rb) pathway, is common in human neoplasia,1 although inactivation of the Rb gene infrequently occurs in patients with hematologic malignant disease.2 The p16 and p15 genes, which encode cyclin-dependent kinase inhibitors, have been recognized as tumor suppressor genes in solid tumors and hematologic neoplasms.3-8 Deletions of p16, p15, or both are commonly found in acute lymphoblastic leukemia (ALL) but infrequently observed in acute myeloid leukemia (AML).3-5,9-11 Intragenic mutations in p16 andp15 are only rarely detected in ALL or AML.12,13The p16/p15 deletion correlates with a high risk of relapse or death in patients with ALL.14 15
Methylation of the p15/p16 promoter is associated with loss of transcription in neoplasia.3,7,8,16 Frequent p15methylation was demonstrated in leukemia cell lines and primary acute leukemias with use of Southern blot analysis.3 However,p16 methylation has only rarely been observed in primary acute leukemias.10,11,17 In acute leukemias, p16 is predominantly inactivated by homozygous deletion and p15 is mainly inactivated by methylation. Differential use of p16 or p15 protein in a selective manner has been implicated, and this is supported by the fact that p16 andp15 genes are regulated by means of distinct pathways.17p16 expression is regulated partly by pRb protein, while Rb expression is in turn negatively controlled by p53 protein.18-20 There is evidence that p16 inactivation is closely associated with aberrant p53 expression, suggesting a collaborating role for p16 in apoptosis.21,22 Conversely, p15 expression appears to be independent of pRb but is regulated by the extracellular growth inhibitors interferon-α and transforming growth factor-β (TGF-β).23,24 Thus, loss of p15 function in acute leukemias might lead to an escape from normal growth control by TGF-β in bone marrow.3
Multiple chromosomal translocations that aberrantly activate or deregulate tyrosine kinase or transcription factor genes are remarkably specific for hematopoietic cells arrested at definitive differentiation stages in acute leukemias.25-27 The oncogenic fusion genes may work synergistically with altered tumor suppressor genes in the multistep process of leukemogenesis. Although the fusion transcripts may serve as diagnostic and prognostic markers for monitoring minimal residual disease (MRD), early relapse, and response to therapy, they are detectable at only low rates in specific morphologic subtypes of ALL and AML.25-27 In contrast, aberrant p15methylation is frequently found in adult and childhood ALL and AML.3,10,11,17 Thus, p15 methylation has potential value as a molecular prognostic marker in acute leukemias. For this application, methylation-specific polymerase chain reaction (MSP) is sensitive and specific in detecting epigenetic changes within CpG dinucleotides that are critical sites for gene silencing.28
In this prospective investigation, we used MSP to analyze p15and p16 promoter methylation in patients with adult and childhood acute leukemias of multiple French-American-British (FAB) classification subtypes and studied the association of p15methylation with several prognostic variables, including chromosomal abnormalities, fusion transcripts, and other known risk factors. Also, we examined cell-free p15 methylation in blood plasma to investigate its possible biologic implications in relation to the behavior of leukemic blasts. To assess whether p15 methylation could be applied as a molecular prognostic marker for detecting MRD or early relapse, we sequentially monitored p15 methylation status in peripheral blood from patients with acute leukemias at diagnosis and during follow-up. The prognostic relevance of p15 methylation was also evaluated by correlating p15 methylation status with morphologic disease stage and overall survival. The MSP analysis forp15 may create exciting possibilities for risk assessment, early detection of MRD and relapse, and disease monitoring.
Patients and methods
Patients and controls
We recruited 52 adults (age range, 17-80 years) with AML (n = 38), ALL (n = 13, including 1 of T-lineage), or acute biphenotypic leukemia (ABL) (n = 1) and 27 children (age range, 2-15 years) with AML (n = 4), ALL (n = 21, including 1 of T-lineage), or ABL (n = 2). Patients were classified into FAB subtypes in accordance with morphologic and cytochemical findings and immunophenotypes. Overall, there were 42 patients with AML (M1, 4 patients; M2, 13; M3, 3; M4, 10; M5, 6; M6, 3; and M7, 3), 34 patients with ALL (L2 of T-lineage, 2 patients; L1 of B lineage, 18; and L2 of B lineage, 14), and 3 patients with ABL. Bone marrow aspirates (n = 54) and peripheral blood samples (n = 25) were collected from all 79 patients at diagnosis of acute leukemia. Treatment regimens for all patients were standardized.
Forty-five peripheral blood samples were collected sequentially from 13 patients at diagnosis and during follow-up. Bone marrow aspirates and trephine biopsy specimens from these patients were examined morphologically to determine disease status. Control peripheral blood samples (n = 10) and bone marrow aspirates (n = 2) were obtained from 12 healthy donors. The plasmacytoma-derived HS-Sultan cell line (American Type Culture Collection [ATCC] CRL-1484) served as a methylated control. The positive control was the chronic myeloid leukemia-derived K562 cell line (ATCC CCL-243), which expressedMBCR-ABL transcripts fused at major breakpoints.
Cytogenetic studies
Cytogenetic studies using Giemsa banding were done with unstimulated cultures of bone marrow aspirates obtained at diagnosis. After overnight incubation, cultures were synchronized with fluorodeoxyuridine, and direct chromosome preparations were made. The cytogenetic results were described by using the International System for Cytogenetic Nomenclature.29
DNA extraction
Peripheral blood was centrifuged at 3000g and plasma was collected from a tube containing EDTA. DNA was extracted from 400 μL of plasma by using the QIAamp blood kit (Qiagen, Hilden, Germany). Buffy coats were isolated from peripheral blood or bone marrow, and genomic DNA was extracted by using standard sodium dodecyl sulfate-proteinase K treatment and phenol-chloroform-isoamylalcohol extraction.
MSP and Southern blot analysis
Bisulfite treatment of DNA converts unmethylated cytosine residues into uracil,28,30 but methylated cytosine residues remain unmodified. Therefore, methylated and unmethylated DNA sequences can be distinguished by using sequence-specific polymerase chain reaction (PCR) primers. We conducted bisulfite conversion by using the CpGenome DNA modification kit (Intergen, New York, NY). Bisulfite-treated buffy coat DNA (1 μg) or extracted plasma DNA was amplified by using p15MF/p15MR and p16MF/p16MR primer sets specific for the methylatedp15 and p16 sequences, respectively (Table1).28 All bisulfite-treated DNA samples were also amplified by using p15UF/p15UR and p16UF/p16UR primer sets specific for the unmethylated p15 and p16sequences, respectively. Any unconverted DNA was amplified by using p15WF/p15WR and p16WF/p16WR primer sets specific for the wild-typep15 and p16 sequences, respectively.
Primer sequences for methylation-specific polymerase chain reaction and reverse transcriptase-polymerase chain reaction and probe sequences for Southern blot analysis
| Primer . | Sequence . |
|---|---|
| p15MF | 5′ GCG TTC GTA TTT TGC GGT T 3′ |
| p15MR | 5′ CGT ACA ATA ACC GAA CGA CCG A 3′ |
| p15UF | 5′ TGT GAT GTG TTT GTA TTT TGT GGT T 3′ |
| p15UR | 5′ CCA TAC AAT AAC CAA ACA ACC AA 3′ |
| p15WF | 5′ CGC ACC CTG CGG CCA GA 3′ |
| p15WR | 5′ AGT GGC CGA GCG GCC GG 3′ |
| p16MF | 5′ TTA TTA GAG GGT GGG GCG GAT CGC 3′ |
| p16MR | 5′ GAC CCC GAA CCG CGA CCG TAA 3′ |
| p16UF | 5′ TTA TTA GAG GGT GGG GTG GAT TGT 3′ |
| p16UR | 5′ CAA CCC CAA ACC ACA ACC ATA A 3′ |
| p16WF | 5′ CAG AGG GTG GGG CGG ACC GC 3′ |
| p16WR | 5′ CGG GCC GCG GCC GTG G 3′ |
| mBCRABLF | 5′ ACC ATC GTG GGC GTC CGC AA 3′ |
| MBCRABLF | 5′ GAA GTG TTT CAG AAG CTC CTC C 3′ |
| BCRABLR | 5′ TTG GTT TGG GCT TCA CAC CAT TCC 3′ |
| AF4MLLF | 5′ GAC TAT CGA CAG CAG CAG ACC TT 3′ |
| AF4MLLR | 5′ TCT TCT TCT TTG TGG GTT TG 3′ |
| AML1ETOF | 5′ AGC CAT GAA GAA CCA GG 3′ |
| AML1ETOR | 5′ AGG CTG TAG GAG AAT GG 3′ |
| p15MP | 5′ TAG GC/TG TTT TTT TTT AGA AGT AAT TTA GG 3′ |
| p16MP | 5′ GAG TAG TAT GGA GTT TTC GGT TGA TTG GTT G 3′ |
| BCRABLP | 5′ TGT GAT TAT AGC CTA AGA CCC GGA GCT TTT C 3′ |
| AF4MLLP | 5′ ACT CAG GGT GAT AGC TGT TTC G 3′ |
| AML1ETOP | 5′ GTC TTC ACA TCC ACA GGT GAG TCT 3′ |
| Primer . | Sequence . |
|---|---|
| p15MF | 5′ GCG TTC GTA TTT TGC GGT T 3′ |
| p15MR | 5′ CGT ACA ATA ACC GAA CGA CCG A 3′ |
| p15UF | 5′ TGT GAT GTG TTT GTA TTT TGT GGT T 3′ |
| p15UR | 5′ CCA TAC AAT AAC CAA ACA ACC AA 3′ |
| p15WF | 5′ CGC ACC CTG CGG CCA GA 3′ |
| p15WR | 5′ AGT GGC CGA GCG GCC GG 3′ |
| p16MF | 5′ TTA TTA GAG GGT GGG GCG GAT CGC 3′ |
| p16MR | 5′ GAC CCC GAA CCG CGA CCG TAA 3′ |
| p16UF | 5′ TTA TTA GAG GGT GGG GTG GAT TGT 3′ |
| p16UR | 5′ CAA CCC CAA ACC ACA ACC ATA A 3′ |
| p16WF | 5′ CAG AGG GTG GGG CGG ACC GC 3′ |
| p16WR | 5′ CGG GCC GCG GCC GTG G 3′ |
| mBCRABLF | 5′ ACC ATC GTG GGC GTC CGC AA 3′ |
| MBCRABLF | 5′ GAA GTG TTT CAG AAG CTC CTC C 3′ |
| BCRABLR | 5′ TTG GTT TGG GCT TCA CAC CAT TCC 3′ |
| AF4MLLF | 5′ GAC TAT CGA CAG CAG CAG ACC TT 3′ |
| AF4MLLR | 5′ TCT TCT TCT TTG TGG GTT TG 3′ |
| AML1ETOF | 5′ AGC CAT GAA GAA CCA GG 3′ |
| AML1ETOR | 5′ AGG CTG TAG GAG AAT GG 3′ |
| p15MP | 5′ TAG GC/TG TTT TTT TTT AGA AGT AAT TTA GG 3′ |
| p16MP | 5′ GAG TAG TAT GGA GTT TTC GGT TGA TTG GTT G 3′ |
| BCRABLP | 5′ TGT GAT TAT AGC CTA AGA CCC GGA GCT TTT C 3′ |
| AF4MLLP | 5′ ACT CAG GGT GAT AGC TGT TTC G 3′ |
| AML1ETOP | 5′ GTC TTC ACA TCC ACA GGT GAG TCT 3′ |
PCR was conducted by using the GeneAmp DNA amplification kit and AmpliTaq Gold polymerase (Perkin Elmer, Foster City, CA). Thirty-five cycles were used for buffy-coat DNA and 55 cycles for plasma DNA.30 PCR products were analyzed by agarose gel electrophoresis and ethidium bromide staining. Each sample was analyzed in duplicate. The identity of the PCR product for the methylatedp15 or p16 sequence was confirmed by nonradioactive Southern blot analysis using a p15MP or p16MP probe labeled at the 3′ end with digoxigenin (Table 1).31
Sensitivity of MSP
HS-Sultan, which was previously shown to have p15 andp16 methylation on Southern blot analysis, served as a methylated control for MSP.7 30 To determine the sensitivity of the MSP, HS-Sultan DNA was serially diluted in water, mixed with normal blood cell DNA, converted with bisulfite, and then amplified with MSP. For the methylated p15 sequence, the lower detection limit was 0.25 ng or 1 ng of HS-Sultan DNA in 1000 ng of normal blood cell DNA with use of 55 or 35 PCR cycles. For the methylated p16 sequence, the lower detection limit was 0.05 ng or 0.2 ng of HS-Sultan DNA in 1000 ng of normal blood cell DNA with use of 55 or 35 PCR cycles. The sensitivity of MSP for p15/p16reached 2.5 × 10−4 to 5 × 10−5.
RNA extraction
Reverse transcriptase-PCR (RT-PCR) and Southern blot analysis
Total RNA (1-2 μg) was denatured at 65°C for 2 minutes and annealed at 37°C for 10 minutes with 1 μg of random primers (GIBCO-BRL, Gaithersburg, MD).31 Complementary DNA (cDNA) was synthesized at 37°C for 1 hour by using Moloney murine leukemia virus reverse transcriptase (GIBCO-BRL). PCR was conducted by using primers specific for mBCR-ABL or MBCR-ABL (fused at minor or major breakpoints), AF4-MLL, or AML1-ETOfusion transcripts (Table 1). β2-microglobulin cDNA was amplified as a control to ensure that high-integrity RNA was reverse transcribed in each reaction. The thermal profile included initial denaturation at 95°C for 12 minutes, followed by 40 cycles at 95°C for 1 minute, 58°C for 1 minute, and 72°C for 1 minute and final extension at 72°C for 10 minutes. Each sample was analyzed in duplicate. The identity of the PCR product was verified by nonradioactive Southern blot analysis using a BCRABLP, AF4MLLP, or AML1ETOP probe.31
Statistical analysis
The correlation between p15 methylation status and known risk factors and the association of p15 methylation status in buffy coats from bone marrow or peripheral blood with methylation positivity in blood plasma were assessed by the Fisher exact test or χ2 test. The association between p15 methylation status and morphologic disease stage was analyzed by using the McNemar test. Overall survival durations in different subsets of adults with acute leukemia with or without p15 methylation at diagnosis were compared by using the Kaplan-Meier method and log-rank test.
Results
Aberrant p15 methylation in adult and childhood acute leukemias of nearly all morphologic subtypes
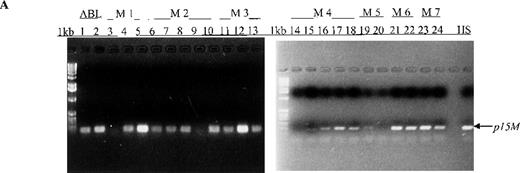
In this prospective analysis of bone marrow and peripheral blood cells, p15 methylation was detected at diagnosis by MSP in 58% (46/79) of patients with adult or childhood acute leukemias of nearly all the FAB subtypes (Figure 1, panel A). As shown in Table 2, p15methylation was found in 64% (27/42) of patients with AML, 50% (17/34) of patients with ALL, and 67% (2/3) of patients with ABL. Using MSP, we found higher p15 methylation frequencies in patients with the M3 (3/3 patients), M4 (8/10), M2 (10/13), or M7 (2/3) subtypes than in those with the M1 (2/4 patients), M6 (1/3), or M5 (1/6) subtypes (Figure 2). Comparable frequencies of p15 methylation were found in patients with L1 (8/18 patients) and L2 (8/14) of B lineage and T-ALL of the L2 subtype (1/2). p15 methylation was not detected in blood cells or bone marrow from 12 healthy control subjects.
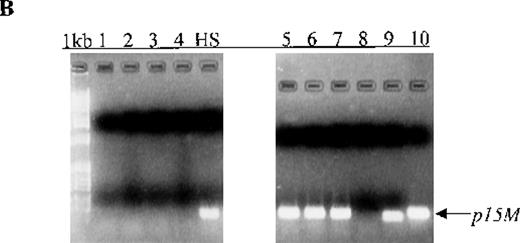
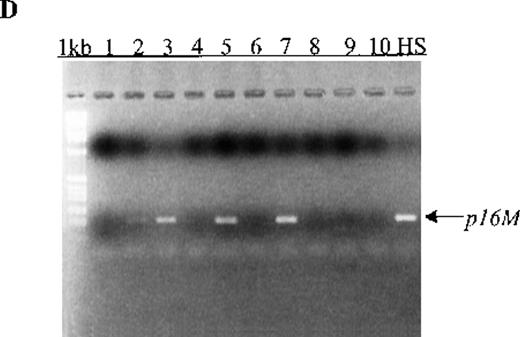
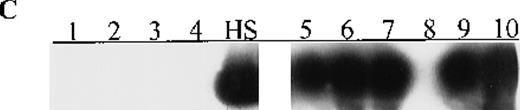
Aberrant p15 methylation in adult and childhood acute leukemias.
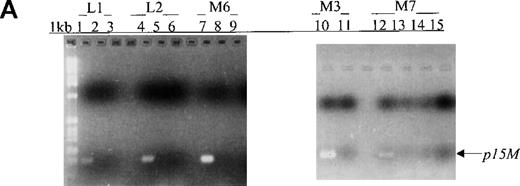
(A) p15 methylation patterns in acute biphenotypic leukemia (ABL; lanes 1 and 2) and acute myeloid leukemia (AML; subtypes M1, M2, M3, M4, M5, M6, and M7 in lanes 3-24). DNA was amplified by using p15MF and p15MR primers. HS-Sultan DNA (HS) served as a methylated control. The 1-kilobase (kb) ladder is the molecular-weight standard. (B, C) Aberrantly methylated p15 sequences in blood plasma from patients with acute leukemia on ethidium bromide-stained agarose gels and Southern blots. Plasma DNA samples from patients withoutp15 methylation in blood cells or bone marrow are shown in lanes 1 to 4; samples from patients with p15 methylation in blood cells or bone marrow are shown in lanes 5 to 10. (D) p16methylation status in patients with AML (lanes 1-5) and acute lymphoblastic leukemia (ALL; lanes 6-10). DNA was amplified by using p16MF and p16MR primers.
Aberrant p15 methylation in adult and childhood acute leukemias.
(A) p15 methylation patterns in acute biphenotypic leukemia (ABL; lanes 1 and 2) and acute myeloid leukemia (AML; subtypes M1, M2, M3, M4, M5, M6, and M7 in lanes 3-24). DNA was amplified by using p15MF and p15MR primers. HS-Sultan DNA (HS) served as a methylated control. The 1-kilobase (kb) ladder is the molecular-weight standard. (B, C) Aberrantly methylated p15 sequences in blood plasma from patients with acute leukemia on ethidium bromide-stained agarose gels and Southern blots. Plasma DNA samples from patients withoutp15 methylation in blood cells or bone marrow are shown in lanes 1 to 4; samples from patients with p15 methylation in blood cells or bone marrow are shown in lanes 5 to 10. (D) p16methylation status in patients with AML (lanes 1-5) and acute lymphoblastic leukemia (ALL; lanes 6-10). DNA was amplified by using p16MF and p16MR primers.
Frequency of p15 and p16 methylation in patients with adult and childhood acute myeloid (AML), lymphoblastic (ALL), and biphenotypic (ABL) leukemias
| . | AML . | ALL . | ABL . | |||
|---|---|---|---|---|---|---|
| Adult . | Childhood . | Adult . | Childhood . | Adult . | Childhood . | |
| p15 methylation | 23/38 | 4/4 | 9/13 | 8/21 | 1/1 | 1/2 |
| p16 methylation | 7/30 | 0/2 | 1/6 | 0/8 | 0/1 | 0/1 |
| . | AML . | ALL . | ABL . | |||
|---|---|---|---|---|---|---|
| Adult . | Childhood . | Adult . | Childhood . | Adult . | Childhood . | |
| p15 methylation | 23/38 | 4/4 | 9/13 | 8/21 | 1/1 | 1/2 |
| p16 methylation | 7/30 | 0/2 | 1/6 | 0/8 | 0/1 | 0/1 |
Values are numbers of patients.
Frequency of aberrant p15 methylation in multiple morphologic subtypes of adult and childhood acute leukemias.
Samples are from patients with ABL (2/3 patients had p15methylation), AML (M1 [2/4 patients], M2 [10/13], M3 [3/3], M4 [8/10], M5 [1/6], M6 [1/3], and M7 [2/3] subtypes), or ALL (L1 [8/18 patients] and L2 [8/14] subtype of B-lineage and T-ALL of L2 subtype [1/2]).
Frequency of aberrant p15 methylation in multiple morphologic subtypes of adult and childhood acute leukemias.
Samples are from patients with ABL (2/3 patients had p15methylation), AML (M1 [2/4 patients], M2 [10/13], M3 [3/3], M4 [8/10], M5 [1/6], M6 [1/3], and M7 [2/3] subtypes), or ALL (L1 [8/18 patients] and L2 [8/14] subtype of B-lineage and T-ALL of L2 subtype [1/2]).
Aberrant p15 methylation was found at similarly high frequencies in adults with AML (61%; 23/38) and adults with ALL (69%; 9/13). Of note, the p15 methylation rate in patients with childhood AML (100%; 4/4 patients) was significantly higher than that in patients with childhood ALL (38%; 8/21; P = .039 by Fisher exact test). Similarly high frequencies of p15methylation were found in adult and childhood AML. In contrast, the rate of p15 methylation was apparently higher in adult ALL than in childhood ALL (P = .078 by χ2 test).
White blood cell (WBC) counts were similar in patients with and without p15 methylation (P = .96 by χ2test). In addition, bone marrow blast counts of patients with and without p15 methylation were not significantly different (P = .54 by χ2 test).
Cell-free methylated p15 sequences in blood plasma from patients
We also examined p15 methylation status in peripheral blood plasma obtained at diagnosis from 16 patients with acute leukemia (9 with AML and 7 with ALL). We detected methylated p15 sequences in plasma from 92% (11/12) of patients who had identical methylation in blood cells or bone marrow (Figure 1, panels B and C). In contrast,p15 methylation was not detected in plasma from 10 healthy subjects or any of the 4 patients without p15 methylation in blood cells or bone marrow. The association of p15 methylation status in blood cells or bone marrow with methylation positivity or negativity in plasma was significant (P = .003 by Fisher exact test).
Concomitant p16 and p15 methylation in adult acute leukemias of particular subtypes
Aberrant p16 methylation was not detected in bone marrow or peripheral blood cells from 11 patients with childhood AML, ALL, or ABL or 12 healthy control (Table 2). Among 37 adults with acute leukemias,p16 methylation was detected at diagnosis in 7 (M2 or M4 subtype) of 30 patients with AML (23%) and in 1 (L2 of B lineage) of 6 patients with ALL (17%) but not in the patient with ABL (Figure 1, panel D). Notably, p16 methylation occurred concomitantly withp15 methylation in the 8 adults with acute leukemias. Seven of the 8 patients were ≥ 50 years old and had WBC counts of < 50 × 109/L (median WBC count, 7.6 × 109/L). The other patient, who was 31 years old and had the M4 subtype, had a WBC count of 207 × 109/L. Of these 8 patients, 3 (M2 or M4 subtype) died of leukemia within 3 months after diagnosis.
p15 methylation, karyotypes, and fusion transcripts
Among 35 patients with adult or childhood AML, ALL, or ABL for whom cytogenetic information was available, 82% (14/17) of patients carrying unmethylated p15 alleles had normal karyotypes or hyperdiploidies associated with good prognosis (Table3). In contrast, 44% (8/18) of patients with p15 methylation at diagnosis had chromosomal deletions, the inversion inv(16)(p13q22), or one of the following translocations: t(9;22)(q34;q11), t(4;11)(q21;q23), t(7;22)(p15;q23), t(15;17)(q22;q12), or t(8;21)(q22;q22) (P = .088 by χ2 test). The mBCR-ABL and AF4-MLLtranscripts were detected by RT-PCR in 6% (1/16) and 8% (1/13), respectively, of patients with ALL (Table 3). AML1-ETOtranscripts were found in 6% (2/32) of patients with AML (Table 3). In contrast to the low rates of detection of these fusion transcripts,p15 methylation appeared to be more applicable as a molecular marker in 56% (27/48) of the same patients. Both methylatedp15 alleles and BCR-ABL transcripts were detected in blood cells from a patient with ALL in morphologic remission, suggesting the presence of MRD (Table 3).
Karyotypic analysis and detection of fusion transcripts by reverse transcriptase-polymerase chain reaction in bone marrow or peripheral blood from patients with adult or childhood acute leukemias with and without p15 methylation at diagnosis
| . | Normal Karyotype . | Hyperdiploidies ≥51 Chromosomes . | Chromosomal Deletions . | Chromosomal Inversions . | Chromosomal Translocations . | Karyotyping Not Done . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL (n = 6) . | AML (n = 4) . | ABL (n = 1) . | ALL (n = 12) . | AML (n = 1) . | ALL (n = 1) . | AML (n = 1) . | ABL (n = 1) . | ALL (n = 1) . | AML (n = 1) . | ALL (n = 4) . | AML (n = 2) . | AML (n = 2)3-150 . | |
| p15 methylation | 2 | 4 | 0 | 4 | 0 | 1 | 0 | 1 | 0 | 1 | 33-151 | 2 | 1 |
| No p15 methylation | 4 | 0 | 1 | 8 | 1 | 0 | 1 | 0 | 1 | 0 | 13-152 | 0 | 1 |
| . | Normal Karyotype . | Hyperdiploidies ≥51 Chromosomes . | Chromosomal Deletions . | Chromosomal Inversions . | Chromosomal Translocations . | Karyotyping Not Done . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL (n = 6) . | AML (n = 4) . | ABL (n = 1) . | ALL (n = 12) . | AML (n = 1) . | ALL (n = 1) . | AML (n = 1) . | ABL (n = 1) . | ALL (n = 1) . | AML (n = 1) . | ALL (n = 4) . | AML (n = 2) . | AML (n = 2)3-150 . | |
| p15 methylation | 2 | 4 | 0 | 4 | 0 | 1 | 0 | 1 | 0 | 1 | 33-151 | 2 | 1 |
| No p15 methylation | 4 | 0 | 1 | 8 | 1 | 0 | 1 | 0 | 1 | 0 | 13-152 | 0 | 1 |
Both patients had AML1-ETO fusion transcripts.
One of the 3 patients had mBCR-ABL fusion transcripts.
The patient had AF4-MLL fusion transcripts.
Sequential monitoring of p15 methylation in patients
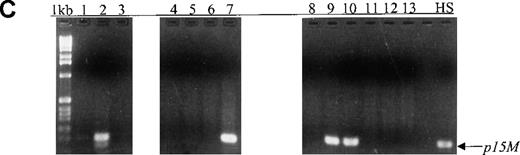
During a median follow-up period of 8 months, we used MSP for sequential monitoring of p15 methylation status in 43 peripheral blood samples from 12 patients (9 adults and 3 children) with AML, ALL, or ABL (median time to achieve first remission, 38 days). p15 methylation was detected in all 12 blood samples obtained at diagnosis. During follow-up, p15 methylation was not detected in 9 samples from 5 patients (2 with ALL and 3 with AML) and, for 8 samples, the unmethylated status was in concordance with morphologic remission and lack of residual leukemia (Figure3, panels A and B). Among these 5 patients, 1 with the M6 subtype had p15 methylation during the active leukemia stage (Figure 3, panel B, lane 9). In an additional 7 patients (5 with AML, 1 with ABL, and 1 with ALL), p15 methylation was detected in 9 samples, and the methylation status of 7 samples was in agreement with morphologic relapse or active or residual leukemia (Figure 3, panels C and D). Conversely, p15 methylation was not detected in 12 samples from these 7 patients, and the unmethylated status of 11 samples correlated with morphologic remission and lack of residual leukemia.
Sequential monitoring of p15 methylation status in peripheral blood from patients with acute leukemia.
(A, B) Results on ethidium bromide-stained agarose gels and Southern blots for patients with L1, L2, M6, M3, and M7 subtypes at diagnosis (lanes 1, 4, 7, 10, and 12) or morphologic remission. The adult with the M6 subtype in whom active leukemia developed had methylatedp15 sequences (panel B, lane 9). The child with the M7 subtype had persistent unmethylated p15 status largely associated with morphologic remission and lack of residual leukemia for 3 months (lanes 13-15). (C, D) Results on agarose gels and Southern blots for patients with M2, ABL, and M4 subtypes at morphologic relapse, during active leukemia or residual leukemia, or in remission. HS-Sultan DNA (HS) served as a methylated control. The 1-kb ladder is the molecular-weight standard. The adult with M2 subtype had p15 methylation at diagnosis and relapse (lanes 1-2). The adult with ABL had p15methylation in association with morphologic relapse (lane 7) and the subsequent stage of active leukemia. p15 methylation was detected at morphologic remission in the adult with M4 subtype, who had relapse 2 weeks later (lanes 9-10).
Sequential monitoring of p15 methylation status in peripheral blood from patients with acute leukemia.
(A, B) Results on ethidium bromide-stained agarose gels and Southern blots for patients with L1, L2, M6, M3, and M7 subtypes at diagnosis (lanes 1, 4, 7, 10, and 12) or morphologic remission. The adult with the M6 subtype in whom active leukemia developed had methylatedp15 sequences (panel B, lane 9). The child with the M7 subtype had persistent unmethylated p15 status largely associated with morphologic remission and lack of residual leukemia for 3 months (lanes 13-15). (C, D) Results on agarose gels and Southern blots for patients with M2, ABL, and M4 subtypes at morphologic relapse, during active leukemia or residual leukemia, or in remission. HS-Sultan DNA (HS) served as a methylated control. The 1-kb ladder is the molecular-weight standard. The adult with M2 subtype had p15 methylation at diagnosis and relapse (lanes 1-2). The adult with ABL had p15methylation in association with morphologic relapse (lane 7) and the subsequent stage of active leukemia. p15 methylation was detected at morphologic remission in the adult with M4 subtype, who had relapse 2 weeks later (lanes 9-10).
Overall, 91% (39/43) of blood samples from 12 patients with AML, ALL, or ABL with p15 methylation at diagnosis showed an excellent concordance between p15 methylation status and morphologic disease stage (P = 1.00 by McNemar test). There was discordance between methylation positivity and morphologic remission status in 2 cases. p15 methylation was detected at morphologic remission in an adult with the M4 subtype who indeed had relapse 2 weeks later (Figure 3, panels C and D, lanes 9 and 10). Both methylated p15 alleles and BCR-ABL transcripts were detected in a patient with childhood ALL at morphologic remission, suggesting the presence of MRD (Table 3). Discordance between unmethylated status and the presence of residual leukemia was found in another 2 patients at an assessment time. One of these patients, a child with the M7 subtype, subsequently had persistent unmethylatedp15 status in association with morphologic remission and lack of residual leukemia for 3 months (Figure 3, panels A and B, lanes 13-15). The other patient, an adult with ABL, was reexamined 9 days later and p15 methylation was found in association with morphologic relapse and the subsequent stage of active leukemia (Figure3, panels C and D, lanes 6 and 7). In a child with M2 transformed from myelodysplastic syndrome (MDS), p15 methylation was not detected at diagnosis of AML but was found at early relapse.
Overall survival of adults with acute leukemias
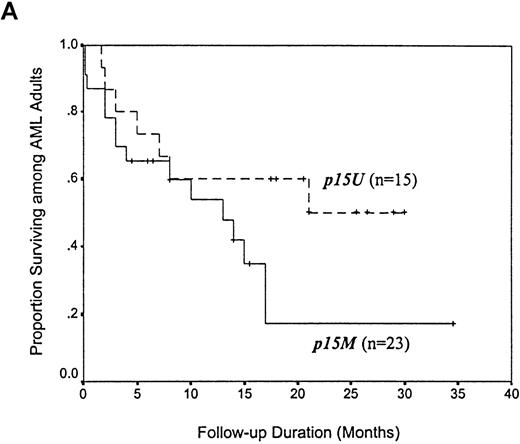
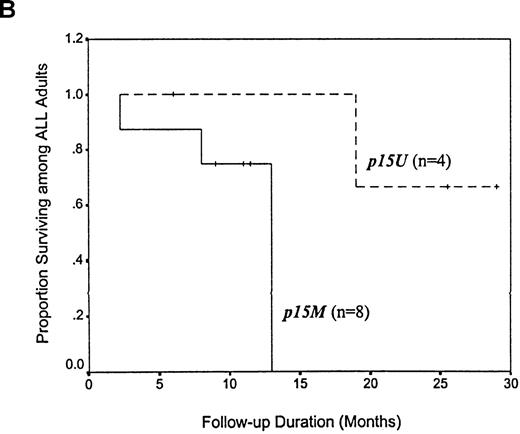
Among 38 adults with AML, the median survival time for the 23 patients with p15 methylation at diagnosis was 13 months, which was apparently shorter than the median survival time of 21 months for the 15 patients with unmethylated p15 alleles (P = .145 by Kaplan-Meier method and log-rank test; Figure4, panel A). There appeared to be a trend toward improved survival after 21 months among the patients with AML who had unmethylated p15 alleles. Among 12 adults with ALL, the median survival time for 8 patients with p15 methylation at diagnosis was also 13 months, which represented a > 2-fold reduction compared with the 29-month survival time for 4 patients with unmethylated p15 alleles (P = .079 by Kaplan-Meier method and log-rank test; Figure 4, panel B).
Kaplan-Meier survival curves for 38 adults with AML and 12 adults with ALL, according to p15 methylation category.
Twenty-three patients with AML and 8 with ALL had methylation(p15M); 15 patients with AML and 4 with ALL had unmethylated status (p15U). Panel A shows AML patients; B, ALL patients.
Kaplan-Meier survival curves for 38 adults with AML and 12 adults with ALL, according to p15 methylation category.
Twenty-three patients with AML and 8 with ALL had methylation(p15M); 15 patients with AML and 4 with ALL had unmethylated status (p15U). Panel A shows AML patients; B, ALL patients.
Discussion
For nearly all the FAB subtypes, our studies using MSP demonstrated aberrant p15 promoter methylation in a significant proportion of patients with adult or childhood AML, ALL, or ABL. We clearly depicted p15 methylation patterns in many subtypes arising in different lineages and differentiation stages. With MSP, we confirmed that frequent p15 methylation is likely a crucial event in acute leukemias. Our findings suggest that p15 methylation arises universally and de novo during leukemic transformation and progression in hematopoietic progenitors developing in the myeloid/lymphoid pathway (B or T lineage) or in primitive stem cells with multilineage potential. Lack of correlation between p15methylation and WBC and bone marrow blast counts might provide evidence suggesting that p15 methylation is one of the early events in leukemogenesis.
Using MSP, we observed comparable p15 methylation frequencies in L1 and L2 (B lineage) and T-ALL (L2) subtypes. Among patients with AML, we found higher p15 methylation frequencies in those with M2, M3, or M4 subtypes than in those with M1, M5, M6, or M7 subtypes. Frequent p15 methylation at the myeloblastic/granulocytic, promyelocytic, or myelomonocytic differentiation stage suggests an interplay of p15 methylation with oncogenic fusion genes found consistently in M2, M3, and M4 subtypes: AML1-ETO,PML-RARα, and CBFβ-MYHII. In support of this notion, we observed that 44% of patients with p15 methylation had chromosomal deletions or translocations, including t(8;21)(q22;q22) and t(15;17)(q22;q21), or inv(16)(p13q22). In the immature leukemia subgroup (M1) and in megakaryoblastic leukemia (M7), the intermediate frequency of p15 methylation is possibly associated with inconsistent and diverse chromosomal abnormalities. Interestingly,p15 methylation frequency in acute erythroleukemia (M6) is similar to that in MDS, a closely related disorder.33 Of note, p15 methylation, which rarely occurs in monoblastic/monocytic leukemia, might be inversely associated with chromosome 11q23 abnormalities commonly found in the M5 subtype. The possibility that this relation exists is supported by the fact thatp15/p16 homozygous deletion was not detected in 44 patients with acute leukemia who had MLL rearrangements.34
The rate of p15 methylation detected by Southern blot analysis is slightly higher than that detected by our MSP in adult AML of nonspecified subtypes (88% compared with 61%).17The difference could be related to patient characteristics, FAB subtypes, or the techniques used for analysis. Incomplete DNA digestion with restriction enzymes might be a source of false positivity, even in normal samples.8-11 In fact, aberrant p15/p16methylation has not been detected in normal bone marrow by MSP or bisulfite sequencing,17,35 and we had no false-positive results in healthy subjects. A comparison of our MSP results with previously reported Southern blotting data11,17 shows similarly high frequencies of p15 methylation in patients with adult AML (61% compared with 88%), adult ALL (69% compared with 71%), childhood AML (100% compared with 67%), and ABL (67% by MSP). Much lower frequencies of p15 methylation were observed among patients with childhood ALL (38% compared with 43% to 48%). It is noteworthy that homozygous deletions of p16, p15, or both are common in childhood ALL but rare in childhood AML and adult AML and ALL (B lineage).4,5,9-11,13-15 17 These data suggest an important role for p16 deletion in leukemogenesis of childhood ALL. Conversely, p15 methylation may be more crucial in the pathogenesis of childhood AML and adult AML and ALL.
Detection of methylation abnormalities in the plasma, serum, or sputum of patients with cancer may be important for the noninvasive diagnosis of cancer.30,36,37 Consistent with the presence ofN-ras mutations or rearranged IgH DNA in blood plasma or serum of patients with acute leukemia,38,39 we detected cell-free methylated p15 sequences with MSP in blood plasma of 92% of patients with acute leukemia who had the identical alteration in blood cells or bone marrow. The mechanism of release of circulating leukemic DNA is unclear but may be related to cellular turnover, necrosis, or apoptosis. The biologic implication of p15methylation in blood plasma and its association with the biologic behavior of leukemic blasts would be clarified by using a quantitative approach.40
Aberrant p16 methylation has previously been found in only 2 patients with T-ALL and 1 patient with childhood AML.10,11,17,35 Abrogation of both p16 andp15 by means of homozygous deletions was reported in 34% of cases of T-ALL and 19% of cases of childhood ALL of B lineage.10,11,17 Dual inactivation of p16 andp15 by homozygous deletion and methylation, respectively, has been described exclusively in 16% of cases of T-ALL with high proliferative indexes and a poor prognosis but in none of 52 cases of childhood ALL of B lineage.10,11,17 In 37 adults with acute leukemia, we found, for the first time, concomitant p16 andp15 methylation in 8 patients with particular subtypes (M2, M4, and L2 of B lineage). Seven of these patients were ≥ 50 years old; the eighth, a 31-year-old patient with the M4 subtype, had a WBC count of > 200 × 109/L. Three of these 8 patients died of leukemia within 3 months after diagnosis. Concomitant p16and p15 methylation has rarely been detected in T-ALL (1/82 cases) and childhood AML (1/27).10,11 17 Consistent with these findings, we did not find p16 methylation in 11 patients with childhood ALL, AML, or ABL. Possibly, p16 methylation may play a unique role in adult AML and ALL of particular subtypes.
To augment selective growth advantage, p16 methylation may act in concert with p15 methylation in adult acute leukemias. Concurrent p15 and p16 methylation has also been reported in glioma, myeloma, non-Hodgkin's lymphoma, Burkitt's lymphoma, and mantle cell lymphoma.3,6,7,17,41 In different tumor types, p16 methylation was detected during tumor progression and associated with metastasis or invasive phenotype.36,42-44 Additional p16 inactivation would not only promote cell proliferation but would also contribute to immortalization and inhibition of apoptosis.45 The prognostic importance of concomitant p16 and p15methylation in adult acute leukemias deserves further investigation in a larger series of patients.
Interestingly, deletions of p16, p15, or both orp15 hypermethylation were not found in 25 patients with childhood t(1;19)-positive ALL but occurred frequently in 3 of 4 patients with childhood t(17;19)-positive ALL.46 47Nevertheless, the interrelations among p15 methylation and other chromosomal abnormalities have not been addressed. In this study, we found that 82% of patients with acute leukemia carrying unmethylated p15 alleles had normal karyotypes or hyperdiploidies associated with favorable prognosis. In contrast, 44% of patients with p15 methylation had chromosomal deletions, inversion, or translocations with prognostic implications. This observation suggests an interplay between the shared p15epigenetic defect and aberrant oncogene expression affected by subtype-specific translocations during leukemogenesis.
Detection of MRD and early relapse has relied mainly on the analysis of fusion transcripts with diagnostic and prognostic implications.25-27 However, BCR-ABL,AF4-MLL, and AML1-ETO transcripts detectable in 8% of cases of ALL or AML would be applied as molecular markers only in a small proportion of patients. It is evident that p16/p15deletion correlates with poor prognosis in patients with acute leukemia.14 15 Unfortunately, it is difficult to detect MRD by assessing gene deletions or inconsistent mutations in peripheral blood that consists of normal nucleated cells. On the other hand,p15 methylation that is detectable in 56% of the same patients appears to be a more useful molecular marker in childhood and adult acute leukemias. Highly specific and sensitive MSP (2.5 × 10−4-5 × 10−5for p15/p16), as opposed to radioactive Southern blot hybridization, should be more applicable for methylation analysis among the low percentage of marrow blasts and particularly useful for detecting MRD or early relapse.
In an attempt to follow the clinical course of patients withp15 methylation, we initiated the first longitudinal study using MSP by sequentially monitoring p15 methylation status in peripheral blood samples from 12 patients with acute leukemia. We found excellent concordance between p15 methylation status and morphologic disease stage in 91% (39/43) of blood samples. This finding suggests that p15 methylation may putatively be a specific molecular abnormality largely associated with disease recurrence. Of note, p15 methylation was detected very early in morphologic remission in an adult with AML who indeed had relapse 2 weeks later. Also, both methylated p15 alleles andBCR-ABL transcripts were detected during morphologic remission in a child with ALL, thereby suggesting the presence of MRD. Therefore, early detection of p15 methylation during apparent remission may indicate an increased risk of disease recurrence. Sequential monitoring of p15 methylation status may also be useful for anticipating the stage of clinical remission as reflected by the unmethylated status.
Acquisition of homozygous deletions of p16, p15, or both or p15 hypermethylation after diagnosis of ALL was detected by Southern blot analysis at relapse in patients with childhood ALL.48 With its higher sensitivity, MSP can detect small leukemic subclones possessing p15 epigenetic changes. In the child we studied who had MDS-transformed ALL (M2 subtype), p15 methylation was not detected at diagnosis of AML but was found at early relapse. We also found concomitant p15and p16 methylation or p15 methylation in adult chronic myeloid leukemia (CML) that progressed to ALL (L1 and L2 subtypes) and in MDS-transformed AML (M5b subtype). These findings are consistent with the acquisition of p16/p15 homozygous deletions during progression of adult chronic T-cell leukemia or lymphoid transformation of CML.49,50 In contrast, we did not find p15methylation in adult CML-transformed AML, in agreement with previous findings.17 These results suggest that p15methylation may play a role in disease progression as well as in leukemogenesis.33
The clinical importance of p16 and p15 deletions in childhood ALL and adult T-ALL is controversial.9,14,15,51Among our 50 adults with AML or ALL, the median survival time for those with p15 methylation was notably shortened (up to > 2-fold) compared with that for patients without methylation. Because our findings with respect to the prognostic relevance of p15methylation were not statistically significant with the limited number of patients studied, we are currently analyzing a larger cohort of patients with longer follow-up. Early detection of aberrant p15methylation may create exciting possibilities for approaching disease monitoring, risk assessment, and effective management of most patients with acute leukemias.33 Furthermore, p15methylation may have important clinical implications for guiding the selection of therapy and monitoring its efficacy. The reversibility of this epigenetic change may also provide the basis for developing novel therapies such as demethylation treatment.52
Acknowledgments
We thank our colleagues in the hematology/BMT laboratories and the hematology and oncology units for support during this project and Eric Wong for helpful advice on statistical analysis of the data.
Supported by research grants 2040505 and 2040507 from the Chinese University of Hong Kong.
Reprints:Margaret H. L. Ng, Hematology Section, Department of Anatomical and Cellular Pathology, Prince of Wales Hospital, The Chinese University of Hong Kong, Shatin, NT, Hong Kong SAR; e-mail:margaretng@cuhk.edu.hk.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

![Fig. 2. Frequency of aberrant p15 methylation in multiple morphologic subtypes of adult and childhood acute leukemias. / Samples are from patients with ABL (2/3 patients had p15methylation), AML (M1 [2/4 patients], M2 [10/13], M3 [3/3], M4 [8/10], M5 [1/6], M6 [1/3], and M7 [2/3] subtypes), or ALL (L1 [8/18 patients] and L2 [8/14] subtype of B-lineage and T-ALL of L2 subtype [1/2]).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/95/6/10.1182_blood.v95.6.1942/6/m_bloo00622002x.jpeg?Expires=1769172994&Signature=ryMuNv-QH4M0dSJ4YIS4gwwV7wmKBpVWEFR1padWuzoKtVGUGcIzMl8vQ678RkvbWOYllCjag4Yonx2pKCaFho7tkvmAVSdzqc3oef0O4tnm0LHD4KhC~mWEhmDSSN32vaeciLGze02bQO~dYvgegvZwMVuzaRFrdMj2I2eEob1EnIos2Enap3GhzzCW1Do1BIvEJX35tlZPxeQbC~yJm9qDsa-P0VND7jqLywE4gfAwfPRK54ykkWoe0O-ND5XAoHRpl2ldKS~qaOWrmsxbS2-8fFZZkUbq0XpbUCeSUGL~3jK8mDILVr6TkTXhxwT-cq~7Ru4gD9Sn-9BarbjtRA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
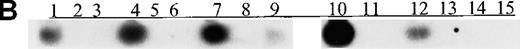

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal