Abstract
Polycythemia vera (PV) is a clonal stem cell disorder characterized by hyperproliferation of the erythroid, myeloid, and megakaryocytic lineages. Although it has been shown that progenitor cells of patients with PV are hypersensitive to several growth factors, the molecular pathogenesis of this disease remains unknown. To investigate the molecular defects underlying PV, we used subtractive hybridization to isolate complementary DNAs (cDNAs) differentially expressed in patients with PV versus normal controls. We isolated a novel gene, subsequently named PRV-1, which is highly expressed in granulocytes from patients with PV (n = 19), but not detectable in normal control granulocytes (n = 21). Moreover, PRV-1 is not expressed in mononuclear cells from patients with chronic myelogenous leukemia (n = 4) or acute myelogenous leukemia (n = 5) or in granulocytes from patients with essential thrombocythemia (n = 4) or secondary erythrocytosis (n = 4). Northern blot analysis showed that PRV-1 is highly expressed in normal human bone marrow and to a much lesser degree in fetal liver. It is not expressed in a variety of other tissues tested. Although PRV-1 is not expressed in resting granulocytes from normal controls, stimulation of these cells with granulocyte colony-stimulating factor induces PRV-1 expression. The PRV-1 cDNA encodes an open reading frame of 437 amino acids, which contains a signal peptide at the N-terminus and a hydrophobic segment at the C-terminus. In addition, PRV-1 contains 2 cysteine-rich domains homologous to those found in the uPAR/Ly6/CD59/snake toxin-receptor superfamily. We therefore propose that PRV-1 represents a novel hematopoietic receptor.
Polycythemia vera (PV) is 1 of 4 diseases termed the myeloproliferative disorders (MPDs).1 Besides PV, this group includes essential thrombocythemia (ET), idiopathic myelofibrosis (IMF), and chronic myelogenous leukemia (CML). All MPDs result from the clonal expansion of a mutant pluripotent hematopoietic stem cell.2-5 PV is characterized by an increased proliferation of all 3 myeloid lineages, which results in an excess production of mature red cells, granulocytes, and platelets.6 Because the disease results from the hyperproliferation of a single aberrant stem cell, the peripheral red blood cells, granulocytes, monocytes, and platelets are clonal in these patients.2 7-9 Although the molecular etiology of PV remains unknown, progress has recently been made in characterizing the malignant cells.
The PV cells are hypersensitive to several hematopoietic growth factors including interleukin-3 (IL-3), granulocyte/macrophage colony-stimulating factor (GM-CSF), stem cell factor (SCF), and thrombopoietin (TPO).10-13 However, regarding erythropoietin (EPO), a long debate had ensued whether PV cells are hypersensitive to or in fact independent of this growth factor. In elegant experiments using a novel serum-free medium devoid of any burst-promoting activity, Correa et al have recently shown that PV erythroid progenitor cells are independent of EPO.14 In addition, these cells are exquisitely hypersensitive to insulin-like growth factor-1 (IGF-1).14 Interestingly, patients with PV also show 4-fold elevated plasma levels of insulin-like growth factor binding protein-1 (IGFBP-1), which, together with IGF-1, stimulates erythroid burst formation in vitro.15
The observation that PV cells are hypersensitive to a large variety of growth factors suggests that signal transduction pathways may be altered in these cells. Moliterno and colleagues have recently reported that platelets from patients with PV displayed impaired tyrosine phosphorylation in response to TPO stimulation, whereas the response to thrombin remained intact.16 The TPO response was also deficient in patients with IMF, but not in patients with a variety of other hematopoietic diseases. The inability to transduce the TPO signal was due to a dramatic reduction or a complete absence of the TPO receptor, c-mpl, in 34 of 34 PV and 13 of 14 IMF patients.16 However, it is at present not clear how a loss of c-mpl expression could contribute either to the growth factor hypersensitivity of PV cells, or, more generally, to the molecular pathology of the disease.
An intriguing observation about PV cells was recently reported. Silva et al showed that PV erythroid precursor cells express the anti-apoptotic protein bcl-xL to a much higher proportion than normal precursor cells (21.8% versus 6.6%).17 In addition, in PV, more mature cells, which normally show no bcl-xL expression, still express high levels of the protein. Hematopoietic growth factors act in part by suppressing apoptosis. IGF-1, in particular, has been shown to suppress apoptosis of erythroid progenitors and myeloid cells.18 19 Thus, perhaps the observed growth factor hypersensitivity of PV cells results from an intrinsic protection from apoptosis, thereby requiring less protection through growth factor stimulation.
Despite these recent advances in characterizing the malignant PV clone, the molecular defect leading to the development of this disease remains unclear. In an attempt to find such a defect, we wished to define differences in gene expression between PV and normal cells. We used subtractive hybridization to clone complementary DNAs (cDNAs) that are either over- or underexpressed in PV cells. We report here the cloning of a novel hematopoietic cell surface receptor, which is strongly overexpressed in cells from 19 of 19 patients with PV and not expressed in 21 of 21 normal controls. We have named this novel receptor polycythemia rubra vera-1 (PRV-1).
Materials and methods
Patients
Peripheral blood samples were obtained from 19 patients with PV as well as 6 patients with ET, 4 patients with CML in chronic phase, 1 patient with IMF, 5 patients with acute myelogenous leukemia (AML), 4 patients with secondary erythrocytosis, and 21 healthy volunteers. The diagnosis of PV and ET was made according to the clinical and laboratory criteria established by the Polycythemia Vera Study Group.20 Patient characteristics are listed in Table1. The study protocol was approved by the local ethics committee and informed consent was obtained from all patients.
List of patients tested for PRV-1 expression
| Patient . | Sex . | Age (y) . | Duration of Disease . | Treatment . | LAP (normal: 10-100) . | Diagnosis . | PRV-1 Expression . |
|---|---|---|---|---|---|---|---|
| WZ | F | 75 | 2 y | Phl | nd | PV | +++ |
| EH | F | 68 | 12 y | Phl, HU, Bu | 84 | PV | +++ |
| HJL | M | 59 | 4 y | Phl | 200 | PV | +++ |
| SR | M | 62 | 7 y | Phl | nd | PV | +++ |
| UW | M | 65 | 10 y | Phl, HU | nd | PV | +++ |
| UD | F | 66 | < 1 y | Phl | 264 | PV | +++ |
| EM | F | 62 | 15 y | Phl, HU, Bu | nd | PV | +++ |
| HL | M | 62 | 2 y | Phl, HU | 400 | PV | +++ |
| HH | M | 65 | < 1 y | Phl | 76 | PV | +++ |
| RH | M | 54 | 4 y | Phl | nd | PV | +++ |
| RS | F | 38 | 14 y | Phl | nd | PV | +++ |
| FB | M | 74 | 10 y | Phl | nd | PV | +++ |
| EW | F | 62 | 16 y | Phl, HU, IFN-α | 192 | PV | +++ |
| SH | M | 43 | 6 y | Phl, ASS | nd | PV | +++ |
| DJ | M | 40 | 6 y | Phl | 105 | PV | +++ |
| IP | F | 60 | 3 y | Phl | 176 | PV | +++ |
| WA | F | 46 | 37 y | Phl | nd | PV | +++ |
| HS | F | 64 | 1 y | HU | 179 | PV | +++ |
| MT | M | 58 | 8 y | Phl, HU | 163 | PV | +++ |
| FW | M | 69 | 10 y | IFN-α | 134 | ET | +++ |
| RW | F | 39 | 4 y | HU, ASS Anag | 33 | ET | + |
| EW | M | 63 | 6 y | HU | nd | ET | − |
| LH | M | 60 | 9 y | HU | nd | ET | − |
| MS | F | 50 | 1 y | HU | 123 | ET | − |
| HZ | F | 73 | 11 y | HU, ASS IFN-α | 2 | ET | − |
| HUB | M | 60 | 12 y | HU, AraC | nd | CML | − |
| BE | F | 80 | 3 y | HU | nd | CML | − |
| RW | F | 54 | 1 y | None | nd | CML | − |
| XR | M | 44 | at dx | None | nd | CML | − |
| HJH | M | 64 | < 1 y | Erythrocyte transfusion | nd | IMF | + |
| VW | M | 21 | 2 y | Multiple polychemo Allo BMT | nd | AML FAB M4 | − |
| FK | F | 78 | at dx | None | nd | AML FAB M1 | − |
| EF | M | 45 | < 1 y | Allo BMT | nd | AML FAB M5 | − |
| VF | M | 61 | at dx | None | nd | AML FAB M5b | − |
| UM | F | 56 | at dx | None | nd | AML FAB M1 | − |
| JMPM | M | 41 | at dx | None | nd | Erythrocytosis | − |
| RB | M | 46 | at dx | None | nd | Erythrocytosis | − |
| JP | M | 78 | at dx | None | nd | Erythrocytosis | − |
| YK | M | 52 | at dx | None | nd | Erythrocytosis | − |
| Patient . | Sex . | Age (y) . | Duration of Disease . | Treatment . | LAP (normal: 10-100) . | Diagnosis . | PRV-1 Expression . |
|---|---|---|---|---|---|---|---|
| WZ | F | 75 | 2 y | Phl | nd | PV | +++ |
| EH | F | 68 | 12 y | Phl, HU, Bu | 84 | PV | +++ |
| HJL | M | 59 | 4 y | Phl | 200 | PV | +++ |
| SR | M | 62 | 7 y | Phl | nd | PV | +++ |
| UW | M | 65 | 10 y | Phl, HU | nd | PV | +++ |
| UD | F | 66 | < 1 y | Phl | 264 | PV | +++ |
| EM | F | 62 | 15 y | Phl, HU, Bu | nd | PV | +++ |
| HL | M | 62 | 2 y | Phl, HU | 400 | PV | +++ |
| HH | M | 65 | < 1 y | Phl | 76 | PV | +++ |
| RH | M | 54 | 4 y | Phl | nd | PV | +++ |
| RS | F | 38 | 14 y | Phl | nd | PV | +++ |
| FB | M | 74 | 10 y | Phl | nd | PV | +++ |
| EW | F | 62 | 16 y | Phl, HU, IFN-α | 192 | PV | +++ |
| SH | M | 43 | 6 y | Phl, ASS | nd | PV | +++ |
| DJ | M | 40 | 6 y | Phl | 105 | PV | +++ |
| IP | F | 60 | 3 y | Phl | 176 | PV | +++ |
| WA | F | 46 | 37 y | Phl | nd | PV | +++ |
| HS | F | 64 | 1 y | HU | 179 | PV | +++ |
| MT | M | 58 | 8 y | Phl, HU | 163 | PV | +++ |
| FW | M | 69 | 10 y | IFN-α | 134 | ET | +++ |
| RW | F | 39 | 4 y | HU, ASS Anag | 33 | ET | + |
| EW | M | 63 | 6 y | HU | nd | ET | − |
| LH | M | 60 | 9 y | HU | nd | ET | − |
| MS | F | 50 | 1 y | HU | 123 | ET | − |
| HZ | F | 73 | 11 y | HU, ASS IFN-α | 2 | ET | − |
| HUB | M | 60 | 12 y | HU, AraC | nd | CML | − |
| BE | F | 80 | 3 y | HU | nd | CML | − |
| RW | F | 54 | 1 y | None | nd | CML | − |
| XR | M | 44 | at dx | None | nd | CML | − |
| HJH | M | 64 | < 1 y | Erythrocyte transfusion | nd | IMF | + |
| VW | M | 21 | 2 y | Multiple polychemo Allo BMT | nd | AML FAB M4 | − |
| FK | F | 78 | at dx | None | nd | AML FAB M1 | − |
| EF | M | 45 | < 1 y | Allo BMT | nd | AML FAB M5 | − |
| VF | M | 61 | at dx | None | nd | AML FAB M5b | − |
| UM | F | 56 | at dx | None | nd | AML FAB M1 | − |
| JMPM | M | 41 | at dx | None | nd | Erythrocytosis | − |
| RB | M | 46 | at dx | None | nd | Erythrocytosis | − |
| JP | M | 78 | at dx | None | nd | Erythrocytosis | − |
| YK | M | 52 | at dx | None | nd | Erythrocytosis | − |
M indicates male; F, female; age: age at time of blood sampling; duration of disease, expressed in years (y) at time of blood sampling; at dx, at the time of diagnosis; treatment, treatment prior to or at the time of blood sampling; Phl, phlebotomy; HU, hydroxyurea; ASS, salicylic acid (aspirin); Bu, busulfan; INF-α, interferon alpha; Anag, anagrelide; AraC, cytosine arabinoside; Allo BMT, allogenic bone marrow transplant; LAP, leukocyte alkaline phosphatase, normal range 10-100; nd, not determined; PV, polycythemia vera; ET, essential thrombocythemia; CML, chronic myelogenous leukemia; AML, acute myelogenous leukemia; erythrocytosis, secondary erythrocytosis; PRV-1 expression: +++ indicates strong PRV-1 expression, + indicates weak PRV-1 expression, − indicates no PRV-1 expression.
Separation of cells
Heparinized blood from patients and healthy controls was used as a source of granulocytes and mononuclear cells. Granulocytes were purified by dextran sedimentation followed by Ficoll-Paque (Pharmacia Biotech, Uppsala, Sweden) separation.21Erythrocytes were eliminated by hypotonic lysis (0.2% NaCl for 30 seconds). This method consistently yielded granulocyte preparations with more than 98% purity as judged by visual inspection of Wright-Giemsa-stained slide preparations. Peripheral blood mononuclear cells from patients with AML or CML were isolated by Ficoll-Paque separation.
RNA isolation and Northern blots
Total cellular RNA was harvested using an acidic phenol extraction (Trizol, GIBCO/BRL, Rockville, MD). Either 5 μg or 10 μg of RNA was analyzed in a Northern blot. Alternatively, Multiple Tissue Northern Blots (Clontech, Palo Alto, CA) were used. The blots were hybridized in ExpressHyb Hybridization Solution (Clontech) at 68°C. PRV-1 and actin cDNAs were labeled using the Prime-It-II labeling kit (Stratagene, La Jolla, CA) and α-32P-dCTP (Amersham, Uppsala, Sweden). The blots were washed 3 times for 10 minutes in 2× sodium chloride sodium citrate (SSC), 0.05% sodium dodecyl sulfate (SDS) at room temperature and twice at 50°C for 20 minutes in 0.1 × SSC, 0.1% SDS.
After the first hybridization, membranes were reprobed with a 1.2-kb Pstl-fragment of the human beta-actin gene to control for equal loading of the RNA.
Isolation of poly(A)+RNA and subtractive hybridization
Poly(A)+RNA was isolated using the Fast Track 2.0 Kit (Invitrogen, Carlsbad, CA). Double-stranded cDNA was synthesized using the PCR-Select cDNA Subtraction Kit (Clontech) at the manufacturer's recommendation. Subtractive hybridization was performed with cDNA from PV granulocytes pooled from 5 patients (MT, WZ, IP, FB, and DJ, Table1) and cDNA from normal control granulocytes using the PCR-Select cDNA Subtraction Kit (Clontech) according to the manufacturer's recommendation. The subtracted cDNA was cloned directly into the pCR 2.1 vector (TA Cloning Kit, Invitrogen).
Western blots
Total cell extracts were prepared using a high-salt detergent buffer (Totex) as previously described.22 Cell extracts (30 μg) were boiled in Laemmli sample buffer and subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting as described.22 Primary polyclonal antibodies were raised by injection of 2 hapten coupled synthetic peptides into rabbits. These peptides encode amino acids 13 to 25 and 368 to 383 of the predicted mature PRV-1. The purified rabbit serum was used at a dilution of 1:500. Bound antibody was decorated with peroxidase conjugated secondary antibody (goat antirabbit IgG, Amersham). The immunocomplexes were detected using ECL Western blotting reagents (Amersham). Exposure to Kodak XAR-5 films was performed for 5 to 10 seconds.
Immunohistochemistry
The bone marrow biopsy was decalcified, paraffin embedded, and stained as previously described.23-25 Serial sections were stained as follows: (1) with a mouse polyclonal antiserum raised against native PRV-1 by DNA vaccination (Genovac GmbH, Freiburg, Germany); for vaccination, the 1.4-kb PRV-1 coding region cloned into pCDNA3.1 (Invitrogen) was used; (2) enzymatically for naphthol-AS-D-chloroacetate esterase; or (3) with an antibody against hemoglobin (DAKO, Hamburg, Germany).
Results
Cloning of PRV-1
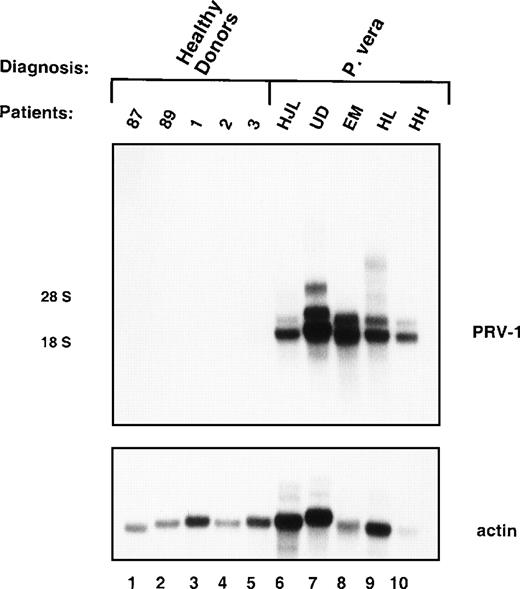
Messenger RNA (mRNA) from granulocytes of 5 PV patients (MT, WZ, IP, FB, and DJ, Table 1) was compared to normal granulocyte mRNA in a subtractive hybridization. Five clones were obtained, which encoded different overlapping fragments of the same cDNA. Northern blot hybridization to granulocyte RNA from 5 patients with PV and 5 healthy volunteers showed that this cDNA hybridized to 2 distinct RNAs, 2.1 kb and 3.1 kb in size, in all 5 PV patients, but showed no hybridization to normal granulocyte RNA (Figure 1). RNA from a total of 19 patients with PV and 21 normal controls has been analyzed to date. In Northern blots, peripheral blood granulocytes from all 19 PV patients displayed strong hybridization to the cDNA probe, whereas none of the controls showed a detectable signal (data not shown).
Expression of PRV-1 in peripheral blood granulocytes from patients with PV and normal controls.
Granulocytes were isolated from peripheral blood to more than 98% purity. Total RNA was extracted and a Northern blot prepared using 10 μg RNA. Lanes 1-5 healthy volunteer donors, lanes 6-10 patients with PV. (Top) The membrane was probed with a 1.1-kb fragment of the PRV-1 cDNA. The positions of the 18S and 28S ribosomal RNAs are indicated. (Bottom) The membrane was stripped and reprobed with a 1.2-kb fragment of the human β-actin cDNA.
Expression of PRV-1 in peripheral blood granulocytes from patients with PV and normal controls.
Granulocytes were isolated from peripheral blood to more than 98% purity. Total RNA was extracted and a Northern blot prepared using 10 μg RNA. Lanes 1-5 healthy volunteer donors, lanes 6-10 patients with PV. (Top) The membrane was probed with a 1.1-kb fragment of the PRV-1 cDNA. The positions of the 18S and 28S ribosomal RNAs are indicated. (Bottom) The membrane was stripped and reprobed with a 1.2-kb fragment of the human β-actin cDNA.
Structure of the PRV-1 protein
From 3 clones, a complete cDNA was constructed. (The nucleotide sequence has been submitted to the GenBank/EBI Data Bank with the accession number AF 146747.) Homology searches with several databases revealed that the sequence represents a novel previously uncharacterized cDNA, which we subsequently named polycythemia rubra vera-1 (PRV-1). This sequence encodes an open reading frame of 437 amino acids (Figure 2), which contains the following features: (1) an N-terminal signal sequence of 21 amino acids, (2) 2 highly homologous cysteine-rich domains of 188 amino acids, and (3) a highly hydrophobic C-terminal sequence.
The protein sequence of PRV-1.
The 2 homologous cysteine-rich regions are shown aligned to each other. Identical amino acids are shown in red, conserved cysteine residues in bold red. Similar amino acids are depicted in blue. The position of the leader peptide, the carboxy-terminal hydrophobic region as well as the ω-sites, potential sites of GPI-anchor attachment, are shown. The postulated cysteine-disulfide bridges are indicated by brackets.
The protein sequence of PRV-1.
The 2 homologous cysteine-rich regions are shown aligned to each other. Identical amino acids are shown in red, conserved cysteine residues in bold red. Similar amino acids are depicted in blue. The position of the leader peptide, the carboxy-terminal hydrophobic region as well as the ω-sites, potential sites of GPI-anchor attachment, are shown. The postulated cysteine-disulfide bridges are indicated by brackets.
The presence of a signal peptide suggests that the protein is imported into the endoplasmic reticulum and destined either for insertion into the plasma membrane or for secretion from the cell.
The 2 cysteine-rich domains show homology to the previously described uPAR-domains, found in cell surface receptors of the uPAR/Ly6/CD59/snake toxin-family of proteins (Figure3).26 These domains contain 8 to 10 cysteine residues spaced at conserved distances from each other.28 PRV-1 contains 6 of these cysteines with the conserved spacing (Figure 3).
Alignment of PRV-1 to members of the uPAR/Ly6/CD59/snake toxin family.
The protein sequences were retrieved from the Swissprot database and aligned to PRV-1 according to the model of Kieffer et al.27 Identical amino acids are shown in red, conserved cysteine residues in bold red. Similarities shared only between PRV-1 and the snake toxin family are shown in blue. Numbers preceding the sequences indicate the first amino acid aligned.
Alignment of PRV-1 to members of the uPAR/Ly6/CD59/snake toxin family.
The protein sequences were retrieved from the Swissprot database and aligned to PRV-1 according to the model of Kieffer et al.27 Identical amino acids are shown in red, conserved cysteine residues in bold red. Similarities shared only between PRV-1 and the snake toxin family are shown in blue. Numbers preceding the sequences indicate the first amino acid aligned.
The C-terminal hydrophobic sequence could encode a transmembrane domain. However, the 12 amino acids predicted to form the transmembrane domain are too short for spanning the membrane and would leave a cytoplasmic tail of only 7 amino acids. All members of the uPAR/Ly6/CD59 family of proteins described so far are attached to the membrane via a glycosyl phosphatidylinositol anchor (GPI-anchor).28-31 It seems likely, therefore, that the hydrophobic sequence at the C-terminus of PRV-1 encodes a signal for the addition of a GPI-link.
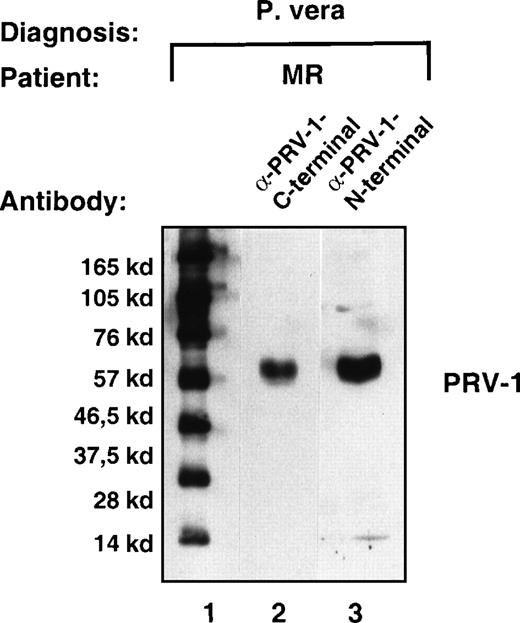
To verify that the polypeptide predicted from the amino acid translation of the cDNA was indeed synthesized in PV cells, we raised antibodies against 2 peptides in the predicted primary sequence. These peptides encode amino acids 13 to 25 and 368 to 383 of the predicted mature PRV-1. Total cell extracts were prepared from purified granulocytes of a PV patient (MR). The extracts were separated by SDS-PAGE and subjected to Western blotting with antibodies raised against the 2 peptides. Both antibodies recognized a single polypeptide of approximately 60 kd (Figure 4, lanes 2 and 3). The apparent molecular weight of the detected polypeptide is 14 kd larger than the weight calculated from the amino acid sequence. This observation can be explained by the existence of 3 potential glycosylation sites in the PRV-1 sequence (N-46, N-189, and N-382). The addition of sugar residues could account for the additional weight observed. Many GPI-linked proteins including uPAR are modified by glycosylation.28
Expression of the PRV-1 protein in granulocytes from a patient with PV.
Granulocytes were isolated from peripheral blood to more than 98% purity. Total cell extracts were prepared and subjected to SDS-PAGE. The proteins were blotted onto a nylon membrane and decorated with either the antibody raised against the C-terminal peptide (amino acids 368-383, lane 2) or the antibody raised against the N-terminal peptide (amino acids 13-25, lane 3). Immune complexes were detected by decoration with peroxidase-conjugated secondary antibody and visualized by chemoluminescence. Lane 1 shows a molecular size marker. The film was exposed for 5 seconds.
Expression of the PRV-1 protein in granulocytes from a patient with PV.
Granulocytes were isolated from peripheral blood to more than 98% purity. Total cell extracts were prepared and subjected to SDS-PAGE. The proteins were blotted onto a nylon membrane and decorated with either the antibody raised against the C-terminal peptide (amino acids 368-383, lane 2) or the antibody raised against the N-terminal peptide (amino acids 13-25, lane 3). Immune complexes were detected by decoration with peroxidase-conjugated secondary antibody and visualized by chemoluminescence. Lane 1 shows a molecular size marker. The film was exposed for 5 seconds.
Because the structure of PRV-1 suggests that it is expressed on the cell surface, we co-stained granulocytes of a PV patient with an antibody against native PRV-1 and an antibody against CD13, a myeloid marker. Fluorescence-activated cell sorter analysis revealed that all PV granulocytes express PRV-1 on their surface (data not shown).
PRV-1 expression in MPDs
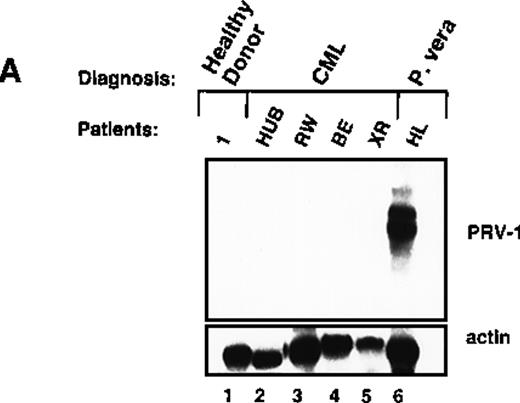
Because PV is 1 of 4 diseases collectively termed the MPDs, we investigated whether overexpression of PRV-1 is unique to PV or whether it also occurs in the other MPDs. Peripheral blood mononuclear cells were purified from 4 patients with chronic phase CML. Morphologically, these cells represent various stages of granulocyte precursors. RNA was prepared and hybridized in a Northern blot with a 1.1-kb fragment of the PRV-1 cDNA (Figure 5A). There was no PRV-1 expression in CML mononuclear cells from 4 different patients (Figure 5A). Hybridization to a β-actin cDNA confirmed the presence of similar amounts of RNA in all samples.
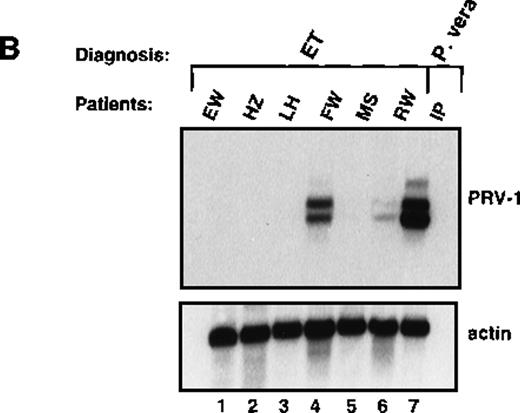
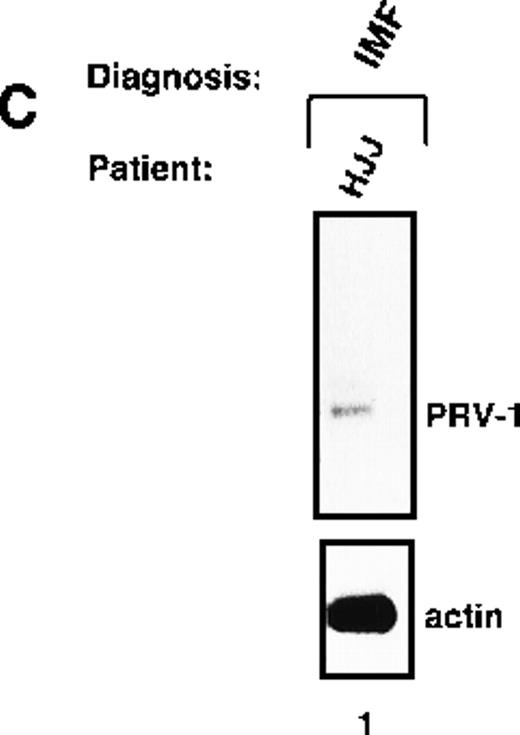
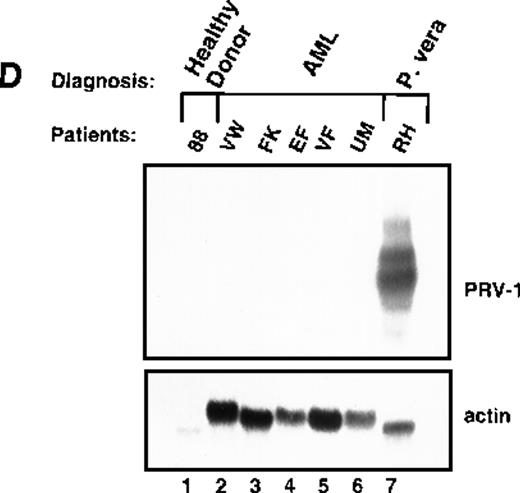
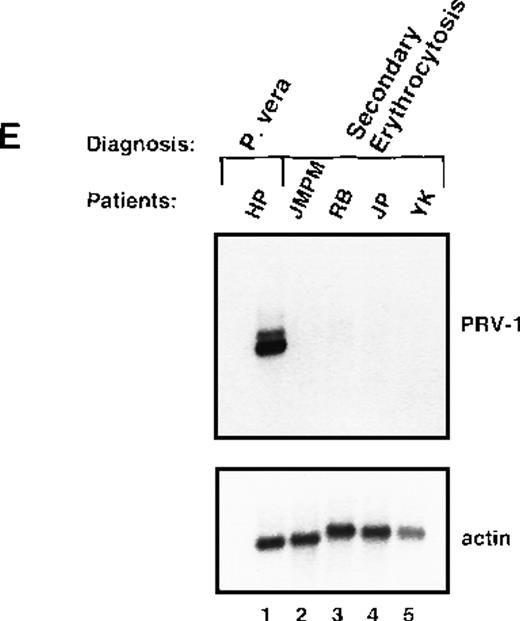
Expression of PRV-1 in other MPDs.
Mononuclear cells were isolated from peripheral blood of patients with CML (A) or AML (D). Granulocytes were isolated from peripheral blood of patients with ET (B), IMF (C), secondary erythrocytosis (E), PV (A, B, D, and E) or healthy volunteers (A and D). Total RNA was extracted and Northern blots prepared using 10 μg RNA. (Top) The membrane was probed with a 1.1-kb fragment of the PRV-1 cDNA. (Bottom) The membrane was stripped and reprobed with a 1.2-kb fragment of the human β-actin cDNA.
Expression of PRV-1 in other MPDs.
Mononuclear cells were isolated from peripheral blood of patients with CML (A) or AML (D). Granulocytes were isolated from peripheral blood of patients with ET (B), IMF (C), secondary erythrocytosis (E), PV (A, B, D, and E) or healthy volunteers (A and D). Total RNA was extracted and Northern blots prepared using 10 μg RNA. (Top) The membrane was probed with a 1.1-kb fragment of the PRV-1 cDNA. (Bottom) The membrane was stripped and reprobed with a 1.2-kb fragment of the human β-actin cDNA.
Likewise, granulocytes were purified from 6 patients with ET and the RNA hybridized to the PRV-1 cDNA (Figure 5B). Four of the patients showed no PRV-1 expression (Figure 5B, lanes 1, 2, 3, and 5). Two patients (FW and RW, Figure 5B, lanes 4 and 6) showed PRV-1 expression, 1 (RW) weakly, the other (FW) strongly.
Peripheral granulocytes were isolated from 1 patient with IMF. This patient showed weak PRV-1 expression in a Northern blot (Figure 5C).
White blood cells from patients with PV frequently display a “left shift,” morphologically less mature cells appear in the peripheral blood. If PRV-1 is normally expressed on immature granulocytes, but its expression is down-regulated during maturation, its expression on PV cells could simply result from the immaturity of the cells. The observation that PRV-1 is not found on CML-mononuclear cells, which represent all stages of granulocytic differentiation, argues against this hypothesis. Nevertheless, we wished to investigate additional immature myeloid cells. We therefore isolated peripheral blast cells from 5 patients with AML. RNA was prepared and analyzed for PRV-1 expression in a Northern blot (Figure 5D). None of the 5 AML patients showed any PRV-1 expression.
Polycythemic states are classified as either primary conditions, which include PV, or as secondary conditions. The latter category is also called secondary erythrocytosis and describes reactive processes. We investigated whether PRV-1 expression discriminates between PV and secondary erythrocytosis by analyzing PRV-1 expression in granulocytes from 4 patients with secondary erythrocytosis. None of these patients showed PRV-1 expression (Figure 5E). In addition, we investigated whether PRV-1 expression correlates with an elevated leukocyte alkaline phosphatase (LAP) score. Patients with PV frequently display elevated LAP scores, whereas the LAP is decreased in patients with CML. We observed no correlation between LAP score and PRV-1 expression. Three patients who express PRV-1 (HH, RW, and EH; Figure 1, lane 10, Figure5B, lane 6, and Table 1) have LAP scores in the normal range (10-100). Conversely, 1 patient who does not express PRV-1 (MS, Figure 5B, lane 5) has an elevated LAP score. In fact, her LAP score is higher than the score of patient DJ, who does express PRV-1. We therefore conclude that PRV-1 expression does not correlate with an elevated LAP score.
PRV-1 expression in normal tissues
We wished to determine the expression pattern of PRV-1 in normal tissues. A Northern blot containing 2 μg mRNA from various hematopoietic tissues was probed with the PRV-1 cDNA (Figure6). PRV-1 expression was very strong in bone marrow and a slight expression was detected in fetal liver (Figure6, lanes 5 and 6). The other tissues including spleen, lymph node, thymus, and peripheral blood leukocytes did not express PRV-1 (Figure6, lanes 1-4). In addition, several other tissues including heart, brain, kidney, testis, ovary, small intestine, and skeletal muscle did not express PRV-1 (data not shown). These data indicate that PRV-1 is selectively expressed in human bone marrow.
Expression of PRV-1 in hematopoietic tissues.
(Top) A Northern blot containing 2 μg of polyA+RNA from the indicated tissues (Clontech) was probed with a 1.1-kb fragment of the PRV-1 cDNA. (Bottom) The membrane was stripped and reprobed with a 1.2-kb fragment of the human β-actin cDNA.
Expression of PRV-1 in hematopoietic tissues.
(Top) A Northern blot containing 2 μg of polyA+RNA from the indicated tissues (Clontech) was probed with a 1.1-kb fragment of the PRV-1 cDNA. (Bottom) The membrane was stripped and reprobed with a 1.2-kb fragment of the human β-actin cDNA.
PRV-1 expression in bone marrow
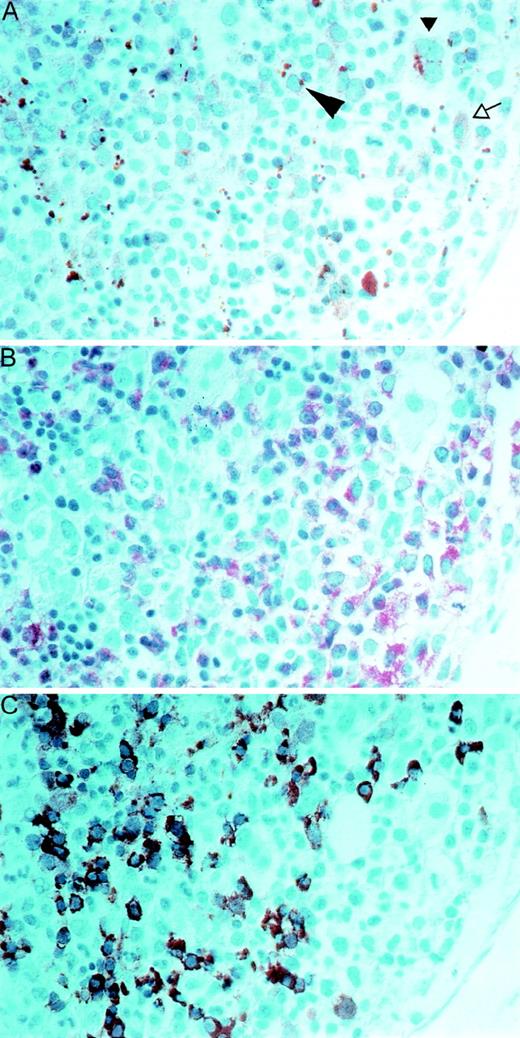
To specify which cell types in the bone marrow express PRV-1, we performed immunohistochemistry (Figure 7). Paraffin sections of a bone marrow biopsy from a patient with PV were stained with a polyclonal antibody directed against the native PRV-1 protein (Figure 7, panel A). In addition, serial sections were stained for naphthol-AS-D-chloroacetate esterase (see Figure 7, NACE, panel B) and with an antibody against hemoglobin (Figure 7, panel C). Comparison of these differently stained serial sections allows identification of the PRV-1–positive cells. The anti-PRV-1 antibody stains early erythroblasts, which have a basophilic cytoplasm, as well as megakaryocytes, promyelocytes, and myelocytes. In early erythrocytes, the anti-PRV antibody stains a solitary paranuclear apparently globular structure, which is located at a site similar to the Golgi zone (large arrowhead, Figure 7, panel A).
PRV-1 expression is stimulated by granulocyte colony-stimulating factor and GM-CSF
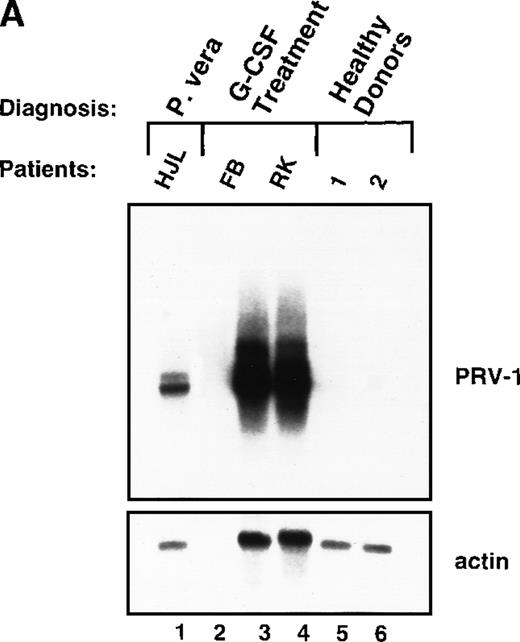
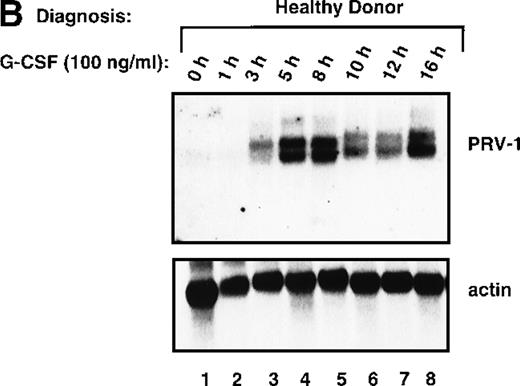
In PV, bone marrow progenitor cells are hyperproliferating, thereby producing the elevated cell counts recorded in the peripheral blood. A hyperproliferation of the myeloid lineage, however, can also be induced in healthy volunteers, for example, when these individuals act as donors for peripheral stem cell transplantation. To increase the number of circulating hematopoietic progenitor cells, these donors are stimulated with 10 μg/kg body weight of recombinant human G-CSF twice daily for 4 days. We investigated whether hyperproliferation of the bone marrow in these healthy donors also leads to PRV-1 expression in the peripheral granulocytes. Granulocytes were purified from 2 individuals treated with granulocyte colony-stimulating factor (G-CSF) and the RNA analyzed by Northern blot (Figure8A). Both G-CSF–treated donors showed a very strong PRV-1 expression (Figure 8A, lanes 3 and 4). This observation prompted us to investigate whether PRV-1 expression can be stimulated by G-CSF in normal, mature peripheral granulocytes as well. Granulocytes from healthy volunteer donors were purified and treated in vitro with 100 ng/mL G-CSF for various times. PRV-1 expression was analyzed by Northern blot (Figure 8B). Stimulation with G-CSF for as little as 3 hours induced PRV-1 expression in mature granulocytes. Interestingly, the larger 3.1 kb mRNA appeared first, after 3 hours of stimulation (Figure 8B, lane 3), whereas the smaller 2.1 kb mRNA was only seen after 5 hours of G-CSF stimulation (Figure 8B, lane 4). These 2 RNA species arise from alternative polyadenylation (data not shown). Stimulation of normal peripheral granulocytes with 100 ng/mL GM-CSF also induces PRV-1 expression with kinetics similar to G-CSF (data not shown).
Immunohistochemical staining of paraffin sections of a bone marrow biopsy from a patient with PV.
Serial sections were stained with a polyclonal antibody to native PRV-1 (panel A), to naphthol-AS-D-chloroacetate esterase (NACE, panel B), or with an antibody to hemoglobin (panel C). In panel A, the large arrowhead points to an early erythroblast, the small arrowhead points out a megakaryocyte, and the open arrow shows a promyelocyte. A 1:400 magnification is shown.
Immunohistochemical staining of paraffin sections of a bone marrow biopsy from a patient with PV.
Serial sections were stained with a polyclonal antibody to native PRV-1 (panel A), to naphthol-AS-D-chloroacetate esterase (NACE, panel B), or with an antibody to hemoglobin (panel C). In panel A, the large arrowhead points to an early erythroblast, the small arrowhead points out a megakaryocyte, and the open arrow shows a promyelocyte. A 1:400 magnification is shown.
Expression of PRV-1 in granulocytes from healthy volunteers treated with G-CSF in vivo and in vitro.
Granulocytes were isolated from peripheral blood of untreated and G-CSF-treated healthy volunteers and a patient with PV. Total RNA was extracted and a Northern blot prepared using 10 μg RNA. (A) Lane 1 patient with PV, lane 2 empty, lanes 3 and 4 healthy volunteers treated with G-CSF in vivo, and lanes 5 and 6 untreated healthy volunteer donors. (B) Lane 1 unstimulated granulocytes; lanes 2-8, granulocytes stimulated in vitro for the indicated times with 100 ng/mL G-CSF. (Top) The membrane was probed with a 1.1-kb fragment of the PRV-1 cDNA. (Bottom) The membrane was stripped and reprobed with a 1.2 kb fragment of the human β-actin cDNA.
Expression of PRV-1 in granulocytes from healthy volunteers treated with G-CSF in vivo and in vitro.
Granulocytes were isolated from peripheral blood of untreated and G-CSF-treated healthy volunteers and a patient with PV. Total RNA was extracted and a Northern blot prepared using 10 μg RNA. (A) Lane 1 patient with PV, lane 2 empty, lanes 3 and 4 healthy volunteers treated with G-CSF in vivo, and lanes 5 and 6 untreated healthy volunteer donors. (B) Lane 1 unstimulated granulocytes; lanes 2-8, granulocytes stimulated in vitro for the indicated times with 100 ng/mL G-CSF. (Top) The membrane was probed with a 1.1-kb fragment of the PRV-1 cDNA. (Bottom) The membrane was stripped and reprobed with a 1.2 kb fragment of the human β-actin cDNA.
Discussion
We describe the cloning of a novel cell surface receptor, named PRV-1 (polycythemia rubra vera-1), which under physiologic conditions is selectively expressed in human bone marrow. The amino acid sequence of PRV-1 shows that it is a novel protein most closely related to members of the uPAR/Ly6/CD59 family of cell surface receptors. Overall amino acid identity between members of this family is low, ranging between 20% and 30%.30 However, the family is defined by the presence of cysteine-rich domains, which contain 8 to 10 cysteine residues spaced at conserved distances.28 PRV-1 contains 2 cysteine-rich domains that are highly homologous to each other, showing 42% identity over 146 amino acids (Figure 2). In each of these domains, 6 cysteines are spaced precisely like those found in the uPAR domains (Figure 3). At first sight, the remaining cysteines found in the classical uPAR domain appear to be missing. However, all uPAR domains described to date contain a “signature sequence,” a conserved sequence found around the last 2 C-terminal cysteine residues of the domain (cysteines 7 and 8, or 9 and 10, depending on the number of cysteines in the domain).26 32 This signature sequence consists of 7 residues with the sequence CXXDXCN. This sequence is also found twice in PRV-1 (amino acids 104-111 and amino acids 294-300; amino acids 104-111 contain 1 additional amino acid resulting in the sequence CXXXDXCN). However, instead of occurring at the C-terminus of the cysteine-rich domain, where this sequence is found in all other uPAR/Ly6/CD59 family members, in PRV-1 it is found N-terminal to the other cysteines.
The genomic structure of several uPAR genes has been elucidated.32-34 In all cases, the uPAR domain is encoded by 2 separate exons, an intron occurring in the middle of the domain. We have determined the genomic structure of PRV-1 and found that the cysteine-rich domains are also encoded by 2 separate exons (data not shown). When the 2 exons encoding the PRV-1 cysteine- rich domains and the 2 exons encoding the first domain of uPAR (exons 3 and 4 of uPAR) are compared, the N-terminal PRV-1 exon is similar to uPAR exon 4, whereas the C-terminal PRV-1 exon is similar to uPAR exon 3. It appears therefore, that an exon switching has occurred in PRV-1, such that the cysteines surrounded by the signature sequence are now located N-terminal to the other cysteines.
In the first, N-terminal domain of the uPAR receptor, the precise location of the disulfide bonds has been elucidated.28 It has been shown that the 1st and the 5th cysteines, the 2nd and the 3rd cysteines, and the 4th and the 6th cysteines in this domain form disulfide bridges. Furthermore, cysteine 7 and 8, which are surrounded by the signature sequence, have also been shown to bind in the first uPAR domain.28 Therefore, the relocation of these 2 cysteines to the N-terminus of the PRV-1 domain may not disrupt disulfide bridge formation (Figure 2). However, it is likely that the tertiary structure formed by the PRV-1 domain is different from that of other uPAR/Ly6/CD59 domains.
Members of the uPAR/Ly6/CD59 family of receptors fulfill a diverse set of functions. For example, CD59 protects cells from autologous lysis by binding the C5-C8 complex, thereby preventing the formation of a membrane attack complex.35,36 TSA, the thymic shared antigen, plays a role during the positive selection of T cells and lineage commitment to the CD4 or CD8 pathways.30 The recently cloned RoBo-1 protein, which is expressed selectively in bone, has been implicated in bone growth and remodeling.37Although these receptors are tethered to the cell membrane via a lipid anchor, they nonetheless participate actively in signal transduction.38 Several groups have shown that uPAR interacts with and can activate protein tyrosine kinases including hck, fyn, lyn, fgr, and lck.39-41 In addition, uPAR has been shown to activate the JAK/STAT pathway.40 42 It is therefore likely that PRV-1 also mediates signal transduction in bone marrow cells.
The first cysteine-rich domain of uPAR (uPAR-1), which shares the greatest homology with the PRV-1 domains, because it also contains only 8 cysteine residues, has been shown to contain the ligand binding activity of uPAR.43 uPAR binds its ligand, uPA, via an epithelial growth factor–like domain (EGF domain) within uPA.44 It is therefore possible that the ligand for PRV-1 may also contain an EGF domain, perhaps also constitutes a growth factor. We are currently conducting experiments to identify the PRV-1 ligand.
This study demonstrated that PRV-1 is overexpressed in the peripheral blood granulocytes of 19 of 19 patients with PV and not expressed in 21 of 21 healthy controls (Figure 1 and Table 1). Moreover, PRV-1 is not expressed in 4 of 4 patients with CML (Figure 5A), 5 of 5 patients with AML (Figure 5D), or 4 of 4 patients with secondary erythrocytosis (Figure 5E). PRV-1 expression was detected in the single patient with IMF (Figure 5C) and 2 of 6 patients with ET (Figure 5B). In a bone marrow biopsy, the patient with IMF displayed hyperplastic erythropoiesis and granulopoiesis as well as increased numbers of megakaryocytes. Thus, all 3 lineages appear to be hyperproliferating in this patient. Interestingly, both patients with ET positive for PRV-1 had slightly elevated peripheral leukocyte counts at the time of blood sampling. Patient RW had 13 000 leukocytes/μL on the day of sampling and had consistently shown leukocyte counts exceeding 12 000/μL for the past 4 months. She had shown detectable levels of PRV-1 mRNA on Northern blots in 3 different blood samples during this time. Patient FW had 17 000 leukocytes/μL on the day of blood sampling. The other 4 patients with ET consistently showed leukocyte counts below 10 000/μL.
It has been previously reported that some patients who initially present with thrombocytosis but normal hematocrit and are therefore diagnosed with ET, subsequently progress to PV.45 Shih and Lee have reported that of 30 patients presenting with idiopathic marked thrombocytosis (platelets > 1000 × 109/L) and a normal or reduced hemoglobin, 11 were found to fulfill the diagnostic criteria for PV 2 to 45 months after initial evaluation.45At the initial presentation, the bone marrow from these 11 patients had shown endogenous erythroid colony formation in the absence of added erythropoietin. Shih and Lee have therefore proposed that endogenous colony formation may be used in the early identification of PV.45 It remains to be seen whether patients PW and RW progress to a diagnosis of PV. If this is the case, PRV-1 overexpression may likewise prove helpful in distinguishing ET from PV. A sensitive reverse transcriptase–polymerase chain reaction assay for PRV-1 has been established, which can be used to screen for PRV-1 overexpression. Together with clinical data, PRV-1 expression may thus be useful in establishing a diagnosis of PV. The consistency with which this receptor is overexpressed in PV suggests that it may play a role in the pathophysiology of this MPD.
Acknowledgments
The authors would like to thank Brigitte Schneider and Gesa Santos for excellent technical assistance. We gratefully acknowledge very helpful and stimulating discussions with Prof Dr J. Prchal and Dr F. Schriever. Our sincere thanks go to Prof Dr K. Geiger for his continuing support. A very special thank you to the helpful staff of the hematology clinic and the therapy ward. Thanks also to the Photo Center at the University Hospital Freiburg for excellent photographic support. This article is dedicated to Irmgard Pahl, mother of the senior author; her diagnosis with polycythemia vera sparked this research.
Reprints:Heike L. Pahl, Department of Experimental Anaesthesiology, University Hospital Freiburg, Center for Tumor Biology, PO Box 1120, 79106 Freiburg, Germany; e-mail:pahl@uni-freiburg.de.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal