Abstract
The locus underlying hereditary resistance to the anticoagulant warfarin (symbol in the rat, Rw) was placed in relation to 8 positionally mapped gene-anchored microsatellite loci whose positions were known in the genome maps of the rat, mouse, and human.Rw segregated with the markers Myl2 (zero recombinants) and Itgam, Il4r, andFgf2r (one recombinant each) during linkage analysis in a congenic warfarin- and bromadiolone-resistant laboratory strain of rats. Comparative ortholog mapping between rat, mouse, and human placedRw onto mouse chromosome 7 at about 60 to 63 cM and onto one of the human chromosomes 10q25.3-26, 12q23-q24.3, and 16p13.1-p11.
Introduction
Warfarin, or 3-(α-acetonylbenzyl)-4-hydroxycoumarin, and its structural relatives are tied to human health and economics. Hereditary resistance to warfarin (locus symbol, Rw), and to several of its related compounds, has been observed in humans and rodents, foremostRattus norvegicus, R rattus, and Musspp.1,2 The mechanism of warfarin resistance in humans, rats, and mice is thought to have orthologous genetic underpinnings; that is, in all species mutations within the same enzyme complex (presumably a vitamin K epoxide reductase) mediate resistance.3
Resistance is a concern both in human medicine and rodent control. In human medicine, warfarin is a frequently used orally administered anticoagulant substance.1 Resistance may cause failure of oral anticoagulant therapy, which may lead to thrombosis and stroke. Rodent pest control worldwide relies largely on the use of warfarin-based rodenticides. The evolution of resistance has led to locally severe management problems resulting in public health concerns and economic loss.2,4 The gene is also of specific interest because it is a central yet-to-be isolated enzyme of the vitamin K cycle.3,5 6
Rw has been recently mapped with anonymous microsatellite markers and one gene-anchored locus (Myl2) in the rat.7 An approximately 6-cM spanning interval on rat chromosome 1 (Chr 1), located between the microsatellitesD1Rat193 and D1Rat130, defined the approximate map position of the Rw gene. However, Myl2 is not well suited for comparative mapping either in the mouse or human. It has no mapped ortholog in the mouse other than the synthetic probe,Mylpf, which hybridized to mouse chromosome 7 (Chr 7). In humans, the use of Myl2 is misleading as anchor because any of the 5 chromosomes 9, 10, 11, 12, and 16 may be partially homologous to the 6-cM interval. Thus, it is currently impossible to even tentatively place Rw in the human genome map.
Here, a warfarin- and bromadiolone-resistant congenic strain of laboratory rats (Rattus norvegicus) is used to placeRw in relation to 8 genes whose locations were known in the genomic maps of the rat, mouse, and human.8 We also use the recently published radiation hybrid map of the rat to delineate regions of homology with the mouse and human.9
Study design
Origin of rats and resistance tests
A previously described laboratory strain was used that was derived from a cross between a wild-caught male rat from the Münsterland area in Germany, homozygous resistant to the anticoagulants warfarin and bromadiolone, with a Wistar albino susceptible (WAS) female.7,10 All rats were tested for warfarin and bromadiolone resistance with the blood clotting response (BCR) method.7,10,11,12 The observed segregation pattern in the back-cross population was compatible with the hypothesis that resistance to both compounds was due to the same dominant gene or, alternatively, 2 tightly linked genes.7 Genetic typing was done on 67 offspring of the seventh back-crossing generation.
Mapping with microsatellite loci
About 50 ng of phenol/chloroform purified DNA13 was used as template for polymerase chain reaction (PCR) amplification of the gene-anchored microsatellite markers D1Smu2 (adrenergic receptor α-2a, Adra2a), D1Smu4 (cytochromeP-450 2e1 ethanol indicuble; Cyp2e1),D1Smu5 (fibroblast growth factor receptor 2;Fgfr2), D1Smu6 (interleukin-4 receptor;Il4r), D1Smu7 (sodium channel, nonvoltage gated β, Scnn1b), D1Smu9 (protein kinase C β;Pkcb), and D1Smu10 (integrin α-M;Itgam)—all reference 8—following standard protocols.7,8 Previously generated genotype data for the markers D1Arb18 (myosin light chain, skeletal muscle;Myl2), D1Rat193, D1Mit13,D1Mgh21 were included into the analysis for reference.7
The Rw locus was mapped by haplotype analysis14as implemented in Map Manager Classic, version 2.6.5.15Briefly, markers were ordered and Rw integrated such that the total map length and the number of double crossovers were minimized while retaining the positions of published markers.14Linkage was assessed at the 99% confidence level. Map positions were tested with the “find best position” and “rearrange” (at 95% confidence level using χ2 statistics) commands, and LOD scores were deduced with the aid of the likelihood calculator. Recombination fractions less than or equal to 0.25 were directly converted into map distances given as cM ± SE and a 95% confidence interval (95% CI).16
Mapping resources
Most of this analysis used (1) the Mouse Genome Database (MGD), World Wide Web (URL:http://www.informatics.jax.org/), January 2000, at the Jackson Laboratory, Bar Harbor, ME; (2) the Rat Genome Database (RATMAP), World Wide Web (URL:http://www.ratmap.gen.gu.edu), 1998, at the University of Gothenborg, Gothenborg, Sweden; and (3) the UniGene genome resource on human, mouse and rat, World Wide Web (URL:http://www.ncbi.nlm.nih.gov/UniGene), January 2000, at the National Center for Biotechnology Information, National Institutes of Health, Bethesda, MD. As additional resources, see references 8 and 9, and those listed previously.7
Results and discussion
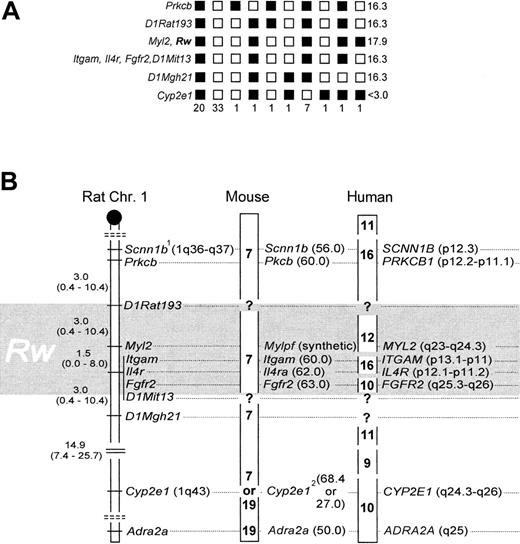
Rw and Myl2 cosegregated without recombination,7 and their linkage position was delineated by one recombinant to the markers Itgam, Il4r, and Fgfr2, and 2 recombinants to D1Rat193, respectively (Figure 1A). To accommodate for a 2% error associated with the BCR method,7,12 and to adjust the mapping resolution according to sample size,14,15 the genomic location of Rw is better described as a 6.0 ± 4.0-cM interval, with an associated 95% CI of 0.8 to 20.8 cM (Figure 1B), which includes Myl2,Itgam, Il4r, and Fgfr2. Data from linkage to coat color,17 microsatellites,7and these gene-anchored loci now independently affirm the map location for Rw in the rat. Fgfr2, Itgam, andIl4r cannot be further resolved with our strain. However, previous work suggests that Fgfr2 is located ∼1.6 cM distal of the latter 2.8Scnn1b was placed as published8 (Figure 1B) because allelic heterogeneity precluded genotype scoring (not shown). Cyp2e1 was weakly associated with Rw (13 recombinants; Figure 1A), andAdra2a was essentially unlinked to Rw (24 recombinants; data not shown).
Genotyping results and comparative mapping of
Rw in rat, mouse, and human. (A) Haplotypes and their respective frequencies. Black boxes represent WAS-derived alleles, white boxes a combination of the wild-derived allele with a WAS allele. Haplotype frequencies are given underneath each column (n = 67). LOD scores are presented on the right panel. (B) The distal q-arm of rat Chr 1 containing Rw, and its orthologs in mouse and human. The map interval containing Rw is shaded in gray. In the rat, map distances and their corresponding 95% CI are given in cM. The centromere is depicted as a black circle, and broken double lines separate linkage groups. In the mouse, chromosomal assignments are noted in boldface, and locus positions are given in cM (in brackets). In humans, chromosomal assignments are denoted as above, but locus positions are as in the cytogenetic human genome map.1 indicates allelic heterogeneity between laboratory and wild strain;2, conflicting information on chromosomal location of Cyp2e1, see references 8 and 9.
Genotyping results and comparative mapping of
Rw in rat, mouse, and human. (A) Haplotypes and their respective frequencies. Black boxes represent WAS-derived alleles, white boxes a combination of the wild-derived allele with a WAS allele. Haplotype frequencies are given underneath each column (n = 67). LOD scores are presented on the right panel. (B) The distal q-arm of rat Chr 1 containing Rw, and its orthologs in mouse and human. The map interval containing Rw is shaded in gray. In the rat, map distances and their corresponding 95% CI are given in cM. The centromere is depicted as a black circle, and broken double lines separate linkage groups. In the mouse, chromosomal assignments are noted in boldface, and locus positions are given in cM (in brackets). In humans, chromosomal assignments are denoted as above, but locus positions are as in the cytogenetic human genome map.1 indicates allelic heterogeneity between laboratory and wild strain;2, conflicting information on chromosomal location of Cyp2e1, see references 8 and 9.
Gene order is highly conserved between rat and mouse within this chromosomal region, and the close spacing of Itgam,Il4ra, and Fgfr2 in both species reflects this.8,9 When these loci are used as anchor, the genetic map interval 60 to 63 cM on mouse Chr 7 emerges as likely location ofwar (Figure 1B). War was previously mapped close to fr (frizzy),18 and its position was extrapolated to the genetic map as 62.5 cM on Chr 7.19Both results are confirmed by our new data. In addition, we have now identified suitable anchor loci to fine-map war by genotyping resistant laboratory mouse strains.
In the examined chromosome segment, homology among rat and human is less preserved than that among rat and mouse.9 Human chromosomes 9, 10, 11, 12, and 16 are partial homologs thereof.9 However, the position of Rw can be narrowed to 10q25.3-q26, 12q23-24.3, and 16p11-13 (Figure 1B). With the available resolution of our strain we cannot discriminate among these 3 possibilities. However, to further narrow the location of Rwin the human genome, we suggest that MYL2, ITGAM,IL4R, and FGFR2 may be useful as anchor markers during genetic typing experiments that use cohorts of patients resistant to warfarin.
In conclusion, the gene-anchored map position of Rw in the rat, mouse, and human will allow for targeted searches for candidate genes and expressed sequence tagged sites on the specified chromosome segments and for initiation of the next positional cloning steps.
Acknowledgments
We thank M. Gitter, H. Naujeck, and E. Kampling for the rearing of animals and technical help. We thank R.K. Wayne for his advice and for providing laboratory facilities for molecular typing.
M.H.K and the project were partially funded by a nonresident tuition fellowship, an OBEE departmental fellowship, an ISOP fellowship (all UCLA), and an NSF dissertation improvement grant.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Michael H. Kohn, Department of Organismic Biology, Ecology, and Evolution (OBEE), University of California at Los Angeles, 621 Charles E. Young Dr South, Los Angeles, CA 90095-1606; e-mail:michaelk@biology.ucla.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal