Abstract
The involvement of the cytokine signaling pathway in oncogenesis has long been postulated. Recently, rearrangements of the gene encoding the tyrosine Janus kinase 2 (JAK2) have been reported in human leukemias indicating a direct JAK-signal transduction and activator of transcription (STAT)-mediated leukemic process. The leukemia-associated TEL-JAK2 fusion protein is formed by the oligomerization domain of the translocated ets leukemia (TEL) protein fused to the catalytic domain of JAK2. TEL-mediated oligomerization results in a constitutive tyrosine kinase activity that, in turn, is able to confer growth factor independence to the murine hematopoietic interleukin-3 (IL-3)-dependent Ba/F3 cell line. Results of the present study indicate that fusion proteins containing the oligomerization domain of TEL and the tyrosine kinase domains of Jak1, Jak2, JAK3, or TYK2 share similar properties and are able to efficiently substitute for the survival and mitogenic signals controlled by IL-3, without concomitant activation of the IL-3 receptor. Electrophoretic mobility shift assays demonstrated Stat5 as the only activated Stat factor in TEL-Jak2- and TEL-Jak1-expressing cells, whereas other Stats, namely Stat1 and Stat3, could be detected in TEL-JAK3-, TEL-TYK2-, and also in TEL-ABL-expressing Ba/F3 cells. High levels of expression of the Stat5-target genes pim-1, osm, and Cis were observed in all the cytokine-independent cell lines. Furthermore, the expression of a dominant negative form of Stat5A markedly interfered with the growth factor independence process mediated by TEL-Jak2 in Ba/F3 cells. Because the BCR-ABL and TEL-PDGFβR oncoproteins also activate Stat5, activation of this factor should be a crucial step in activated tyrosine kinase-mediated leukemogenesis.
The Janus kinases (JAK) are receptor-associated tyrosine kinases involved in intracellular signaling pathways of a wide variety of cytokine and growth factor receptors (for a review, see Ihle et al1). Four members of the JAK kinase family have been identified in mammals: JAK1, JAK2, and TYK2, which are ubiquitously expressed, and JAK3 whose expression is predominant in hematopoietic cells. Structural similarities shared by the JAK proteins define 7 JAK homology regions (JH1-7, Figure 1). Among these, the JH2 region is a pseudocatalytic domain, lacking enzymatic activity but assumed to serve as a regulatory region for the JAK activity and as an anchoring region for substrates and regulatory proteins. The JH1 region located at the carboxy-terminus is endowed with tyrosine kinase activity. JAKs are transiently activated following ligand binding and they subsequently phosphorylate multiple substrates, including the JAKs themselves, the cytoplasmic domain of cytokine-receptors, adapter proteins, tyrosine phosphatases, and the signal transducers and activators of transcription (STAT). STATs form a family of cytoplasmic transcription factors that comprise 7 proteins in mammals (Stat 1-4, 5A, 5B, and 6). STATs are activated by the JAKs, form homodimers or heterodimers, and translocate to the nucleus where they participate to transcriptional regulation (for reviews, see Darnell2 and Pellegrini and Dusanter-Fourt3).
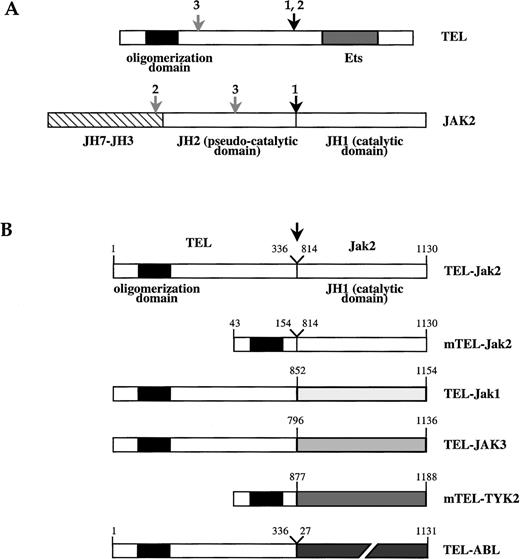
Schematic structure of TEL-JAK fusion proteins.
(A) Schematic of TEL and JAK2 proteins. The TEL oligomerization domain is represented by a hatched box. Arrows indicate the location of the fusion points characterized in human leukemias, as reported in work by Lacronique et al7 and Peeters et al.8 1,: T ALL; 2, atypical CML; 3, pre-B ALL; JH, JAK homology region. (B) TEL-JAK fusion proteins constructed by linking the 336 amino-terminal amino acids of human TEL to the tyrosine kinase domain of murine Jak2 (TEL-Jak2), murine Jak1 (TEL-Jak1), and human JAK3 (TEL-JAK3). Amino acids 53 to 154 of TEL are fused to the corresponding JH1 domains in mTEL-Jak2 and mTEL-TYK2 (human TYK2) constructs. The fusion points in the different chimera are indicated by an arrow. The human TEL-ABL fusion has been previously reported.9 Amino acids numbering on the JH1 part of TEL-JAK fusions refers to the sequences of the wild type JAKs.
Schematic structure of TEL-JAK fusion proteins.
(A) Schematic of TEL and JAK2 proteins. The TEL oligomerization domain is represented by a hatched box. Arrows indicate the location of the fusion points characterized in human leukemias, as reported in work by Lacronique et al7 and Peeters et al.8 1,: T ALL; 2, atypical CML; 3, pre-B ALL; JH, JAK homology region. (B) TEL-JAK fusion proteins constructed by linking the 336 amino-terminal amino acids of human TEL to the tyrosine kinase domain of murine Jak2 (TEL-Jak2), murine Jak1 (TEL-Jak1), and human JAK3 (TEL-JAK3). Amino acids 53 to 154 of TEL are fused to the corresponding JH1 domains in mTEL-Jak2 and mTEL-TYK2 (human TYK2) constructs. The fusion points in the different chimera are indicated by an arrow. The human TEL-ABL fusion has been previously reported.9 Amino acids numbering on the JH1 part of TEL-JAK fusions refers to the sequences of the wild type JAKs.
Deregulation of the JAK-STAT signaling pathway has often been associated with cellular transformation, although its relevance to oncogenic processes was not clearly established. For example, constitutive activation of STATs and JAKs is a recurrent observation in cell lines transformed by viruses and oncogenes and in human leukemic blood samples (reviewed in Garcia and Jove4). Furthermore, Stat3 is required in v-src-mediated transformation of fibroblasts.5,6 A direct link between JAK deregulation and malignant transformation has recently been established by the rearrangement of the JAK2 gene as the result of specific chromosomal translocations in human leukemias. These translocations result in the fusion of various parts of JAK2 to the amino-terminal region of the Translocated Ets Leukemia (TEL) protein, whose gene is frequently rearranged in human malignancies.7,8 The resulting fusion proteins always include the JAK2 catalytic domain. The NH2-terminal Conserved Region (NCR) of TEL has been shown to mediate homotypic oligomerization of the TEL-Jak2 protein leading to constitutive tyrosine kinase activity. The fusion protein exhibits transforming properties, as judged by its ability to relieve the growth factor dependency of murine hematopoietic Ba/F3 cells.7 Similar properties have been shown to be critical for the transforming properties of several neoplasia-associated tyrosine kinases fused to TEL, such as TEL-ABL9 and TEL-PDGFβR.10 11These activated kinases are similarly supposed to mimic growth factor action on cellular processes, mainly proliferation and inhibition of apoptosis.
We studied TEL-JAK chimeric proteins consisting of the catalytic domains of Jak1, Jak2, JAK3, and TYK2 fused to the NCR domain of TEL. Oligomerization of the JAK kinase domains through the NCR domain resulted in constitutive tyrosine kinase activity of TEL-JAK chimeras and in the activation of their transforming properties, similar to those of the TEL-Jak2 fusion. Stat5 activation was observed in cells transformed by all TEL-JAK kinases and shown to be essential to the mitogenic properties of TEL-Jak2.
Materials and methods
Plasmids
The TEL-Jak2 fusion complementary DNA (cDNA) has been described previously.7 The mTEL-Jak2 fusion (see Figure 1) was constructed from TEL-Jak2 by polymerase chain reaction (PCR)-mediated amplification of the appropriate regions of human TEL (amino acids 43-154) linked to the 318 COOH-terminal amino acids of murine Jak2. The TEL-Jak1 and TEL-JAK3 cDNAs were constructed by PCR-mediated amplification to encode the 336 NH2-residues of TEL and the 302 and 341 COOH-terminal residues of murine Jak1 and human JAK3, respectively. The mTEL-TYK2 construct was obtained by splitting together a mTEL-Jak2 clone and a PCR-generated fragment spanning the 311 COOH-terminal residues of human TYK2. PCR products were subsequently showed to be devoid of mutations by sequencing. Jak1, JAK3, and TYK2 cDNAs were gifts from H. Beug, J. J. O'Shea, and S. Pellegrini, respectively. The human TEL-ABL cDNA was kindly provided by D. G. Gilliland. TEL-ABL and TEL-JAK cDNAs were subcloned into the retroviral pBabeNeo vector for stable transfections in Ba/F3 cells.
Cell culture, interleukin-3 independence, and transfections
Ba/F3 cells were grown in RPMI 1640 medium containing 10% fetal bovine serum and 5% WEHI conditioned medium as a source of interleukin-3 (IL-3). For IL-3 stimulation, Ba/F3 cells were starved for 5 hours in Iscove modification of Dulbecco's minimum essential medium containing 0.4% bovine serum albumin and 20 μg/mL iron-saturated transferrin, then stimulated with 10 ng/mL of recombinant murine IL-3 (R and D Systems, Abingdon, UK) for 10 minutes. Deprived Ba/F3 cells were maintained in the same medium for 15 hours. Stably TEL-JAK-transfected Ba/F3 cells were obtained by electroporation (Bio-Rad Gene Pulsar, Ivry-sur-Seine, France) and selected for transformants with 1 mg/mL neomycin. For IL-3-independent proliferation assays, G418-resistant Ba/F3 transfectants were seeded in IL-3-free medium in 96-well plates as previously reported.7Mock-transfected cells and transfectants harboring the pBabeNeo vector alone were used as controls. The Ba/F3 cell line constitutively expressing a dominant negative form of Stat5A (Stat5AΔ749) from the pRSVNeo vector has been previously reported.12 The TEL-Jak2 cDNA was cloned into the retroviral pBabePuro vector and stably transfected into Ba/F3 cells expressing Stat5AΔ749 from pRSV vector and in control pRSVNeo-Ba/F3 cells. Transfectants were selected in the presence of 1 μg/mL puromycine. For IL-3-independent proliferation assays, Ba/F3 cells expressing either TEL-Jak2 or Stat5AΔ749, or coexpressing Stat5AΔ749+TEL-Jak2 and Stat5AΔ749 cells transfected with the empty pBabePuro vector (Stat5AΔ749+pBabePuro) were washed twice in WEHI-free medium then seeded in the same medium at a density of 105 cells/mL. Proliferation rates of the different cell lines were monitored daily by using a Coulter analyzer (Coultertronics, Margency, France). The growth slope presented is representative of duplicates of 2 independent experiments.
Nuclear extracts and electrophoretic mobility shift assays
Nuclear extracts were made as reported previously.13 For electrophoretic mobility shift assays (EMSA), 20 μL of nuclear extracts from TEL-ABL and TEL-JAK-transformed Ba/F3 cells were incubated with the 32P-labeled m67SIE 5′-CATTTCCCGTAAATC-3′ and bovine β-casein 5′-AGATTTCTAGGAATTCAAATC-3′ STAT binding sites. Eight microliters of nuclear extracts from murine IL-3-stimulated parental Ba/F3 cells were used as controls. Supershift assays were performed by using anti-Stat1 (Transduction Laboratories, Lexington, KY), anti-Stat3 (Santa Cruz Biotechnology, Inc, Le Perray-en Yvelines, France) and anti-Stat5 antibodies.14
Stat4 and Stat6 DNA binding activities were investigated, using the specific FcγRI 5′-AGCATGTTTCAAGGATTTGAGATGTATTTCCCAGAAAAG-3′) and Iε 5′-GTCAA CTTCCCAAGAACAGAA-3′) probes. Extracts of COS7 cells co-transfected with TEL-Jak2 and Stat4 or Stat6 expression vectors (a kind gift of J. R. Darnell) were used as positive controls.
Northern blots
Northern blotting analysis was performed by using 10 μg of total RNA and probes corresponding to murine cDNA fragments for pim-1, osm, cis, c-myc, c-jun, and c-fos labeled with 32P-αCTP by random-priming. Northern blots were normalized by hybridization with a GAPDH probe. Radioactivity was quantified using a Molecular Imager (Bio-Rad).
Cell lysates, immunoprecipitates, and Western blot analyses
For Western blotting of total cell lysates, Ba/F3 cells were lysed in 1% Brij 96, 10 mM Tris/Hcl pH 7.4, 150 mM NaCl, 5mM EDTA, 10% glycerol, 0.02% NaN3, and 1 mM Na3VO4. Whole cell lysates were cleared by centrifugation at 13 000g for 20 minutes at 4°C and 50 μg of cellular protein was separated through 10% sodium dodecyl sulfate (SDS)-polyacrylamide gels, transferred to nitrocellulose, and blotted with an antiphosphotyrosine antibody 4G10 (Upstate Biotechnology, Inc, Souffelweyersheim, France). For IL-3 receptor β subunit (IL-3βR) immunoprecipitation analysis, 20 × 106Ba/F3 cells were lysed in the above buffer and supernatants were incubated 2 hours at 4°C with a rabbit polyclonal anti-IL-3βR antibody (Santa Cruz Biotechnology, Inc.). Immunoprecipitates were separated by 8% SDS-polyacrylamide gels, transferred to nitrocellulose, and blotted with antiphosphotyrosine and anti-IL-3βR subunit antibodies. Expression of stably transfected TEL-JAK and TEL-ABL fusions in Ba/F3 cells was evaluated by immunoprecipitation followed by Western blot analysis, using a rabbit polyclonal immunoserum raised against the amino-terminal part of the human TEL protein (α N-TEL)15 present in all the chimeras. Immunocomplexes were detected using the ECL detection kit (Amersham, Orsay, France). Stripping and probing were performed according to the manufacturer's instructions.
Results
TEL-JAK fusion proteins exhibit constitutive tyrosine kinase activity and confer factor-independent growth to IL-3-dependent Ba/F3 cells
To investigate for potential biologic differences between the tyrosine kinase domains (JH1) of JAK family members, we generated chimeric proteins modeled on the structure of the oncogenic TEL-Jak2 fusion in which the JH1 domains of JAK1, JAK3, and TYK2 were fused to TEL sequences. The schematic structures of these TEL-JAK proteins are summarized in Figure 1. All TEL-JAK fusions include the NCR region of TEL responsible for the homotypic oligomerization of TEL and TEL-derived fusion proteins. To determine if TEL sequences outside of the NCR contribute to transformation, a truncated mTEL-Jak2 fusion was constructed, directly linking the NCR region to the JH1 of Jak2. The murine TEL-TYK2 chimeric protein was constructed similarly to the above fusion. Western blotting analysis using antiphosphotyrosine antibody showed the constitutive phosphorylation of in vitro produced TEL-JAK chimeras (data not shown), a property known to reflect the intrinsic tyrosine kinase activity of the chimeric proteins.7 10These results confirm that oligomerization of the catalytic domains of JAKs through the NCR domain of TEL is sufficient to allow their constitutive activation.
To test for the transforming capacities of these proteins, TEL-JAK fusions were stably expressed in Ba/F3 cells using a pBabeNeo retroviral vector. The truncated mTEL-Jak2 fusion was equivalent to the full-length protein regarding its ability to transform Ba/F3 cells, thus indicating that most of the TEL sequences present in the initial chimera do not significantly contribute to its transforming properties in this model. All TEL-JAK chimera were expressed at similar levels (Figure 2A), tyrosine phosphorylated, and found to confer IL-3 independence to Ba/F3 cells without significant variation of cell growth rates between the different transformed cell lines (data not shown). TEL-JAK-transformed cell lines grew identically when maintained with or without a source of IL-3, indicating that chimeric proteins efficiently substitute for the mitogenic and antiapoptotic signals controlled by the cytokine. Growth factor independence was readily obtained in TEL-ABL-expressing Ba/F3 cells, as previously reported.9
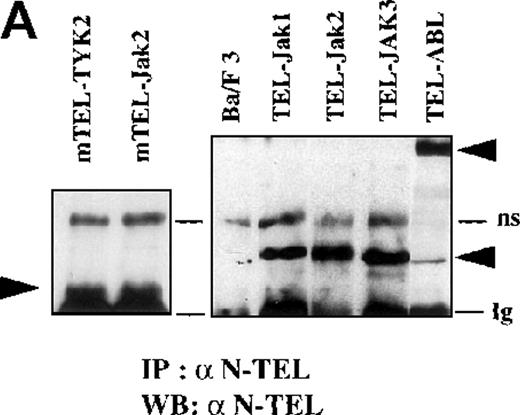
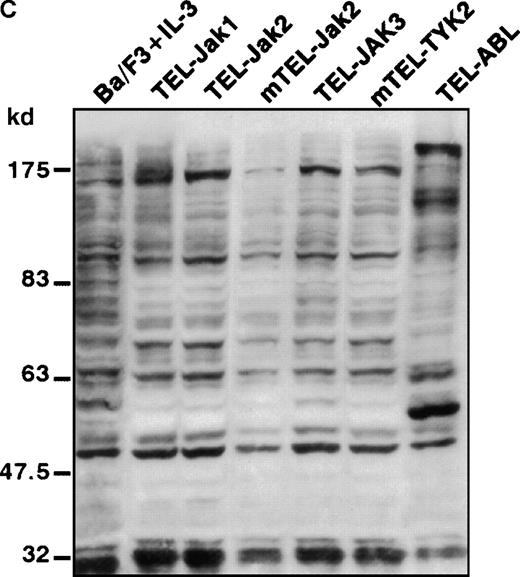
Phosphotyrosine immunoblot analysis of the IL-3 receptor β subunit and of cellular proteins in TEL-JAK- and TEL-ABL-expressing cells.
(A) The expression of stably transfected TEL-JAK and TEL-ABL fusions in the indicated IL-3-independent Ba/F3 cell lines was evaluated by immunoprecipitation (IP) followed by Western blotting (WB) using a polyclonal rabbit immune serum raised against the amino-terminal part (α N-TEL) of the human TEL protein. Immunoprecipitated complexes are indicated by black arrows; ns, nonspecific. Predicted molecular weights of the chimera are as follows: TEL-Jak1, 75.5 kd; TEL-Jak2, 77.7 kd; TEL-JAK3, 79.3 kd; TEL-ABL, 158.8 kd; murine TEL-Jak2, 50.5 kd; murine TEL-TYK2, 49.4 kd. (B) The IL-3βR subunit was immunoprecipitated from total cell lysates of the indicated IL-3-independent Ba/F3 cell lines and analyzed by immunoblotting with an antiphosphotyrosine antibody (α pTyr). The blot was stripped and reprobed with an anti-IL-3βR antibody. (C) Total cell lysates of the indicated IL-3-independent Ba/F3 cells were subjected to Western blotting using an antiphosphotyrosine antibody. As a control, total cell lysates of parental Ba/F3 cells, starved then stimulated with mIL-3, were loaded. Differences in patterns of cellular protein phosphorylations were reproducibly observed between TEL-JAK- and TEL-ABL-expressing Ba/F3 cells. Note that the weaker pattern of phosphorylation seen in the mTEL-Jak2 line is due to underloading of total cell lysates.
Phosphotyrosine immunoblot analysis of the IL-3 receptor β subunit and of cellular proteins in TEL-JAK- and TEL-ABL-expressing cells.
(A) The expression of stably transfected TEL-JAK and TEL-ABL fusions in the indicated IL-3-independent Ba/F3 cell lines was evaluated by immunoprecipitation (IP) followed by Western blotting (WB) using a polyclonal rabbit immune serum raised against the amino-terminal part (α N-TEL) of the human TEL protein. Immunoprecipitated complexes are indicated by black arrows; ns, nonspecific. Predicted molecular weights of the chimera are as follows: TEL-Jak1, 75.5 kd; TEL-Jak2, 77.7 kd; TEL-JAK3, 79.3 kd; TEL-ABL, 158.8 kd; murine TEL-Jak2, 50.5 kd; murine TEL-TYK2, 49.4 kd. (B) The IL-3βR subunit was immunoprecipitated from total cell lysates of the indicated IL-3-independent Ba/F3 cell lines and analyzed by immunoblotting with an antiphosphotyrosine antibody (α pTyr). The blot was stripped and reprobed with an anti-IL-3βR antibody. (C) Total cell lysates of the indicated IL-3-independent Ba/F3 cells were subjected to Western blotting using an antiphosphotyrosine antibody. As a control, total cell lysates of parental Ba/F3 cells, starved then stimulated with mIL-3, were loaded. Differences in patterns of cellular protein phosphorylations were reproducibly observed between TEL-JAK- and TEL-ABL-expressing Ba/F3 cells. Note that the weaker pattern of phosphorylation seen in the mTEL-Jak2 line is due to underloading of total cell lysates.
The TEL-JAK proteins do not significantly phosphorylate tyrosine residues of the IL-3βR chain
Phosphorylation of the tyrosine residues in the intracellular domain of cytokine receptors are known to be controlled by JAK kinases. Because the growth factor independence of Ba/F3 cells expressing TEL-tyrosine kinase fusions could result from the activation of the IL-3 receptor, we analyzed the phosphorylation state of the IL-3βR by phosphotyrosine immunoblotting after immunoprecipitation of cellular extracts (Figure 2B). We failed to detect any consistent IL-3βR phosphorylation in TEL-JAK-expressing cell lines, whereas a constitutive phosphorylation was observed in cells transformed by the TEL-ABL fusion. These results suggest that growth signals derived from TEL-JAK proteins do not involve cooperation with the IL-3βR chain. In contrast, TEL-ABL proteins phosphorylate the IL-3βR chain, but the relevance of this phosphorylation to their transforming capacities is not known. From these observations, one can infer that TEL-JAK proteins have lost some substrate specificity with respect to normal JAK proteins and that TEL-ABL proteins exhibit a wider substrate specificity than TEL-JAKs. In keeping with these results, Western blot analysis of total cell lysates showed a different pattern of phosphorylated protein species between the IL-3-independent TEL-JAK- and TEL-ABL-expressing cells (Figure 2C).
All TEL-JAK fusion proteins induce functional activation of Stat5 factors
To determine if the different TEL-JAK fusions selectively activate members of the Stat family, Stat DNA binding activities were analyzed by EMSA. Nuclear extracts from the TEL-JAK- and TEL-ABL-expressing lines were incubated with probes containing various Stat binding sequences, known to have different affinities for activated Stats. The β-casein probe contains Stat5 binding sites and binds the 2 highly homologous Stat5A and Stat5B factors.16 As a positive control, parental Ba/F3 cells were stimulated with recombinant murine IL-3 (mIL-3). As shown in Figure 3A, nuclear extracts from all TEL-JAK-transformed cell lines showed a specific Stat5 DNA binding activity, consisting of a major protein-DNA complex, which can be supershifted with anti-Stat5 antibodies (Figure3A, compare lanes 4, 7, 10, 13, and 16 to lanes 6, 9, 12, 15, and 18, respectively). Specificity of Stat5-β-casein complexes was confirmed by competitive experiments using unlabeled β-casein probe (Figure 3A, lanes 5, 8, 11, 14, and 17). Nonrelevant competitors did not interfere with the shifted complexes (data not shown). Both Stat5A and Stat5B factors are expressed in Ba/F3 cells but a more pronounced Stat5A DNA binding activity was observed in all transformed Ba/F3 cells. In nuclear extracts from TEL-JAK3 (Figure 3A, lane 13) and mTEL-TYK2 (Figure 3A, lane 16), a faint signal corresponding to a higher mobility complex was observed, similar to the one seen after mIL-3-stimulation (Figure 3A, lane 1). This complex corresponds to Stat1, because the β-casein probe is known to exhibit low affinity for Stat1 factors, which are activated in Ba/F3 cells on IL-3 stimulation.17 Furthermore, supershifting experiments with an anti-Stat1 antibody confirmed the specific interactions of Stat1 with the β-casein probe with these extracts (data not shown). A constitutive activation of Stat5 factors was also seen in TEL-ABL-transformed cells (Figure 3A, lanes 19-21). In addition, Stat5 is activated in TEL-PDGFβR-transformed cells (A.B., unpublished data). Collectively, these results establish the recurrent activation of Stat5 DNA binding activity in Ba/F3 cells for all TEL-fusion proteins studied to date.
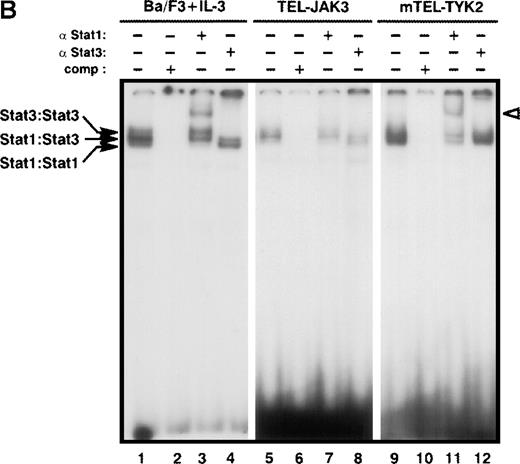
Constitutive activation of Stat factors in TEL-JAK-transformed Ba/F3 cells.
Analyses of Stat DNA-binding activities in IL-3-independent Ba/F3 cells expressing the indicated constructs. As a control for Stat1, Stat3, and Stat5 DNA-binding activities, nontransfected Ba/F3 cells were starved for 5 hours, then stimulated with 10 ng/mL of mIL-3 for 10 minutes. Nuclear extracts were submitted to EMSA using the bovine β-casein (A) and m67SIE (B) probes. The specificity of the shifted complexes was assessed by the addition of a 100-fold molar excess of cold competitors. Complexes supershifted by specific anti-Stat factors antibodies are indicated by open arrows. Note that constitutive Stat1 and Stat3 activations detected in TEL-JAK3-expressing Ba/F3 cells were observed after prolonged exposure.
Constitutive activation of Stat factors in TEL-JAK-transformed Ba/F3 cells.
Analyses of Stat DNA-binding activities in IL-3-independent Ba/F3 cells expressing the indicated constructs. As a control for Stat1, Stat3, and Stat5 DNA-binding activities, nontransfected Ba/F3 cells were starved for 5 hours, then stimulated with 10 ng/mL of mIL-3 for 10 minutes. Nuclear extracts were submitted to EMSA using the bovine β-casein (A) and m67SIE (B) probes. The specificity of the shifted complexes was assessed by the addition of a 100-fold molar excess of cold competitors. Complexes supershifted by specific anti-Stat factors antibodies are indicated by open arrows. Note that constitutive Stat1 and Stat3 activations detected in TEL-JAK3-expressing Ba/F3 cells were observed after prolonged exposure.
Stat1 and Stat3 are activated in Ba/F3 cells transformed by the TEL-JAK3, mTEL-TYK2, and TEL-ABL fusions
Specific activation of Stat1 and Stat3 factors was assessed by using the high affinity m67SIE sequence, derived from the STAT binding motif of the c-fos promoter18 (Figure 3B). This probe allows the formation of 3 DNA-protein species that correspond to homodimers of Stat1, homodimers of Stat3, and heterodimers of Stat1 and Stat3. Constitutive activation of both factors was observed in the TEL-JAK3-, mTEL-TYK2-, and TEL-ABL-expressing cells. Nuclear extracts from TEL-JAK3-transformed Ba/F3 cells showed patterns of protein-DNA complexes and of supershifts with anti-Stat1 and anti-Stat3 antibodies similar to those observed with mIL-3-stimulated cells extracts used as control (Figure 3B, compare lanes 1-4 to 5-8, respectively). Nuclear extracts from the mTEL-TYK2-expressing cells only formed 2 shifted DNA-protein complexes with a high degree of specificity as shown by competition with excess cold m67SIE probe (Figure 3B, compare lanes 9 and 10) and incubation with an anti-Stat1 antibody confirmed the presence of Stat1 factors in the low migrating DNA-protein complex. The presence of 2 shifted complexes indicated that some Stat3 was activated in these extracts and preferentially associated with Stat1 to form Stat1-Stat3 heterodimers, although the use of an anti-Stat3 antibody did not clearly reveal a Stat3 DNA binding activity (Figure 3B, lane 11). It is noteworthy that lower levels of activated Stat1 factors were observed in TEL-JAK3-expressing cells, as compared to mTEL-TYK2-expressing cells (Figure 3B, compare lanes 5 and 9). Minor Stat1 and Stat3 activations were also evidenced in nuclear extracts from Ba/F3 cells expressing the TEL-ABL fusion (data not shown).
Stat4 and Stat6 DNA binding activities were investigated using the specific FcγRI and Iε probes, respectively, but these factors were not found to be activated in TEL-JAK and TEL-ABL-transformed cell lines despite the fact that both are present in Ba/F3 cells (data not shown).
A specific set of cytokine-responsive genes is induced in the TEL-JAK-transformed Ba/F3 cell lines
To further investigate the molecular events leading to growth factor independence conferred by TEL-JAK fusions in the Ba/F3 cellular context, we examined by Northern blot the induction of a series of cytokine-responsive genes, including genes whose products are involved in the control of cellular proliferation. Parental Ba/F3 cells cultured in the presence of WEHI conditioned medium, as a source of growth factors, will be referred to as proliferating cells. The c-myc, c-jun, and c-fos genes are immediate early genes induced in response to IL-3 in Ba/F3 cells19 and the cytokine-driven activation of both c-myc and c-fos transcription has been directly linked to the activation of JAK pathways.20,21 All TEL-JAK-expressing cells exhibited elevated levels of c-myc, c-jun, and c-fos expression, equivalent to those found in proliferating parental cells, indicating that expression of chimeric proteins efficiently substitute for the normal IL-3-mediated mitogenic signals. Thus, it appears that signaling pathways activated by the leukemogenic TEL-Jak2 protein and by the chimeric TEL-JAK fusions studied here can mimic at least some of the events triggered by the cytokine-receptor interaction. In contrast, the expression of TEL-ABL resulted in high expression levels of c-fos, c-jun, and c-myc, because messenger RNA (mRNA) levels were found to be 2, 2.5, and 3 times higher, respectively, as compared to those seen in proliferating parental cells. These findings are in agreement with previous reports indicating that c-myc expression is up-regulated in Ba/F3 cells transformed by the TEL-ABL fusion.22
The constitutive activation of Stat5 in all transformed Ba/F3 cell lines prompted us to examine the induction of several known Stat5-target genes in these cells. Among them, those encoding oncostatin M (osm), the hematopoietic serine/threonine kinase pim-1, and the cytokine-inducible SH2-containing (Cis) protein involved in switching off the cytokine signal by interacting with phosphorylated tyrosine residues on receptor chains,23 are normally activated via Stat5-dependent pathways.14 24-26 As shown in Figure 4, these Stat5-target genes were activated in TEL-JAK- and TEL-ABL-expressing cells, providing further evidence of a functional constitutive Stat5 activation in the cytokine-independent cell lines. Interestingly, although transcriptionnal levels of both osm and pim-1 genes were equivalent in the transformed Ba/F3 cell lines to those of proliferating parental cells, levels of Cis expression were significantly higher. The mechanisms underlying the sustained expression of Cis remain, however, to be established.
Induction of several Stat5-target genes in TEL-JAK- and TEL-ABL- transformed Ba/F3 cells.
Northern blot analyses of total RNA extracted from the indicated cell lines for osm, pim-1, and Cis expression are shown. RNA from Ba/F3 cells cultured in the presence of WEHI conditioned medium (−), starved (starved), and stimulated by mIL-3 (stim) were loaded as controls. The indicated values of Cis induction are expressed relative to the values obtained by proliferating Ba/F3 cells. A GAPDH probe was used as internal standard.
Induction of several Stat5-target genes in TEL-JAK- and TEL-ABL- transformed Ba/F3 cells.
Northern blot analyses of total RNA extracted from the indicated cell lines for osm, pim-1, and Cis expression are shown. RNA from Ba/F3 cells cultured in the presence of WEHI conditioned medium (−), starved (starved), and stimulated by mIL-3 (stim) were loaded as controls. The indicated values of Cis induction are expressed relative to the values obtained by proliferating Ba/F3 cells. A GAPDH probe was used as internal standard.
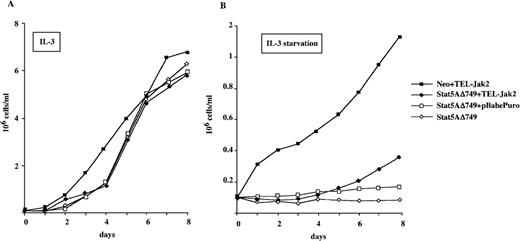
Constitutive expression of a dominant negative form of Stat5A in Ba/F3 cells significantly interferes with the growth factor independence process mediated by TEL-Jak2
To further establish the importance of Stat5 activation for the mitogenic properties of TEL-Jak2, we next analyzed the ability of a Stat5A dominant negative mutant to interfere with TEL-Jak2 transforming properties. The Stat5AΔ749 protein lacks the carboxy terminal transactivation domain of Stat5 and exerts a dominant negative effect on Stat5-responsive gene promoters.27 The pRSVNeo-Stat5AΔ749 Ba/F3 cells12 were stably transfected with the pBabePuro-TEL-Jak2 expressing vector (Stat5AΔ749+TEL-Jak2 transfectants) or with empty vector (Stat5AΔ749+pBabePuro transfectants). TEL-Jak2 cells harboring the pRSVNeo control vector were also established (Neo+TEL-Jak2 transfectants). The numbers of viable cells of the different transfectants were scored daily in the presence or in the absence of IL-3 and are shown in Figure5. The Neo+TEL-Jak2 cells grew efficiently when deprived of IL-3, whereas parental Stat5AΔ749 and Stat5AΔ749+pBabePuro cell lines were unable to proliferate in similar conditions (Figure 5B). The Stat5AΔ749+TEL-Jak2 cells showed poor proliferation rates in the absence of IL-3. These data demonstrate that the expression of a dominant negative Stat5 mutant markedly interferes with the ability of TEL-Jak2 to induce growth factor independence to Ba/F3 cells, strongly suggesting the requirement for Stat5 activation in this process.
The constitutive expression of a dominant negative Stat5A mutant interferes with the growth factor independence conferred by the TEL-Jak2 fusion in Ba/F3 cells.
Ba/F3 cells constitutively expressing the pRSVNeo-Stat5AΔ749 vector (Stat5AΔ749) were stably transfected with TEL-Jak2 (Stat5AΔ749+TEL-Jak2) or the empty expression vector (Stat5AΔ749+pBabePuro). As a control, the parental pRSVNeo Ba/F3 cells were stably transfected by TEL-Jak2 (Neo+TEL-Jak2). The proliferation rates of the indicated Ba/F3 cell lines were scored daily for 8 days in either the presence (A) or absence (B) of WEHI conditioned medium.
The constitutive expression of a dominant negative Stat5A mutant interferes with the growth factor independence conferred by the TEL-Jak2 fusion in Ba/F3 cells.
Ba/F3 cells constitutively expressing the pRSVNeo-Stat5AΔ749 vector (Stat5AΔ749) were stably transfected with TEL-Jak2 (Stat5AΔ749+TEL-Jak2) or the empty expression vector (Stat5AΔ749+pBabePuro). As a control, the parental pRSVNeo Ba/F3 cells were stably transfected by TEL-Jak2 (Neo+TEL-Jak2). The proliferation rates of the indicated Ba/F3 cell lines were scored daily for 8 days in either the presence (A) or absence (B) of WEHI conditioned medium.
Discussion
Chromosomal translocations resulting in TEL-JAK2 fusion proteins have been reported in several human leukemias.7,8 The TEL-JAK2 fusion proteins described to date transform growth factor-dependent cell lines and are leukemogenic in mouse models7,28 (and our unpublished data). Several examples of tyrosine kinases constitutively activated through ectopic oligomerization have been reported in human hematopoietic malignancies, one of the most extensively studied being the BCR-ABL fusion protein in chronic myelogenous leukemia with the Philadelphia chromosome.29 Thus, the identification of signaling molecules regulated by these kinases is essential to elucidate the mechanisms underlying their transforming and leukemogenic properties.
We generated TEL-JAK chimeric proteins by fusing the oligomerization domain of TEL to the catalytic domains (JH1) of Jak1, JAK3, or TYK2. The TEL-JAK fusions were constitutively phosphorylated both in vitro and in vivo, showing that TEL-mediated oligomerization is sufficient to induce their constitutive tyrosine kinase activity. When stably expressed in Ba/F3 cells, all TEL-JAK chimera were able to sustain cytokine-independent cellular growth, indicating that they can substitute for growth factor-mediated signals. These results are somewhat unexpected because, except for JAK2, JAK kinases are presumed to normally signal in pairs when activated under physiologic conditions. Furthermore, the JH2 domain is dispensable for the biologic properties of all chimeras tested, including mTEL-TYK2, whereas it is known to be required for the normal kinase activity of TYK2.30 Activation through oligomerization is, however, not a feature of all JAK tyrosine kinase domains because a TEL-DJAK fusion, containing the JH1 domain of the homologous DrosophilaHopscotch protein, did not exhibit detectable tyrosine phosphorylation in vitro nor transforming properties in Ba/F3 cells (unpublished data). Whether its lack of activity is due to the absence of the JH2, as previously observed,31 or to structural particularities of the DJAK JH1 remains to be determined.
As reported for TEL-Jak2,7 TEL-JAK fusion proteins have a diffuse cytoplasmic location (data not shown) that differs from normal activated JAK proteins, which are associated with the intracellular moiety of cytokine receptors. Accordingly, phosphorylated substrates could differ between the TEL-JAK and JAK proteins. For example, the IL-3βR chain is not detectably tyrosine phosphorylated in Ba/F3 cells expressing the TEL-JAK fusions. Phosphorylation of normal JAK proteins is not seen or inconsistently detected (data not shown).28,32 Consistently, functional studies of artificially activated forms of JAK2 have suggested that the tyrosine kinase domain is not sufficient to interact with and phosphorylate the granulocyte macrophage colony-stimulating factor (GM-CSF) receptor,33 the IL-3 receptor,34 or the growth hormone receptor.35 These observations suggest that TEL-JAK fusions are able to directly activate signaling pathways that lead to cell proliferation or inhibition of apoptosis independently of the cytokine receptor activation or the use of receptor effector domain as accessory signaling chain.
In keeping with this hypothesis, constitutive activation of STAT factors is observed in Ba/F3 cells transformed by the TEL-JAK fusions. A number of studies have suggested that specificity of STAT factors activation in response to cytokine receptor engagement is more related to the nature of the cytokine receptor involved than to the activation of a particular JAK. This notion might, however, not be so absolute because a ligand-independent interaction between the SH2 domain of Stat5 and the JAKs has been reported.36 Also, complexes between the SH2 region of Stat5 and phosphotyrosine residues located in the JH1 region of JAK2 occurred when these proteins are expressed in yeast cells,37 suggesting the possibility of a direct recruitment of Stat5 to the activated kinase, independent of receptor docking. Similarly, the SH2 region of Stat1 has been reported to directly interact with the activated TYK2 kinase.38Moreover, we observed a selective activation of Stat1 and Stat3 in Ba/F3 cells expressing the TEL-JAK3 and mTEL-TYK2 fusions, whose cellular counterparts preferentially activate these factors in a number of cytokine receptor systems. Accordingly, only the JH1 regions of JAK3 and TYK2 possess 2 potential functional Stat3 binding sites centered on Y824 and Y1176, respectively, which conformed to the previously identified Stat3 docking sites on cytokine receptors (YXXQ, preceded by acid amino acids3). In keeping with our results, Ho et al32 recently reported the recurrent activation of Stat5 in Ba/F3 cells expressing TEL-Jak2 fusions, but also constitutive activation of Stat1 and Stat3. In this instance, detection of activated Stat1 and Stat3 factors could be related to higher levels of expression of transforming proteins in Ba/F3 cells than in our model or to differences in Ba/F3 sublines used.
The comparison of Ba/F3 cells expressing either the TEL-JAK or the TEL-ABL fusion proteins reveals several interesting features. In contrast to those expressing TEL-JAK, Ba/F3 cells transformed by the TEL-ABL fusion protein exhibit constitutive phosphorylation of the IL-3βR chain (see Figure 2B) and of endogenous JAK2 protein (data not shown). Similar observations have been reported for hematopoietic cells transformed by the BCR-ABL fusion proteins,39,40 raising the possibility that abrogation of growth factor requirement of hematopoietic cell lines expressing these kinases may, at least partially, involve the activation of cytokine receptors. On the other hand, this phosphorylation could merely reflect the weak substrate specificity of the oncogenic forms of ABL fusion proteins. Alternatively, one cannot exclude that the cytoskeletal-associated localization of TEL-ABL9 allows, therefore, the phosphorylation of membrane-bound proteins by the chimera. In keeping with this hypothesis, the SHP1 phosphatase is heavily tyrosine phosphorylated in TEL-ABL but not detectably in TEL-JAK-expressing Ba/F3 (data not shown). The distinct pattern of tyrosine phosphorylated proteins observed in TEL-JAK- and TEL-ABL-expressing cells (see Figure2C) further underlines the different substrate specificity of these activated TEL-derived fusion kinases.
Transcription of genes whose products are involved in cellular growth regulation also differs between TEL-JAK- and TEL-ABL-expressing Ba/F3. In keeping with our results, high levels of transcription of the early response genes c-myc and c-fos have been reported in Ba/F3 cells expressing TEL-ABL22 and in cell lines expressing BCR-ABL.41 From our data, the cytokine-independent proliferation of TEL-JAK-transformed Ba/F3 cells is not associated with higher level of expression of the early-response c-myc, c-jun, and c-fos genes, because these genes were expressed at levels lower than those observed in TEL-ABL-expressing cells but similar to those observed in exponentially growing parental Ba/F3 cells. Several cytokine receptor models have demonstrated that induction of c-jun and c-fos expression requires not only JAK kinases activation but also the phosphorylation of several tyrosine residues of the activated receptor.42Consistently, the absence of IL-3βR chain phosphorylation may explain the lower level of early gene induction in the TEL-JAK-expressing cells compared to TEL-ABL. Alternatively, interactions of TEL-ABL or TEL-JAK chimera with other signaling molecules known to participate to early gene induction20 21 could differ.
Finally, our results point to a pivotal role for Stat5 activation in the growth factor independence processes. Constitutive activation of STATs, particularly of Stat1, Stat3, and Stat5, is a recurrent observation in hematopoietic cell lines transformed by oncogenes such as v-abl, BCR-ABL, v-mpl, and in leukemogenic blood samples (review in Garcia and Jove4). Stat5 activation is observed in all transformed Ba/F3 cells lines, including those expressing the TEL-ABL (see Figure 3A) and TEL-PDGFβR fusions (unpublished data) and those expressing the various TEL-JAK fusion proteins, suggesting its direct contribution to cellular transformation. Experiments using a dominant negative Stat5A mutant (see Figure 5) strongly support this hypothesis. In this respect, Stat5 can be considered as the prototype of transcription factor implicated in all pleiotropic cytokine-induced cellular responses. It is activated by cytokines that regulate the proliferation and differentiation in all hematopoietic lineages (review in Pellegrini and Dusanter-Fourt3). Stat5 is assumed to regulate genes whose products are involved in the control of cellular growth,43of cell cycle progression including cyclins,44,45p21WAF1/CIP1,46 and possibly p2747 and of apoptosis.12,48,49 Furthermore, the expression of a mutated Stat5 is sufficient to allow growth factor independent proliferation of Ba/F3 cells.50
The constitutive activation of Stat5 factors is, therefore, likely to represent a crucial event in the activated tyrosine kinase-mediated leukemogenic process. The availability of Stat5A- and Stat5B-deficient mice will allow us to establish in vivo the functional role of these factors in the transforming properties of the fusion proteins.
Acknowledgments
The authors thank N. Perrimon for providing the Hopscotch cDNA and the LPH from the Hôpital Saint-Louis (Paris, France) for artwork.
Supported in part by grants from the Association contre le Cancer (ARC), the Ligue Nationale contre le Cancer (LNCC), and the Comité de Paris of the LNCC. R.M. and S.D. are supported by a Ministère de l'Education Nationale, de la Recherche et des Technologies (MENRT) fellowship.
Submitted May 27, 1999; accepted October 28, 1999.
Reprints:O. A. Bernard, U434 INSERM-CEPH, 27 rue Juliette Dodu, 75010 Paris, France; e-mail:olivier.bernard@cephb.fr.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal