Abstract
Some chromosomal translocations in acute leukemias involve the fusion of the trithorax-related protein Mll (also called HRX, All1, Htrx,) with a variety of heterologous proteins. In acute lymphoblastic leukemia associated with the t(4;11)(q21;q23) translocation, the4q21 gene that fuses with Mll is AF4. To gain insight into the potential role of AF4 in leukemogenesis and development, this gene was inactivated by homologous recombination in mice. As expected from the tissue distribution of the AF4 transcript, development of both B and T cells is affected in AF4 mutant mice. A severe reduction of the thymic double positive CD4/CD8 (CD4+/CD8+) population was observed; in addition most double- and single-positive cells expressed lower levels of CD4 and CD8 coreceptors. Most importantly, the reconstitution of the double-positive compartment by expansion of the double-negative cell compartment was severely impaired in these mutant mice. In the bone marrow pre-B and mature B-cell numbers are reduced. These results demonstrate that the function of the mAF4 gene is critical for normal lymphocyte development. This raises the possibility that the disruption of the normal AF4 gene or its association with Mll function by translocation may orient the oncogenic process toward the lymphoid lineage. This represents the first functional study using a knock-out strategy on one of the Mll partner genes in translocation-associated leukemias.
The most common chromosome abnormality among infants with acute lymphoblastic leukemia is a t(4;11)(q2l;q23) and patients with this 4;11 translocation have a very poor prognosis.1,2This genetic rearrangement fuses the Mll/ALL-1/HRX-Htrx gene at 11q23 with the AF4/FEL gene at 4q21.5-7 TheMll gene is involved in different acute leukemias through a series of chromosome translocations5,8,9 and fusion to a variety of genes, most frequently to AF4 andAF9.10 The critical feature of these chromosomal rearrangements is the generation of an in-frame chimeric fusion transcript consisting of 5′-Mll and 3′ sequences of the gene on the partner chromosome.11 The resulting chimeric messenger RNAs (mRNAs) presumably encode chimeric proteins that contribute to the leukemogenic state.12,13 However, it has been shown that hematopoietic precursors are greatly reduced in Mll mutant mice, suggesting that Mll functions as a regulator of the growth of hematopoietic precursors.14
Distinguishing features of Mll alterations include the striking heterogeneity of its recombinations, with more than 30 chromosome partners described in Mll rearrangements among which 17 Mll partner genes have been characterized (for a review see reference 2). Furthermore, each partner is preferentially associated to a different subset of myeloid or lymphoid leukemias.2 Because these genes do not all share the same structural or functional features, their putative roles in Mll activation in a specific hematopoietic lineage still need to be clarified.
The function of the AF4 gene remains poorly understood. The AF4 protein contains a domain with transcriptional activation activity that is consistently retained by Mll-AF4 fusion proteins created by t(4;11) translocations in leukemia15 and shows a nuclear localization, thus supporting a potential role in regulating transcription.16,17 In previous studies, it has been shown that the AF4 transcripts are present in a variety of hematopoietic and nonhematopoietic human cells.18,19 In mice, we and others have previously shown that the mAF4 gene is expressed predominantly in developing thymocytes3 and is strongly expressed in developing and adult lymphoid organs.4 These characteristics support the hypothesis that the AF4 gene might be important for the leukemic phenotype.
To investigate the effects of the mAF4 gene on lymphocyte development, we have inactivated this gene by homologous recombination in the mouse. We show that in the hematopoietic system only the lymphoid compartment is severely affected in mAF4 mutant mice. Our data show that the mAF4 gene affects early events in lymphopoiesis such as precursor proliferation or recruitment, but it is not required for the terminal stages of lymphocyte differentiation.
Materials and methods
Construction of the targeting vector
A XbaI-SalI fragment containing the pGK-neo gene flanked with 2 loxP sites was subcloned in the EcoRV site of pBluescript (pBS) SK vector (Stratagene, San Diego, CA).20 Two DNA fragments from the mAF4 locus were isolated from a λ phage screened from a mouse 129/Ola genomic library: the 5-kb BamHI-HindIII fragment contains part of the 3′ intron 11 region of themAF4 gene; the 3-kb HindIII fragment, from the 5′ region, contains intron 10 and part of exon 11. After filling the ends of restriction sites, the 2 mAF4 homologous fragments were cloned, respectively, in the SmaI and HindIII sites of the modified pBS-neo vector, on each side of the loxP-neo-loxP cassette. This targeting vector pBS-neo-mAF4 was designed to allow the interruption of the exon 11 and the excision of a part of this exon up to the BamHI site in the following intron after homologous recombination in embryonic stem (ES) cells.
Transfection of ES cells and generation of chimeric mice
The E14 (129/Ola) strain ES cells (2×107) were electroporated with 20 μg pBS-neo-MAF4 targeting construct DNA. Twenty-four hours later, cells were positively selected with 400 μg/mL G418. Isolated G418-resistant colonies were picked after 7 days of selection. Homologous recombinants were tested by hybridization of EcoRI-digested genomic DNA using a HindIII fragment (5′ probe) as an external probe. A unique integration event was checked with a neomycin probe. For Northern analysis, RNA was extracted from kidney and expression was checked with a 1.4-kb complementary DNA (cDNA) fragment as a probe.4
Manipulations of the embryos were performed as described.21Two male chimeras (distinguished by the coat color), originating from 2 independent ES clones, gave a germline transmission of the targeted ES cells when mated to BALB/c females. Tail DNA from the F1 offspring were analyzed by Southern blotting for germline transmission of the mAF4 targeted allele. Homozygous mutant mice were obtained by mating the heterozygous mutant mice.
Flow cytometric analysis
Single cell suspensions from thymus were incubated at 1 × 107 cells/mL in 100 μL of staining solution (phosphate-buffered saline [PBS] complemented with 0.2% fetal calf serum [FCS] and 0.2% mouse serum) for 30 minutes on ice with fluorescein isothiocyanate (FITC)-conjugated anti-CD3, -CD69, and H-2Kb, phycoerythrin (PE)-conjugated anti-CD8α (53-6.7) and biotin-conjugated anti-CD4 (H129.19) monoclonal antibodies. After 2 washes in PBS/0.2% FCS solution, the cells were incubated with 50 μL of streptavidin-cy-chrome reagent in staining solution for 30 minutes on ice. The stained cells were washed 3 times and then fixed in 150 μL of 1% paraformaldehyde in PBS. Analysis was performed on a Becton Dickinson FACScan. The monoclonal antibodies and the streptavidin-cy-chrome reagent were purchased from Pharmingen (San Diego, CA).
Glucocorticoid treatments
Two injections were performed at day −2 and day −1 before the time-course experiment. Mice were injected intraperitoneally with 2 mg/20 g hydrocortisone (Roussel, Paris, France).
Results
Generation of mutant mice at the AF4 locus
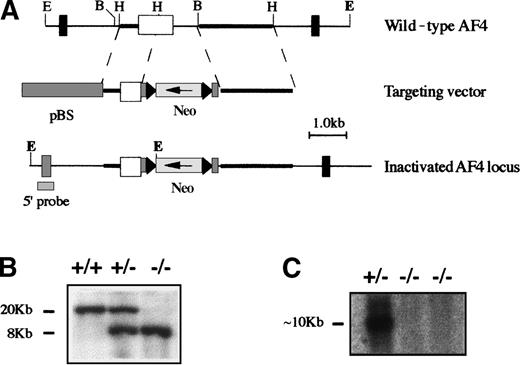
To investigate the effects of the mAF4 gene on lymphocyte development we have inactivated this gene by homologous recombination in the mouse by inserting a neomycin-resistance (neo) gene in reverse orientation to the mAF4 transcription (Figure1A). The targeting vector was electroporated into E14 ES cells and selected by G418. Two independent, homologous recombinant E14 ES cell clones generated several chimeric males, which transmitted the disrupted allele to their offspring (Figure 1B). Male and female mAF4 heterozygous mice appeared normal and fertile and were interbred to generate homozygous mice. From these crosses 25% of the mice observed at birth were of the mAF4−/− genotype indicating that the mutants survive embryogenesis. Genomic Southern blot analysis as well as Northern blot analysis and reverse transcriptase–polymerase chain reaction (RT-PCR, data not shown) confirmed the disruption of the mAF4 gene and the absence of the mAF4 transcript in the homozygous mAF4−/− mice (Figure 1C).
Targeted disruption of the mAF4 gene.
Homologous recombination at the mAF4 locus in ES cells. (A) Structure of the targeting vector and partial restriction map of the mAF4 locus before and after targeted integration. The targeting vector contains the neomycin-resistance (neo) gene flanked by genomic mAF4 sequences. After the recombination event, the neo gene replaces a BamHI-HindIII fragment interrupting exon 11 of the mAF4 gene. E indicates EcoRI; H, HindIII. (B) Southern blot analysis of a representative litter showing alleles from a wild-type (+/+), a heterozygous (+/−), and a homozygous (−/−) animal. Hybridization of genomic DNA with an external probe (5′ probe) reveals a 20-kb EcoRI fragment for the mAF4 wild-type allele and a 8-kb EcoRI fragment corresponding to the inactivated allele. (C) Northern blot analysis of a representative litter showing the mAF4 transcript of RNA (10 μg) extracted from the kidney of a heterozygous mouse (+/−) and from 2 homozygous mice (−/−).
Targeted disruption of the mAF4 gene.
Homologous recombination at the mAF4 locus in ES cells. (A) Structure of the targeting vector and partial restriction map of the mAF4 locus before and after targeted integration. The targeting vector contains the neomycin-resistance (neo) gene flanked by genomic mAF4 sequences. After the recombination event, the neo gene replaces a BamHI-HindIII fragment interrupting exon 11 of the mAF4 gene. E indicates EcoRI; H, HindIII. (B) Southern blot analysis of a representative litter showing alleles from a wild-type (+/+), a heterozygous (+/−), and a homozygous (−/−) animal. Hybridization of genomic DNA with an external probe (5′ probe) reveals a 20-kb EcoRI fragment for the mAF4 wild-type allele and a 8-kb EcoRI fragment corresponding to the inactivated allele. (C) Northern blot analysis of a representative litter showing the mAF4 transcript of RNA (10 μg) extracted from the kidney of a heterozygous mouse (+/−) and from 2 homozygous mice (−/−).
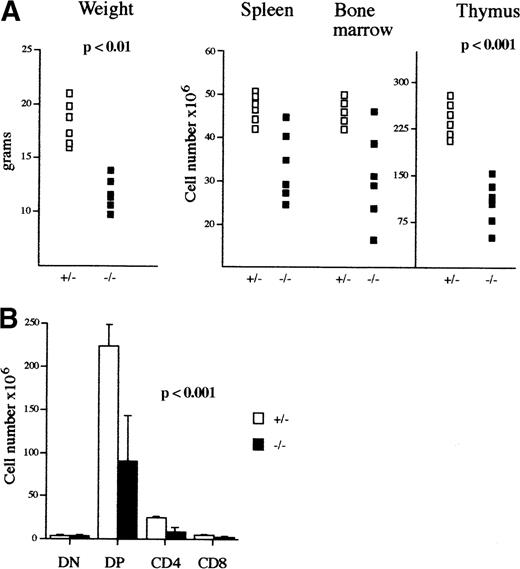
Among the mAF4−/− mice on a mixed 129/BALB/c background, 20% were significantly smaller than the control littermates (P < .01) and had a life span not exceeding 10 days when the reduced size was exaggeratedly marked; otherwise the size retardation was no longer visible after 6 weeks of age. Analysis of lymphoid organs in those mutant mice revealed a significant reduction in thymic cellularity (average 108 cells in mutant mice as compared to an average of 2.3 × 108 in wild-type mice; Table 1), whereas bone marrow and spleen were less affected (Figure 2A; Table1). However, the mutant mice with no growth retardation had a normal lymphocyte compartment.
Lymphoid cell count numbers in mutant and wild-type mice
| . | AF4+/− . | AF4−/− . |
|---|---|---|
| Weight (g) | 18.2 ± 1.70 | 11.83 ± 1.8* |
| Spleen (106) | 47.8 ± 3.65 | 32.16 ± 8.9 |
| Bone marrow (106) | 37.7 ± 10.4 | 27.70 ± 10.3 |
| Thymus | ||
| Thymocytes (106) | 234 ± 36.1 | 100.1 ± 38.2† |
| DN (106) | 3.25 ± 0.37 | 3.47 ± 0.34 |
| DP (106) | 217.17 ± 7.53 | 86.17 ± 13.85† |
| SP CD4+ (106) | 23.33 ± 0.71 | 7.33 ± 1.77† |
| SP CD8+ (106) | 4.55 ± 0.12 | 1.33 ± 0.17† |
| Bone marrow | ||
| B220+ (106) | 13.75 ± 2.58 | 8.5 ± 2.5* |
| CD43+ (106) | 0.89 ± 0.01 | 1.02 ± 0.32 |
| CD43−(106) | 12.33 ± 2.04 | 8.3 ± 1.44* (n = 5) |
| IgM+ (106) | 4.28 ± 0.31 | 4.14 ± 0.66 |
| IgM− (106) | 8.62 ± 0.84 | 4.35 ± 0.82† |
| . | AF4+/− . | AF4−/− . |
|---|---|---|
| Weight (g) | 18.2 ± 1.70 | 11.83 ± 1.8* |
| Spleen (106) | 47.8 ± 3.65 | 32.16 ± 8.9 |
| Bone marrow (106) | 37.7 ± 10.4 | 27.70 ± 10.3 |
| Thymus | ||
| Thymocytes (106) | 234 ± 36.1 | 100.1 ± 38.2† |
| DN (106) | 3.25 ± 0.37 | 3.47 ± 0.34 |
| DP (106) | 217.17 ± 7.53 | 86.17 ± 13.85† |
| SP CD4+ (106) | 23.33 ± 0.71 | 7.33 ± 1.77† |
| SP CD8+ (106) | 4.55 ± 0.12 | 1.33 ± 0.17† |
| Bone marrow | ||
| B220+ (106) | 13.75 ± 2.58 | 8.5 ± 2.5* |
| CD43+ (106) | 0.89 ± 0.01 | 1.02 ± 0.32 |
| CD43−(106) | 12.33 ± 2.04 | 8.3 ± 1.44* (n = 5) |
| IgM+ (106) | 4.28 ± 0.31 | 4.14 ± 0.66 |
| IgM− (106) | 8.62 ± 0.84 | 4.35 ± 0.82† |
Results are expressed as mean ± SD. Two-tailed Student Pvalue.
P < .01;
P < .001.
Phenotypic analysis of mAF4−/− mice.
(A) Body weight (g), lymphoid cell content of spleen, bone marrow, and thymus (×106) of 3-week-old mAF4 mutant (▪) and control (□) mice. (B) Absolute number of thymocyte subpopulations from 3-week-old mAF4−/− and wild-type mice (6 mice per group) stained to reveal CD4 and CD8 distribution.
Phenotypic analysis of mAF4−/− mice.
(A) Body weight (g), lymphoid cell content of spleen, bone marrow, and thymus (×106) of 3-week-old mAF4 mutant (▪) and control (□) mice. (B) Absolute number of thymocyte subpopulations from 3-week-old mAF4−/− and wild-type mice (6 mice per group) stained to reveal CD4 and CD8 distribution.
Analysis of the thymocyte populations in mAF4−/− mutant mice
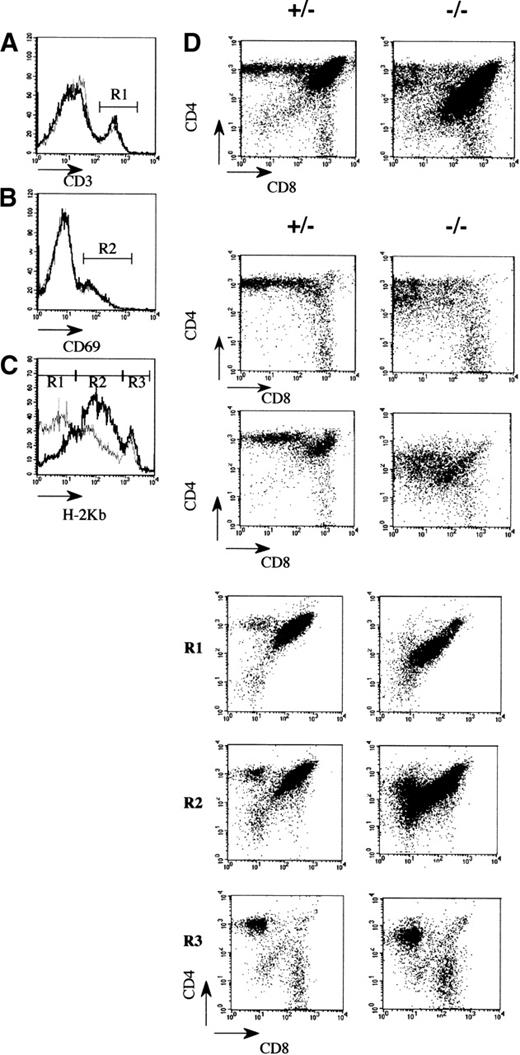
To evaluate which thymic subpopulations were modified, in the affected mutant mice, a flow cytometric analysis using conventional markers was performed. The most striking observation was a reduction in the number of cortical CD4+CD8+ thymocytes (Figure 2B; Table 1). In contrast, the number and the different subsets of CD4−CD8− precursor thymocytes are normal, indicating that the seeding and the early cytokine-dependent expansion phase of T-cell antigen receptor (TCR)-prothymocytes were not significantly affected by the mAF4 mutation.22 Similarly, the presence of mature CD4+ or CD8+ thymocytes indicates that there is no block in thymocyte differentiation despite a 3-fold reduction of CD4+ cells (Figures 2B and 3A; Table 1). This point is further substantiated by the fact that these cells express normal levels of TCR/CD3 molecules (Figure3B) and based on the increased expression of the H-2 class I major histocompatibility complex (MHC) marker (Figure 3C, gate R3) complete their maturation program.23 24 These mature thymocytes colonize efficiently peripheral lymphoid organs in appropriate proportions (data not shown).
Flow cytometric analysis of thymocytes in 3-week-old mAF4 mutant mice.
(A) Distribution of the CD4 and CD8 markers in mutant and control mice. (B) CD3+ cells (gate R1) on CD4 and CD8 thymocytes in mutant versus control mice. Histograms are plotted as overlays of the control thymocytes (black line) and AF4 mutant thymocytes (gray line). (C) H-2Kb distribution on CD4 and CD8 cell populations. Gates are H-2Kb low (R1), H-2Kb intermediate (R2), and H-2Kb high (R3). (D) CD69 distribution on CD4 and CD8 thymocytes. Data are displayed as dot plots with log scale. Data are representative of 6 control mice and 6 mAF4−/− mice.
Flow cytometric analysis of thymocytes in 3-week-old mAF4 mutant mice.
(A) Distribution of the CD4 and CD8 markers in mutant and control mice. (B) CD3+ cells (gate R1) on CD4 and CD8 thymocytes in mutant versus control mice. Histograms are plotted as overlays of the control thymocytes (black line) and AF4 mutant thymocytes (gray line). (C) H-2Kb distribution on CD4 and CD8 cell populations. Gates are H-2Kb low (R1), H-2Kb intermediate (R2), and H-2Kb high (R3). (D) CD69 distribution on CD4 and CD8 thymocytes. Data are displayed as dot plots with log scale. Data are representative of 6 control mice and 6 mAF4−/− mice.
An important difference was a lowered expression (50%) of the CD4 and CD8 coreceptor molecules on a fraction of double and single positive thymocytes (Figure 3A), and this might suggest modifications in the efficiency of T-cell selection or increased susceptibility to apoptosis.25,26 This latter possibility seems unlikely because cultured mAF4−/− thymocytes show a normal rate of spontaneous apoptotic cell death in vitro (data not shown). Then the expression of the CD69 molecule was evaluated as a marker of positive selection.27 As shown in Figure 3D, CD69 is normally expressed by DP thymocytes at the transition toward the SP stage because the preferential expression of 1 coreceptor is detectable on most cells. Interestingly, the lowered CD4 (×5) and CD8 (×1.5) expression is observed on thymocytes undergoing positive selection (Figure 3D, gate R2) and on terminally differentiated thymocytes (Figure 3C, gate R3). In total, the effect of the AF4 mutation mainly concerns the immature CD4+ CD8+H-2 low thymocyte population, which contains the cells undergoing positive selection (Figure 3C, gate R1). Thus, these results indicate that both expansion and selection might be affected in mutant animals.
Glucocorticoid treatments
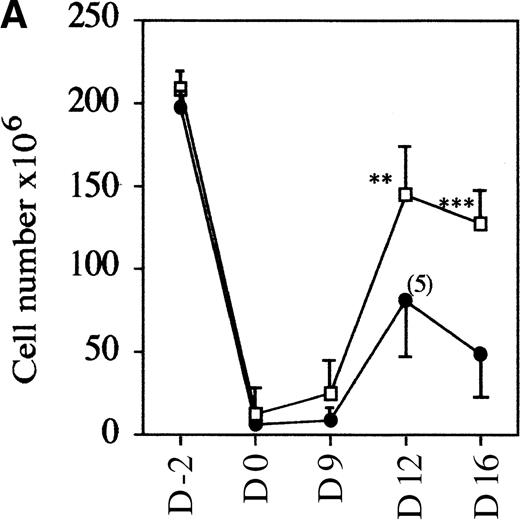
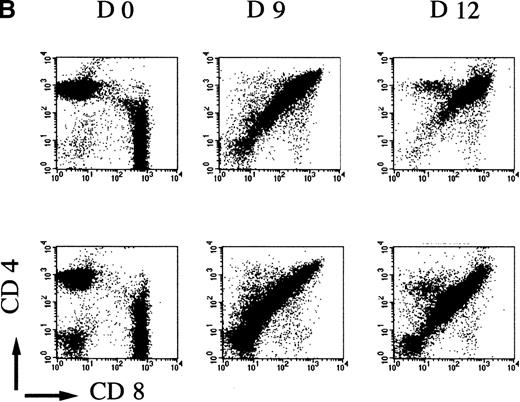
To evaluate if the mutant mice had a normal rate of thymocyte expansion, the potential of the DN cell population to reconstitute the DP compartment was assessed.23 Nonaffected mutant and control wild-type mice were treated with glucocorticoid, known to eliminate the majority of DP but not DN thymocytes.28 29 In a time-course experiment, the kinetics of reconstitution is clearly retarded in mutant versus control mice. Over a 2-week period, first the number of thymocytes recovered per thymus increased up to 1.2 × 108 in wild-type and heterozygote samples compared with an increase to an average of 0.6 × 108 in mAF4−/− mice (Figure4A). This reduction in cell number is accompanied by a marked retardation in maturation, although representatives of all stages of thymocyte differentiation were observed as seen with CD4 and CD8 markers. Indeed, FACS profiles at day 12 in mutant mice are similar to those observed at day 9 in wild-type animals (Figure 4B). These results show that in mutant mice, precursor cells can transit from the immature DN to the most mature SP thymocytes, but generate far fewer T cells.
AF4 is required for normal thymocyte expansion.
Thymus from mAF4−/− glucocorticoid-treated mice show a decrease in the reconstitution of the number of DP cells. (A) Kinetic analysis of thymocyte reconstitution. T-cell counts after glucocorticoids in mutant (•) and control (□) mice over a 2-week period. Data are representative of 6 control mice and 6 mutant mice except when indicated. Two-tailed Student P value is shown, where significant. **P < .05; ***P < .001. (B) FACS analysis was performed on thymocytes of control and mutant mice at day 0 (d0), day 9, and day 12 after the glucocorticoid injections as indicated in “Materials and methods.” T-cell maturation was assayed using the CD4 and CD8 markers. Results shown are representative of 6 separate experiments.
AF4 is required for normal thymocyte expansion.
Thymus from mAF4−/− glucocorticoid-treated mice show a decrease in the reconstitution of the number of DP cells. (A) Kinetic analysis of thymocyte reconstitution. T-cell counts after glucocorticoids in mutant (•) and control (□) mice over a 2-week period. Data are representative of 6 control mice and 6 mutant mice except when indicated. Two-tailed Student P value is shown, where significant. **P < .05; ***P < .001. (B) FACS analysis was performed on thymocytes of control and mutant mice at day 0 (d0), day 9, and day 12 after the glucocorticoid injections as indicated in “Materials and methods.” T-cell maturation was assayed using the CD4 and CD8 markers. Results shown are representative of 6 separate experiments.
Analysis of bone marrow hemopoiesis in AF4−/− mutant mice
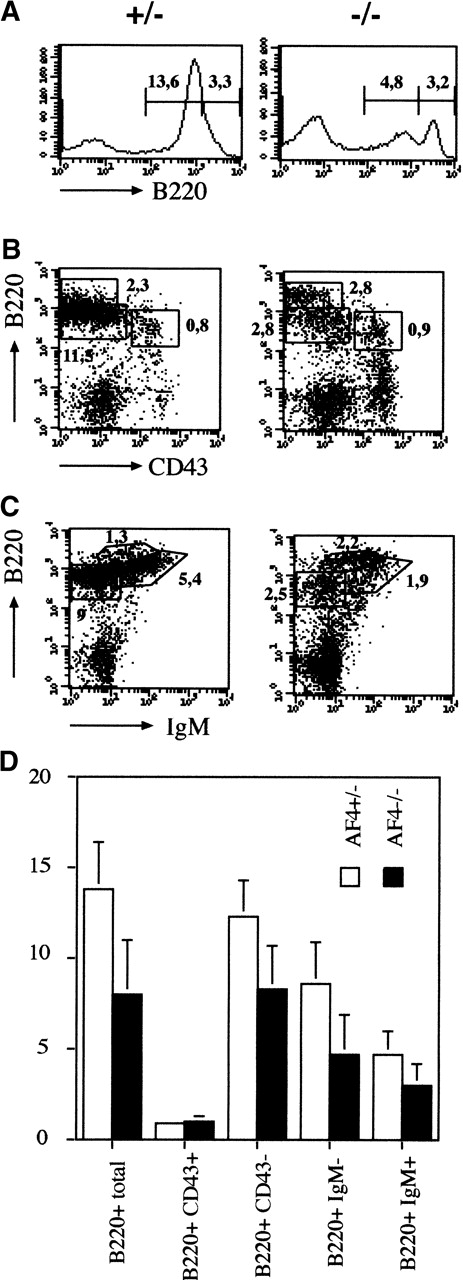
A similar study was performed on the B-lymphocyte lineage in the bone marrow. As shown in Figure 5A and D the size of the B-cell compartment is reduced by 3-fold and this diminution concerns the B220 intermediate population that contains mainly the immature B cells.30 To refine which subset of the B-cell progenitors is specifically affected, the pro-B and pre-B cell compartments were analyzed using the CD43 maturation marker.30 As shown in Figure 5B, the CD43+B220+ pro-B cells are not affected in mutant mice. In contrast B220+ CD43− or B220+IgMlow pre-B cells and mature B220+IgMhigh B cells are reduced without evidence of a block in B-cell differentiation (Figure 5C-D; Table1). These results suggest that the pre-B cell receptor-dependent expansion phase (pre-BCR) is affected in mAF4 mutant mice. On the contrary, the numbers of monocytes (CD11b+) and granulocytes (Gr−1+) were unaltered in the mutant bone marrow. Moreover, all the other hematopoietic compartments were unaltered in mAF4 mutant mice (data not shown).
Analysis of bone marrow hemopoiesis.
(A) Representative flow cytometry analysis of bone marrow B cell. Absolute number of B220+ cells in the bone marrow of mAF4 mutant mice and controls. Cell counts are given (×106). (B) and (C) Representative flow cytometry analysis of bone marrow B-cell–stained populations from 3-week-old mAF4−/− and wild-type mice; cell counts (×106) are indicated in the corresponding quadrant. Mature, immature, and transitional B cells are boxed. Staining was performed with B220, IgM, and CD43. (D) Total B cell numbers in the bone marrow of mAF4 mutant and wild-type mice. The data are the mean of 6 mAF4 null mice and of an equal number of wild-type littermates. The relative SD is shown. For statistical analysis, see Table 1.
Analysis of bone marrow hemopoiesis.
(A) Representative flow cytometry analysis of bone marrow B cell. Absolute number of B220+ cells in the bone marrow of mAF4 mutant mice and controls. Cell counts are given (×106). (B) and (C) Representative flow cytometry analysis of bone marrow B-cell–stained populations from 3-week-old mAF4−/− and wild-type mice; cell counts (×106) are indicated in the corresponding quadrant. Mature, immature, and transitional B cells are boxed. Staining was performed with B220, IgM, and CD43. (D) Total B cell numbers in the bone marrow of mAF4 mutant and wild-type mice. The data are the mean of 6 mAF4 null mice and of an equal number of wild-type littermates. The relative SD is shown. For statistical analysis, see Table 1.
Altogether our results demonstrate that the absence of the mAF4gene specifically affects lymphoid development despite its large domain of expression in the hematopoietic system.
Discussion
We generated mAF4-deficient mice and found that themAF4 gene is not essential for embryonic development. From mAF4 heterozygous mice crosses, 25% of the mice observed at birth were of the mAF4−/− genotype indicating that the mutants survive embryogenesis. In a 129/BALB/c mixed background 20% of the mice showed a pronounced effect on growth and on lymphoid development. This incomplete penetrance could be due to a partial complementation by a related gene. Recently, a novel human gene,LAF4, was isolated from a subtracted cDNA library that showed strong sequence similarity to AF4. AF4 and LAF4are homologous throughout their coding regions. HumanLAF4 hybridized with genes in mouse and chicken, showing that this gene family has been highly conserved during vertebrate evolution.31 In mouse tissues, although the LAF4and mAF4 genes are differentially expressed, the highest levels for both of them are observed in lymphoid tissues. It is thus possible that LAF4 could partially substitute for the mAF4 gene function during lymphoid development.
During early T- and B-lymphoid maturation the expansion of immature lymphocytes depends on the expression of a functional pre-T or pre-B cell receptor. In mutant mice the prolymphoid cell population, which precedes this stage, is not affected. In contrast all the lymphoid-committed populations beyond that critical first checkpoint are reduced in number, inferring that the mAF4 gene controls this expansion phase. This analysis was refined in the case of T-cell development. Two genetic checkpoints regulate thymocyte differentiation and cell proliferation.32 The second checkpoint controls the progression from CD4+CD8+ immature to mature CD4+ or CD8+ thymocytes associated with positive selection.33 In the absence of positive selection, thymocytes undergo apoptosis. Our results indicate that the mAF4 mutation affects the size of the immature double-positive thymocyte subset. We found no evidence that mAF4−/− thymocytes had a higher rate of cell death in vitro (data not shown); in addition, their sensitivity to cell death-inducing factors such as glucocorticoids28,34 is not modified in hydrocortisone-treated mutant mice (Figure 4, day 0). Thus, the most likely possibility would be that the mAF4 gene has a role in the expansion or differentiation of early DP thymocytes. Our experiments using glucocorticoid treatment on mutant mice reveal that the DN population is affected in its potential to generate a normal number of DP and SP thymocytes. Indeed, mAF4−/− thymocytes are depleted of CD4+CD8+TCR−/low thymocytes expressing low levels of H-2 molecules prior to any TCR-dependent selection step (11 × 106 in mutant mice versus 50 × 106 in wild-type mice). Furthermore, we observed a lowered expression of coreceptor molecules. The fact that DP thymocytes express normal levels of the CD69 molecule and become mature CD3high/ H-2high SP thymocytes suggests that positive selection is operational in mAF4 mutant mice. However, the threshold of selection, which is dependent on the expression of high levels of CD4, CD8, and TCR molecules, might be affected35,36; indeed, in mutant mice both CD4 and CD8 coreceptor molecules are down-regulated in CD69+ DP thymocytes and in the fully selected SP thymocytes. Thus the absence of the mAF4 gene might render thymocytes more susceptible to TCR-dependent signal transduction. However, it has been shown using Gal4-AF4 constructs that AF4 is a potent transactivator15; it is then also possible that mAF4 product controls directly the transcriptional rate of different target genes involved in signal transduction including the coreceptor molecules in T-lymphocyte development and similar molecules involved in B-lymphocyte maturation.
The AF4/FEL5,37 gene was first identified in the human by its involvement with Mll in the translocation t(4;11)(q21;q23) in acute mixed-lineage leukemia.5,38 The most frequent reciprocal translocation found in infant leukemias is the t(4;11)(q21;q23); it has a poor prognosis. The leukemic cells express the stem cell marker CD34, HLA-DR, and a pro-B cell marker CD19 as well as the myelomonocytic marker CD15.39,40 These characteristics of the blast cells suggest that the t(4;11) occurs and transforms a multipotential progenitor cell. The mechanism by which the fusion protein is able to induce neoplasia is not fully understood and 2 models are proposed. The first model implies a gain of function of the MLL/AF4 chimeric protein in which the DNA binding domain is fused to a transactivation domain of AF4 altering the regulation of target genes.13Some proposed that the truncation of Mll is sufficient to induce neoplasia.41
A previous in vitro study demonstrated that an HRX-ENL fusion cDNA transduced into a cell population enriched in hematopoietic stem cells confers to the infected cells an enhanced potential to generate myeloid colonies with primitive morphology, demonstrating a direct role for HRX-ENL in the immortalization and leukemic transformation of a myeloid progenitor and supporting a gain-of-function mechanism for HRX-ENL–mediated leukemogenesis.13 Moreover, in other experiments AF9 sequences were fused by homologous recombination12 into the mouse Mll gene so that expression of the Mll-AF9 fusion gene occurred from endogenous Mll promoter, as in t(9;11) found in human leukemia. Chimeric mice carrying the fusion gene developed tumors, which were also restricted to acute myeloid leukemias. Those results demonstrate that the Mll-AF9 fusion protein is oncogenic predominantly in myeloid or myeloid-committed cells despite the large domain of activity of the Mll promoter and its demonstrated activity in B and T cells. Chimeric mice expressing an Mll-LacZ fusion gene42develop acute leukemia similar in phenotype to the Mll-AF9 fusion mice.43 This could indicate that Mll-mediated oncogenesis is on the myeloid pathway by default.44 Then the lymphoid leukemic phenotype may require an additional specific fusion partner. Our results show that the mAF4 gene is important for lymphoid cell development because in mAF4−/− mice only the lymphoid compartment is affected although this gene is expressed in the other hematopoietic cell populations. This emphasizes that the partner gene in translocation-associated leukemias could be an important factor for the lineage of the leukemic phenotype. However t(4;11) positive acute lymphocytic leukemia with a T phenotype is extremely rare. Alternatively, the timing of the Mll translocation during cell commitment could alone influence the lineage of the associated leukemia. Further studies on the function of AF9 or ENL and creating an Mll-mAF4 fusion product in mice should clarify this point.
Acknowledgments
We thank C. Schiff for critical reading of the manuscript. The authors are grateful to G. Warcollier and M. Pontier for helping with the animal facility and to A. Boned for technical assistance.
Supported by grants from CNRS, ARC, and from the Ligue contre le cancer. P.I. is supported by an ARC and an FRM fellowship.
Reprints:Malek Djabali, Centre d'immunologie INSERM-CNRS de Marseille Luminy, Case 906, 13288 Marseille Cedex 9, France; e-mail:djabali@ciml.univ-mrs.fr.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal