Abstract
Using DNA fiber fluorescence in-situ hybridization (FISH) and 3-color interphase FISH, 2 cases of follicular lymphoma were identified in which the BCL2 gene was excised from 18q21 and inserted into the immunoglobulin heavy chain (IGH) locus at 14q32. Both the insertion breakpoint at 14q32 and the deletion breakpoint at 18q21 were cloned using inverse polymerase chain reaction. Sequence analysis showed that the JH sequences were juxtaposed to the 5′-side of BCL2, and the DH sequences were juxtaposed to the 3′-side of BCL2. There were breakpoints at both the JH and DH recombination signal sequences, and N-nucleotides were present at all breakpoint junctions. At theBCL2 locus, the 3′-breakpoints in both cases were localized at exactly the same nucleotide position, 6.2 kilobase downstream of the major breakpoint region, directly adjacent to a complete cryptic recombination signal sequence (RSS) consisting of a heptamer, a nonamer, and a 23–base pair (bp) spacer. The BCL25′-breakpoints were approximately 600 bp upstream of the gene, within the CA repeats. Although less evident than for the BCL23′-breakpoints, cryptic RSSs were also identified at these breakpoints, with a 12-bp spacer. On the basis of structural characteristics of these rearrangements, a model is proposed in which the BCL2 gene is deleted from its locus by recombination activation gene-1/-2 (RAG-1/-2)–mediated excision. The gene is subsequently inserted into the recombiningIGH locus, a process involving the formation of hybrid joints between the IGH coding ends and theBCL2 signal ends.
Introduction
Follicular lymphomas (FLs) are characterized by a t(14;18)(q32;q21), which juxtaposes the enhancers of the immunoglobulin heavy chain (IGH) locus at 14q32 to the BCL2 gene at 18q21. Breakpoints at theIGH locus are located at the J or D segments, and breakpoints at the BCL2 locus are generally located in the major breakpoint region (mbr) in the 3′–untranslated region or downstream of the gene. Using contigs of probes covering theBCL2 gene and the IG loci, we previously performed a fiber fluorescence in-situ hybridization (FISH) study of the BCL2 translocations in FL.1 In 4 of 40 cases, the BCL2 gene was affected by 2 translocation breakpoints on the same allele: 1 at the 5′-side and 1 at the 3′-side of the gene. In 2 cases, the IGH locus was juxtaposed to the 3′-side of BCL2 and the immunoglobulin-λ (IGL) locus to the 5′-side, indicating 2 separate translocation events. In the other 2 cases, the BCL2 3′- and 5′-flanking regions were juxtaposed to each other (3′-BCL2/5′-BCL2). Juxtaposition of the IGH constant genes to the 5′-side and the DH gene region to the 3′-side of BCL2suggested that the BCL2 gene had been excised from its locus at chromosome 18q21 and inserted into the IGH locus at chromosome 14q32, an orientation that is the opposite of the orientation in normal t(14;18). In this report, these variantBCL2 rearrangements are analyzed in more detail and give evidence of a mechanism that involves the recombination activation gene-1/-2 (RAG-1/-2)–mediated excision of BCL2and the formation of hybrid joints between BCL2 andIGH.
Materials and methods
Tissue samples
Frozen tissue of FLs was obtained from the tissue bank of the Department of Pathology, Leiden University Medical Center, Leiden, The Netherlands. We selected cases that were unambiguously diagnosed as FL on the basis of morphology and immunophenotype (CD10+, CD5−, and Bcl2+).1
Probes for fiber FISH
The BCL2 gene was color bar-coded with a set of alternately biotin- and digoxigenin-labeled probes containing 7 cosmids (G2, B1, F6, G4, E6, F11, and H2) and 2 PACs, 210c12 and 15o23. The cosmids were subcloned from YAC YA153A6 (gift of G. A. Silverman, Washington University School of Medicine, Seattle, WA). PAC 210c12 and PAC 15o23 were obtained from the Roswell Park Cancer Center Human PAC Library, Buffalo, NY, as described elsewhere.1The probe set used for detection of the IGH locus has been described in detail previously2 and consisted of cosmids U2-2 and 3/64 (gift of M. J. S. Dyer, Royal Marsden Hospital, Sutton, England), cosmid cosIg6 (gift of T. H. Rabbitts, MRC Center, Cambridge, England), and a plasmid probe specific forCG (Cγ) genes (gift of P. Leder, Boston, MA).
Probes for interphase FISH
BCL2 insertions into the IGH locus were visualized using the following probes: cosmid cosIg6, the 14q-subtelomeric cosmid IgH2 (provided by H. Riethman, The Wistar Institute, Philadelphia, PA), and BCL2 intronic cosmid F6.
FISH and fluorescence microscopy
Preparations of DNA fibers and interphase nuclei were made as described previously.2,3 The probes were labeled by standard nick-translation with biotin-16-dUTP (uridine 5′-triphosphate), digoxygenin-11-dUTP, or fluorescein-12-dUTP (Roche, Basel, Switzerland). The hybridization solution consisted of 30% formamide; 10% dextransulphate; 50 mmol/L sodium phosphate (pH 7.0); 2 times sodium chloride/sodium citrate (SCC), 3 ng/μL each probe; and a 50-fold excess of human Cot-1 DNA. Hybridization and immunofluorescence detection for dual-color FISH were performed as described previously.3,4 For triple-color FISH, immunofluorescence detection was performed essentially according to Wiegant et al.5 Biotin-labeled probes were first detected with avidin-AMCA (Vector Laboratories, Burlingame, CA); then goat–anti-avidin–biotin (Vector); and finally, avidin-AMCA. Digoxigenin-labeled probes were detected with mouse-antidigoxin (Sigma Chemical, St Louis, MO), and sheep-antimouse-digoxigenin and sheep-antidigoxigenin-TRITC (Roche). Fluorescein-labeled probes were detected with rabbit–anti-FITC (fluorescein isothiocyanate) (Dako, Copenhagen, Denmark) and goat-antirabbit-FITC (Vector).
Microscopic images were captured using a COHU 4910 series monochrome CCD camera (COHU, San Diego, CA) attached to a DM fluorescence microscope (Leica, Wetzlar, Germany) equipped with a PL Fluotar 100 times NA 1.30-0.60 objective and I3 and N2.1 filters (Leica) and Leica QFISH software (Leica Imaging Systems, Cambridge, England). The images were processed with Paintshop Pro (JASC, Eden Prairy, CA) and Harvard Graphics (Software Publishing, Santa Clara, CA).
Southern blot analysis
Southern blotting was performed with IGH probe IGHJ6 (provided by J. J. M. van Dongen, Rotterdam, The Netherlands)6; mbr probe pSp65/18-4RH, a 3-kilobase (kb)EcoRI-HindIII fragment (provided by Y. Tsujimoto, Osaka, Japan); 5′-BCL2 probe pB16, a 1.6-kb EcoRI cDNA fragment (provided by Y. Tsujimoto)7; and probe 5′-5′-BCL2, a 450–base pair (bp) polymerase chain reaction (PCR) product located 1 kb upstream from the BCL2 gene (see Figure 2A). The 5′-5′-BCL2 probe was made with the following primers: forward 5′-CCTATTAAGTAAGCCGCTGTGC-3′ (GenBank accession numberX51898; 8-29 bp) and reverse 5′-CGTGTCCACCTGAACACCTAG-3′ (GenBankX51898; 486-466 bp). Genomic DNA was digested with BamHI,EcoRI, HindIII, and BglII (Roche, Pharmacia), size-fractionated, blotted on nylon filters, and hybridized, as described previously.8
Inverse PCR cloning of the 3′-BCL2/5′-BLC2 junction
High-molecular-weight genomic DNA was cut with restriction enzymes, and the product was purified by phenylchloroform, chloroform extraction, and ethanol precipitation. Subsequently, 50-100 ng purified digested DNA was self-ligated in a 100-μL volume containing 54 mmol/L Tris HCl (tris[hydroxymethyl] aminomethane hydrochloride) (pH 7.5), 4.5 mmol/L magnesium dichloride (MgCl2), 0.9 mmol/L diothiothreitol (DTT), 0.1 mmol/L adenosine 5′-triphosphate (ATP), and 1 unit T4 DNA ligase (Roche). We used 10 μL of this reaction for the first PCR, with primer 5′-GAAGTCCTCATCGTGTAGCAC-3′ (forward; GenBank X51898; 329-349 bp) and primer 5′-GACAAGAGGACAAACAAGTTGC-3′ (reverse; GenBank X51898; 119-98 bp). PCR was performed using a touchdown protocol, with the annealing temperature decreasing from 65°C to 60°C. We used 1 μL of this reaction for a second PCR with nested primers 5′-GAGCTGCTGAGTTCTGCATGG-3′ (forward; GenBank X51898; 433-453 bp) and 5′-GCACAGCGGCTTACTTAATAGG-3′ (reverse; Genbank X51898; 29-8 bp).
PCR and sequence analysis
The region between the BCL2 mbr and the 3′-breakpoint region of cases FL4104 and FL5117 was amplified using mbr primer 5′-CCTTTAGAGAGTTGCTTTACGT-3′ (forward; GenBank M13994; 4412-4433 bp) and 3′-BCL2 primer 5′-CCATGTAGATGGTGTTTGAGTG-3′ obtained from inverse PCR and sequencing of the 3′BCL2/5′BCL2junction. The JH 5′-breakpoints were amplified using a JH6 primer 5′-CTAGAGTGGCCATTCTTACCTG-3′ (GenBank X97051; 89841-89820 bp) and a 5′-BCL2 primer 5′-CTGGACCCTTTCTGGCCGTG-3′ (GenBank X51898; 856-837 bp). The DH 3′-BCL2 breakpoints were amplified using a DH3 primer 5′-GGTGAGGTCTGTGTCACTGTGG-3′ and a 3′-BCL2primer 5′-TGAATTTGAGGATAGAAAGTGCC-3′ (GenBank AF204739; 484-506 bp). All PCRs were performed using standard conditions and protocols, except for the mbr 3′-BCL2 PCR, which was performed according to a long-range PCR protocol using the Expand Long Template PCR System (Roche). The PCR products were directly sequenced using the Big Dye Terminator sequencing kit (Perkin Elmer Biosystems, Foster City, CA) and an ABI PRISM 377 automated sequencer (Perkin Elmer Biosystems). The sequences were compared with the GenBank database using the Basic Logical Alignment Search Tool (BLAST) program with CENSOR, a database of repetitive sequences.9
Results
BCL2 insertion into the IGH locus in 2 cases of FL
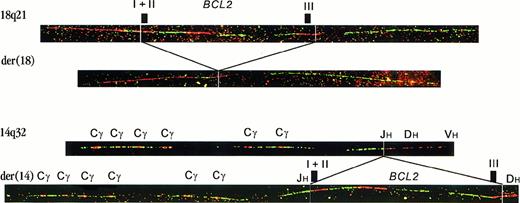
In 2 cases of FL (from a series of 401), fiber FISH with BCL2 and IGH bar codes revealed copies of 18q21 with deletion of the entire BCL2 gene (Figure1). In the same preparations, we found copies of the BCL2 gene that had the immunoglobulin constant region juxtaposed to their 5′-side, while part of cosmid U2-2, representing the DH region, was present at the 3′-side. These results suggested that the BCL2 gene had been excised from its original location on chromosome 18 and inserted into the IGHlocus on chromosome 14. Using 3-color interphase FISH with a probe for the BCL2 gene, a probe for the IGH constant region, and a subtelomeric chromosome 14 probe, we were able to colocalize these 3 probes and confirm the presence of a BCL2insertion into the IGH locus (not shown).
Fiber FISH with bar codes of probes for the
BCL2 gene and IGH locus.The top fiber shows a hybridization pattern representing a germlineBCL2 allele. The second fiber is a rearrangement pattern, as observed in case FL4104. It represents the derivative chromosome 18 from which the BCL2 gene was deleted. The third fiber is the hybridization pattern of a germline IGH allele. The position of Cγ genes and JH, DH, and VH regions is indicated.2 The last fiber shows an insertion of theBCL2 gene into the IGH locus. The CHgene region is juxtaposed to the 5′-side of the BCL2 gene. At the 3′-side, part of cosmid U2-2, containing most of the DH region, is present (labeled in red). Because in this image the most 3′-BCL2 cosmid is also in red, the breakpoint junction is not visible. Hybridization of these probes in different colors confirmed the juxtaposition of cosmid U2-2 to the BCL2 gene (not shown).
Fiber FISH with bar codes of probes for the
BCL2 gene and IGH locus.The top fiber shows a hybridization pattern representing a germlineBCL2 allele. The second fiber is a rearrangement pattern, as observed in case FL4104. It represents the derivative chromosome 18 from which the BCL2 gene was deleted. The third fiber is the hybridization pattern of a germline IGH allele. The position of Cγ genes and JH, DH, and VH regions is indicated.2 The last fiber shows an insertion of theBCL2 gene into the IGH locus. The CHgene region is juxtaposed to the 5′-side of the BCL2 gene. At the 3′-side, part of cosmid U2-2, containing most of the DH region, is present (labeled in red). Because in this image the most 3′-BCL2 cosmid is also in red, the breakpoint junction is not visible. Hybridization of these probes in different colors confirmed the juxtaposition of cosmid U2-2 to the BCL2 gene (not shown).
Localization of the BCL2 breakpoints
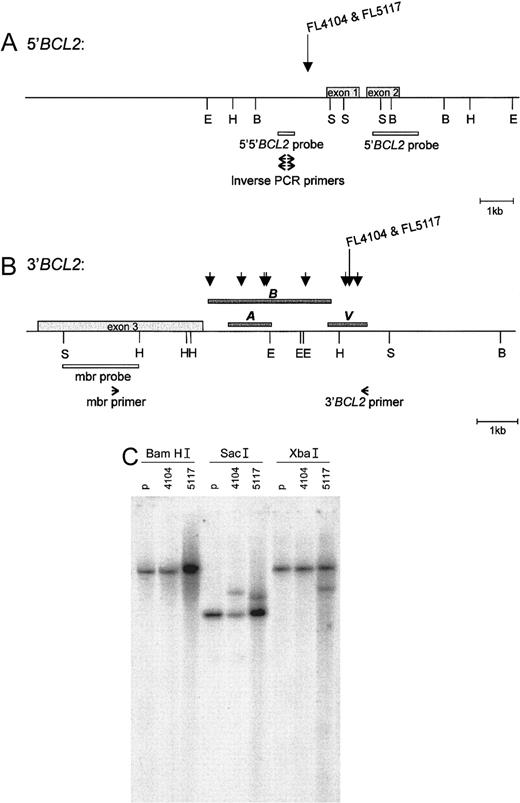
Southern blots of BamHI, EcoRI, andHindIII digests of tumor DNA were hybridized with the 5′-BCL2 probe (pB16) and the 5′-5′-BCL2 probe (Figure2A). Rearranged bands were observed in several digests: FL4104 with the 5′-BCL2 and 5′-5′-BCL2 probes: BamHI, EcoRI, and HindIII; FL5117 with a 5′-BCL2 probe: HindIII; and FL5117 with a 5′-5′-BCL2 probe: BamHI. The bands obtained with the 5′-5′-BCL2 probe were different from those obtained with the 5′-BCL2 probe. Because the enzymes used did not cut in the region between the 2 probes, in both tumors, the breakpoint should have been located between these 2 probes. Hybridization with the mbr probe (Figures 2B,C) showed rearranged bands in tumor DNA digested with SacI (both FL 4104 and FL 5117) and XbaI (FL 5117), but not withEcoRI and HindIII, enzymes which normally detect mbr breakpoints. This suggested that the 3′-BCL2 breakpoints were not located within the mbr, but were several kb 3′ of the gene.
5′-
BCL2 and 3′-BCL2 breakpoint regions. The breakpoints of FL4104 and FL5117 are indicated in the (A) 5′-BCL2 and (B) 3′-BCL2breakpoint regions. The BamHI, EcoRI,HindIII, and SacI restriction sites are indicated with B, E, H, and S, respectively, according to published maps.10,35 (A) The position of the 5′-5′-BCL2 probe, the 5′-BCL2 probe, and the primers used for inverse PCR cloning of the 5′-BCL2/3′-BCL2 junction. (B) The position of the mbr probe and primers used for long-distance PCR of the novel 3′-BCL2 region. Short arrows indicate t(14;18) breakpoints reported by Akasaka et al.10 Dark gray bars indicate the regions sequenced by us (bar V), by Akasaka et al10 (bar A), and by Buchonnet et al (GenBank AF217803) (bar B). The position of our sequence relative to the restriction map was based on the distance from the mbr as determined by the long-range PCR, the overlap with the sequence published by Buchonnet et al, and the presence of aHindIII site. (C)The Southern blot results obtained after digestion of DNA obtained from both lymphomas and control placenta (p) with BamHI, SacI, and XbaI and after hybridization with the BCL2 mbr probe. The localization of theBamHI and SacI restriction sites, as well as the position of the BCL2 mbr probe, is shown in panel B. No rearranged bands were seen in the BamHI digests, but 2 and 1 rearranged bands, respectively, were seen in the SacI andXba1 digests.
5′-
BCL2 and 3′-BCL2 breakpoint regions. The breakpoints of FL4104 and FL5117 are indicated in the (A) 5′-BCL2 and (B) 3′-BCL2breakpoint regions. The BamHI, EcoRI,HindIII, and SacI restriction sites are indicated with B, E, H, and S, respectively, according to published maps.10,35 (A) The position of the 5′-5′-BCL2 probe, the 5′-BCL2 probe, and the primers used for inverse PCR cloning of the 5′-BCL2/3′-BCL2 junction. (B) The position of the mbr probe and primers used for long-distance PCR of the novel 3′-BCL2 region. Short arrows indicate t(14;18) breakpoints reported by Akasaka et al.10 Dark gray bars indicate the regions sequenced by us (bar V), by Akasaka et al10 (bar A), and by Buchonnet et al (GenBank AF217803) (bar B). The position of our sequence relative to the restriction map was based on the distance from the mbr as determined by the long-range PCR, the overlap with the sequence published by Buchonnet et al, and the presence of aHindIII site. (C)The Southern blot results obtained after digestion of DNA obtained from both lymphomas and control placenta (p) with BamHI, SacI, and XbaI and after hybridization with the BCL2 mbr probe. The localization of theBamHI and SacI restriction sites, as well as the position of the BCL2 mbr probe, is shown in panel B. No rearranged bands were seen in the BamHI digests, but 2 and 1 rearranged bands, respectively, were seen in the SacI andXba1 digests.
Cloning of the 5′-BCL2/3′-BCL2junctions
To clone the 5′-BCL2/3′-BCL2 junction at derivative chromosome 18 in both tumors, an inverse PCR was performed with primers 5′-side of the supposed location of the 5′-BCL2breakpoints (Figure 2A) on self-ligation products of tumor DNA digested with several restriction enzymes. In FL4104, inverse PCR on aHindIII digest yielded a product of approximately 2 kb. In FL5117, the PCR products were obtained with Sau3AI (2 kb) and TaqI (1.5 kb). Sequencing of the products in FL4104 and FL5117 showed that the breakpoints were 639 bp and 596 bp upstream of the BCL2 gene. Both breakpoints were present in 2 different CA repeats (Figure 3; Genbank X51898). The sequence that was juxtaposed to the 5′-BCL2 region was for a large part identical in FL4104 and FL5117, indicating that the 3′-BCL2 breakpoints of FL4104 and FL5117 were located close to each other. The sequence did not show homology to knownBCL2 breakpoint regions.
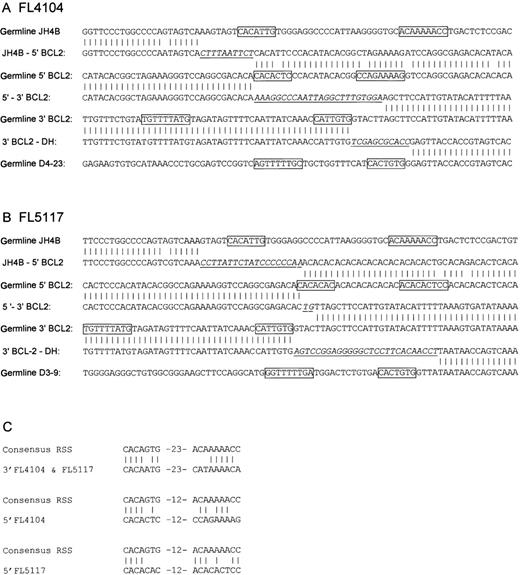
DNA sequences at the breakpoints in FL4104 and FL5117.
In each case, (A) FL4101 and (B) FL5117, 3 breakpoints were present: the 5′-BCL2/JH junction, the 3′-BCL2/DH junction, and the deletion breakpoint joining the BCL2 5′- and 3′-flanking regions. The breakpoints are aligned with the germline sequences of JH, DH, 5′-BCL2, and 3′-BCL2. Germline sequences are from the following GenBank accession numbers: DH, X97051; 5′-BCL2, X51898; and 3′-BCL2,AF204739. JH sequences are from GenBank, accession number J00256 and from V BASE36; N-nucleotides are underlined and in italics. Natural and cryptic RSS heptamers and nonamers are boxed. All sequences were obtained from 2 sequence reactions in different orientations. (C) An alignment of the cryptic RSS elements in FL4104 and FL5117 with the consensus IGH RSS. In both cases, the sequence of the cryptic RSS in the JH4 5′-BCL2 fusion product has a few mismatches with the published germline sequence. Possibly these mismatches represent somatic mutations acquired after the translocation. Therefore, the germline sequence has been aligned with the consensus RSS.
DNA sequences at the breakpoints in FL4104 and FL5117.
In each case, (A) FL4101 and (B) FL5117, 3 breakpoints were present: the 5′-BCL2/JH junction, the 3′-BCL2/DH junction, and the deletion breakpoint joining the BCL2 5′- and 3′-flanking regions. The breakpoints are aligned with the germline sequences of JH, DH, 5′-BCL2, and 3′-BCL2. Germline sequences are from the following GenBank accession numbers: DH, X97051; 5′-BCL2, X51898; and 3′-BCL2,AF204739. JH sequences are from GenBank, accession number J00256 and from V BASE36; N-nucleotides are underlined and in italics. Natural and cryptic RSS heptamers and nonamers are boxed. All sequences were obtained from 2 sequence reactions in different orientations. (C) An alignment of the cryptic RSS elements in FL4104 and FL5117 with the consensus IGH RSS. In both cases, the sequence of the cryptic RSS in the JH4 5′-BCL2 fusion product has a few mismatches with the published germline sequence. Possibly these mismatches represent somatic mutations acquired after the translocation. Therefore, the germline sequence has been aligned with the consensus RSS.
To determine whether the sequence cloned by inverse PCR was indeed from the 3′-side of the BCL2 gene, fiber FISH was performed on normal cells with a 3′-BCL2 cosmid probe, the mbr probe labeled in green, and the 1.5-kb FL5117-TaqI inverse PCR product labeled in red. The inverse PCR product gave a hybridization signal a few kb 3′-side of the mbr (not shown). Based on this observation, PCR was performed according to a long-range PCR protocol, with a forward primer just 5′-side of the mbr and a reverse primer 456 bp 3′-side of the 3′-BCL2 breakpoints of both FL4104 and FL5117. The PCR was performed on normal placenta DNA and on cosmid F11 containing the 3′-end of the BCL2 gene. A product of 7 kb was obtained with both templates, indicating that the breakpoints were located approximately 6.5 kb 3′-side of the mbr, as estimated upon gel analysis. Using this PCR product as a template, 1022 bp of sequence surrounding the breakpoints was determined (GenBank AF204739). Sequence analysis showed that the breakpoints were located 2 bp from each other within a MER1-like repeat unit named Charlie2 (according to the CENSOR database). The sequence showed no similarity with a previously identified breakpoint region far 3′-side of the mbr.10 The presence of a HindIII site 193 bp 5′-side of the breakpoints and the presence of a 66-bp overlap with a recently published sequence of the 3′-MBR region of BCL2 (GenBank AF217803; 2852-2918 bp) enabled us to map both breakpoints 6.2 kb 3′-side of the MBR ofBCL2 (Figure 2B). At the 5′-BCL2/3′-BCL2 junction, N-nucleotides were present in both cases (Figure 3).
Cloning of the 3′-BCL2/DH and 5′-BCL2/JH junctions
On the fiber FISH level, the 3′-side of BCL2colocalized with cosmid U2-2, which covered most of the DH region. To clone the breakpoints, PCRs were performed with a primer 5′-side of the 3′-BCL2 breakpoints and 1 of 3 different primers specific for the 5′-recombination signal sequences of the D2, D3, and D7-27 (DHQ52) families, respectively.11 Products of 1600 bp (FL4104) and 500 bp (FL5117) were obtained with the D3-specific primer. Sequence analysis with a primer in the BCL2 breakpoint region revealed a juxtaposition of the BCL2 gene to D4-23 in FL4104 and a juxtaposition to D3-9 in FL5117. N-nucleotides were present at the breakpoint junctions (Figure 3). In addition, comparison with the 5′-BCL2/3′-BCL2 junctions and the germline sequence showed that 6 and 4 nucleotides of the sequence had been deleted in FL4104 and FL5117, respectively. Taking these deletions into account, the BCL2 breakpoints at the 3′-BCL2/DH junction were in both cases at the same nucleotide position. On the 3′-side, the breakpoints were flanked by a cryptic IG recombination signal sequence (RSS) consisting of a heptamer and a nonamer separated by a 23-bp spacer. The RSS shows the identity of 11 of 16 nucleotides to the consensus IGRSS12 (Figure 3C). The heptamer showed only 1 mismatch, and the pentamer contained 5 matches including adenosines at positions 5, 6, and 7 of the nonamer, in which most of the nonamer's RSS function resides.12 13
To amplify the junction between the 5′-BCL2 andJH locus, a PCR was performed on FL4104 and FL5117 with a primer 3′-side of the JH genes and another immediately 3′-side of the 5′-BCL2 breakpoints of both tumors. Products of 1100 bp (FL4104) and 1000 bp (FL5117) were obtained. Sequence analysis revealed that in both cases, the BCL2 gene was juxtaposed to the 5′-side of the JH4 gene, and N-nucleotides were present at the breakpoint junctions (Figure 3). In both cases a cryptic RSS with a 12-bp spacer was identified flanking the 5′-BCL2 breakpoint (Figure 3). Although less similar to the consensus RSS, both shared the first 4 nucleotides of the heptamer (CACA) with the consensus. In FL4104, adenosines were present at positions 5, 6, and 7 of the nonamer. In FL5117, the nonamer was less well defined and contained part of the CA repeat; still 6 of 9 bp were identical to the consensus sequence. The presence of these cryptic RSS elements strongly suggested that the BCL2 deletion had been mediated by the RAG-1 and RAG-2 proteins.
Discussion
We identified 2 cases of FL with excision of the BCL2gene from 18q21 and insertion of the gene into the IGHlocus. Sequencing of the BCL2/IGH junctions revealed that the IGH breakpoints were at the JH and DH RSS elements, and N-nucleotides were present at all breakpoints. This demonstrates that the insertion of BCL2 into IGHtook place during V(D)J recombination in precursor B cells, just like t(14;18) found in most other FLs.
In the case of the usual t(14;18), the mechanism of chromosome breakage at 14q32 is clearly a RAG-1/-2–mediated recombination, because IGH breakpoints are consistently located at RSSs. A normal RSS consists of conserved heptamer and nonamer sequences separated by a spacer of either 12 or 23 bp in length. The consensus sequence is CACAGTG-12/23-ACAAAAACC,12 with the nonamer being less conserved than the heptamer. Apart from these natural RSSs, RAG-1/-2–mediated cleavage occasionally occurs at sites other than theIG or T-cell receptor (TCR) VDJgenes.14 A comparison of these so-called cryptic RSS elements shows a great variety of sequences. The only feature shared by all these sequences is a CAC trinucleotide at the first 3 positions of the heptamer, with frequently an A at the fourth position. Experiments with plasmid substrates have also suggested that the CAC trinucleotide is the only indispensable part of the RSS, although nucleotides at other positions can influence recombination efficiency by several orders of magnitude. An example is the presence of one or more adenosines at positions 6, 7, and 8 of the nonamer, on which most of the nonamer's activity depends.12 13
Rearrangements involving cryptic RSSs at non-IG, non-TCR loci have been described in T-cell leukemia and in normal T cells, include deletions of HPRT,15,16TAL1,17,18 and MTS1genes19 and t(7;9),20 t(1;7),21and t(10;14).22 In the deletion cases, cryptic RSSs, at least consisting of the sequence CACA, were present at the deleted side of both deletion breakpoints in the following configuration: 5′-break-CACA…TGTG-break-3′. Two breakpoint regions, at theMTS1 and HPRT loci, were located in or at the border of a CA repeat. Other translocations with disrupted CA repeats have been described in a variety of leukemias and lymphomas.23-25 Strikingly, the 5′- and 3′-breakpoint configuration of the BCL2 gene excision in the present FL cases was identical to the above described RAG-1/-2–mediated deletions of TAL1, HPRT, and MTS1. This strongly suggests that the BCL2 gene has also been deleted from its locus by the RAG-1/-2–mediated recombination.
Noteworthy, the BCL2 excision breakpoints in the 2 cases described here were not located at the common t(14;18) breakpoint clusters. Although the latter has been extensively searched for possible cryptic RSS sequences, even the CAC trinucleotide has not been found at the breakpoint sites. It is therefore unlikely that normal t(14;18) breakage of chromosome 18q21 is induced by RAG-1/-2–mediated cleavage. Recently, a model for RAG-1/-2–mediated induction of the BCL1 and BCL2 breakpoints, which does not require RSS elements to be present at the oncogene locus, was proposed. In cell-free systems, it was demonstrated that blunt-ended DNA fragments in complex with truncated RAG-1 and RAG-2 proteins can act as transposons by invading intact plasmid DNA, thereby resulting in either complete insertion of the blunt-ended fragment or in a single-sided strand exchange.26 27
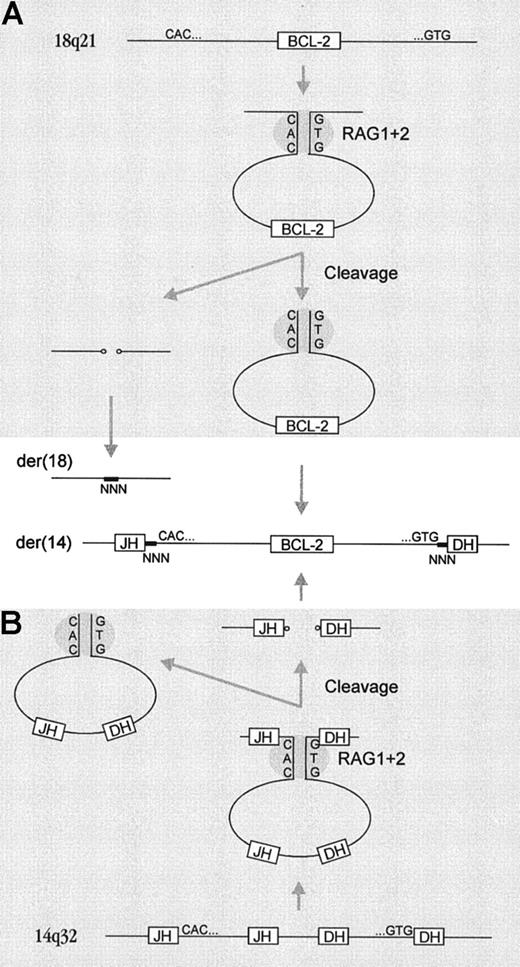
As a mechanism for insertion of the excised BCL2 gene into chromosome 14q32 in the present FL cases, one might initially think of this strand invasion model. The breakpoints at the IGH locus were, however, located at DH and JH RSS heptamer borders, with deletion of intervening sequences. It is therefore much more likely that the synaptic complex formation and excision of the DJ sequences occurred prior to the insertion event. This would imply that the RSS signal ends of the excised BCL2 gene were ligated to the DH and JH coding ends of the IGH locus, thereby forming a hybrid joint. A model for the transposition of BCL2 to theIGH locus shows excision of the BCL2 gene followed by diffusion to its target localization and insertion in theIgH locus (Figure 4). In a variant of this model, both BCL2 ends are not excised synchronously but metachronously, and both BCL2signal ends are subsequently excised and fused to the IgH coding ends. According to this variant model, the BCL2 gene is not freely diffusing from one to the other chromosome; however, this model needs a complex sequence of events at both sides of theBCL2 and IGH loci.
Mechanism for insertion of the
BCL2 gene into the IGHlocus. (A) Cryptic RSSs at both sides of the BCL2 gene form a synaptic complex followed by excision of the BCL2gene. The hairpin ends of the remaining 18q21 locus are opened, processed, and ligated. (B) At the 14q32/IGH locus, excision of DH and JH sequences takes place, but without ligation of the coding ends. The coding end hairpins are opened and processed, and they finally form hybrid joints with the excised BCL2 gene. The hairpins are indicated with small circles; N-nucleotide insertions are indicated with thick lines and “NNN”; and RSSs are symbolized by CAC for forward or GTG for reverse.
Mechanism for insertion of the
BCL2 gene into the IGHlocus. (A) Cryptic RSSs at both sides of the BCL2 gene form a synaptic complex followed by excision of the BCL2gene. The hairpin ends of the remaining 18q21 locus are opened, processed, and ligated. (B) At the 14q32/IGH locus, excision of DH and JH sequences takes place, but without ligation of the coding ends. The coding end hairpins are opened and processed, and they finally form hybrid joints with the excised BCL2 gene. The hairpins are indicated with small circles; N-nucleotide insertions are indicated with thick lines and “NNN”; and RSSs are symbolized by CAC for forward or GTG for reverse.
In normal B and T cells, hybrid joints can be found in the form of inversions in the V(D)J region.28,29 In cell-free systems, hybrid joints can be produced by a mixture of truncated RAG proteins and HMG1.30 Furthermore, their formation in vivo was shown to be independent of Ku86,31 in contrast to both signal joint and coding joint formation. On the basis of these results it was proposed that the mechanism of hybrid joint formation is RAG-1/-2–mediated strand invasion of the coding end hairpin by the blunt signal end in the synaptic complex, essentially identical to the RAG-1/-2–mediated insertion reactions observed in cell-free systems. However, a mechanism in which an intact coding end hairpin is directly attacked by the signal end would predict hybrid joints without N-nucleotides and without deletions because both are the result of coding end processing after opening of the hairpin. In the case of ourBCL2 insertions, as well as in previously studied hybrid joints,32 extensive N-regions and deletions were present at the hybrid joints. This implies that opening of the coding end hairpin and subsequent processing by terminal deoxynucleotidyl transferase (TdT) and exonucleases preceded the joining reaction. Therefore, the actual joining must have involved 2 blunt ends, which is not compatible with the strand invasion hypothesis. This suggests that the joining reaction was mediated by double-stranded DNA (dsDNA) repair proteins, just like normal coding joint and signal joint formation.
The presently described cases of FL have a distinct genomic configuration compared with FL with the usual t(14;18). Whereas concurrent 5′- and 3′-BCL2 breakpoints have been described previously,33 34 our cases contained a unique 3′-breakpoint 6.2 kb from the mbr in a region not previously sequenced. This implies that this breakpoint may have been missed in previous studies, especially because the configuration described will not lead to a cytogenetically detectable t(14;18). Furthermore, in contrast to the present cases, earlier studies did not indicate any resemblance to the RSS elements in both the mbr and minor breakpoint cluster (mcr) region of BCL2, and even the CAC trinucleotide was not found, indicating that these t(14;18) breakpoints are not directly mediated by RAG-1/-2. For these breakpoints, the RAG-1/-2–mediated DNA strand–attack mechanism described above may be an attractive model.
Supported by the Dutch Cancer Society, grant 95-1047.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Philip M. Kluin, Department of Pathology, Leiden University Medical Center, PO Box 9600, Gebouw 1, L1-Q 2300RC Leiden, The Netherlands; e-mail: pkluin@pat.azl.nl.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal