Abstract
Dendritic cells (DCs) are professional antigen-presenting cells in the immune system and can be generated in vitro from hematopoietic progenitor cells, DC precursors, and monocytes in peripheral blood. Serial analysis of gene expression (SAGE) was conducted in lipopolysaccharide (LPS)-stimulated mature and activated DCs (MADCs) derived from human blood monocytes. A total of 31 837 tag sequences from an MADC cDNA library represented 10 962 different genes, and these data were compared with SAGE data for monocyte-derived immature DCs (IMDCs). Many of the genes, such as germinal center kinase–related protein kinase, cystatin F, interferon (IFN)-α–inducible protein p27, EBI3, HEM45, actin-bundling protein, ELC, DC-LAMP, serine/threonine kinase 4, and several genes in expressed sequence tags, were differentially expressed in MADCs, and those encode proteins related to cell structure, antigen-processing enzymes, chemokines, and IFN-inducible proteins. The profile of MADCs was also compared with that of LPS-stimulated monocytes. The Epstein-Barr virus–induced gene 3 and IFN-α–inducible protein p27 are newly identified to be specifically and highly expressed in MADCs, but not in LPS-stimulated monocytes. The comprehensive identification of specific genes expressed in human IMDCs and MADCs should provide candidate genes to define heterogeneous subsets as well as the function and maturation stage of DCs.
Introduction
Dendritic cells (DCs) play a pivotal role in the immune system by processing and presenting antigens to CD4+naive T cells.1 DCs have also been reported to be involved in the direct induction of CD8+ cytotoxic T cells,2 immunoglobulin production by B cells,3 and T-cell tolerance.4,5 DCs, which are found in virtually every tissue and fluid, are mostly immature DCs expressing a chemokine receptor, CCR1 or CCR6, and are able to capture antigens. Once activated by inflammatory stimuli, DCs mature and start to express a chemokine receptor, CCR7, and also produce chemokines that recruit various types of leukocytes, including immature DCs at the inflammatory site. These mature and activated DCs (MADCs) are consequently recruited via the lymphatics to the T-cell–rich areas of secondary lymphoid organs to stimulate CD4+ naive T cells6-8 by SLC produced in the lymphatic endothelium and high endothelial venules.
DCs undergo a series of events leading to irreversible maturation and ending with apoptotic cell death.9 During their migration, DCs undergo further changes of phenotype and function, including loss of antigen uptake and processing and increase of accessory function. DC maturation can be influenced by a variety of factors, notably microbial and inflammatory mediators. Whole bacteria, the gram-negative microbial cell-wall component lipopolysaccharide (LPS),10monocyte-conditioned medium,11 and cytokines such as interleukin (IL)-1 and tumor necrosis factor-α (TNF-α) all stimulate DC maturation. Ceramide, which is induced by maturation signals, can shut down antigen capture by the DCs.12Mature DCs express high levels of the NF-kB family of transcriptional control proteins,13,14 which regulate the expression of many genes encoding immune and inflammatory proteins. During maturation of DCs, the expression of CD83, CD80, CD86, and CD49 is kept, but that of CD1a is reduced.10 15
DCs in lymphoid and nonlymphoid organs vary in their surface markers and functions and therefore have different names, such as Langerhans cells in the epidermis; interdigitating DCs in lymph nodes; interstitial DCs in the heart, lung, kidney, and intestine; and thymic DCs in the thymus. DCs are thought to belong to a lineage distinct from monocytes and macrophages. However, it has been reported that macrophages and DCs share a common progenitor.16,17 Human DCs have been generated from CD34+ precursor cells isolated from cord blood18 and bone marrow19 in the presence of granulocyte-macrophage colony-stimulating factor (GM-CSF) and TNF-α. Moreover, blood monocytes cultured with GM-CSF and IL-4 differentiate into nonadherent CD1a+CD14low cells with morphologic and functional characteristics of DCs.20,21 However, the process of differentiation from monocytes to DCs has not yet been explored systematically. To clarify this point, we previously performed serial analysis of gene expression (SAGE) in immature DCs (IMDCs) derived from human blood monocytes.22 Many of the genes that were differentially expressed in IMDCs were identified to be genes encoding proteins related to cell structure, cell motility, and chemokines. In this study, to further understand the characteristics of DC differentiation, we have applied the SAGE method to human MADCs. The SAGE method has proved to be a powerful means for the quantitative cataloging and comparison of expressed genes in the cells or tissues from various physiologic, developmental, and pathologic states.23-27
Materials and methods
Preparation of cells
Peripheral blood mononuclear cells (PBMCs) were isolated from venous blood drawn from normal healthy volunteers at the Tokyo Metropolitan Red Cross Blood Center.22 Briefly, PBMCs were isolated by centrifugation on a Ficoll-Metrizoate density gradient (d = 1.077, Lymphoprep; Nycomed, Oslo, Norway) and suspended in RPMI 1640 medium containing 7.5% heat-inactivated fetal calf serum (FCS; GIBCO/LIFE Technologies, Tokyo, Japan), 100 μg/mL streptomycin, and 100 U/mL penicillin G. The FCS contained 3 pg of LPS per milliliter as assessed by a Limulus amebocyte lysate. PBMCs were incubated with anti-CD14 monoclonal antibody (mAb) bound with microbeads, and monocytes were isolated by passing the PBMCs through a magnetic cell separation system (MACS; Miltenyi Biotec, Germany) with column type VR. These cell suspensions were then aliquoted into plastic tissue culture plates and incubated for 30 minutes at 37°C in 5% CO2 to obtain the highly purified cells. More than 99% of the cells were judged to be monocytes by morphology, by positive staining for CD14 (LeuM3; Becton Dickinson, San Jose, CA) in flow cytometric analysis, and by nonspecific esterase staining.
Phenotyping with monoclonal antibodies
The expression of leukocyte cell surface markers and cytoplasmic antigens was assessed. The cells were sequentially incubated with optimal concentrations of biotinylated anti-CD86 (2331; PharMingen, San Diego, CA) and anti-CD1a mAbs (HI 149; PharMingen), followed by fluorescein isothiocyanate (FITC)-labeled streptavidin and by phycoerythrin (PE)-conjugated goat anti-mouse IgG, respectively, or directly stained with FITC-labeled anti–HLA-DR (4C3; PharMingen), mouse anti-CD80 (MAB104; Coulter, Fullerton, CA), CD83 (HB15a; Coulter), and PE-labeled anti-CD14 (M5E; PharMingen) mAbs. To block nonspecific FcR-mediated binding of mAbs, cells were incubated for 60 minutes at 4°C with normal goat serum (Cedarlane Laboratories Ltd, Hornby, Ontario, Canada) before staining. For all experiments, irrelevant control mAbs of the same IgG isotype and second step controls were included. Stained cells were fixed with 1% paraformaldehyde (Sigma), and analyses were performed using the EPICS XL (Beckman Coulter, Hialeah, FL).
SAGE protocol
mRNAs of MADCs were purified from a mixture of total RNA from 6 donors. Monocytes were incubated with IL-4 (100 U/mL; Ono Pharmaceutical Co, Ltd, Japan) or GM-CSF (500 U/mL; Kirin Brewery Co, Ltd, Japan) in RPMI 1640 medium containing 7.5% FCS in 5% CO2 at 37°C for 7 days, then incubated with 100 μg/mL of LPS for 2 days. Total RNA from these cells was isolated by direct lysis in RNAzol B (Cinna/Biotex Laboratories, USA). Poly(A)+ RNA was isolated using the FastTrac mRNA purification kit (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. SAGE was done as described.23,24 28 SAGE libraries were generated using 1.5 μg poly(A)+ RNA, which was converted to cDNA with a BRL synthesis kit (GIBCO BRL, Rockville, MD) with the inclusion of primer biotin-labeled 5′-T18-3′. The cDNA was cleaved with the restriction enzyme NlaIII, and the 3′-terminal cDNA fragments were bound to streptavidin-coated magnetic beads (Dynal, Oslo, Norway). After ligation to oligonucleotides containing recognition sites for BsmF1, the linked cDNAs were released from the beads by digestion with BsmF1. The released tags were ligated to one another, concatenated, and cloned into theSphI site of pZero 1.0 (Invitrogen). Colonies were screened with polymerase chain reaction (PCR) using M13 forward and M13 reverse primers. PCR products containing inserts of longer than 400 bp were sequenced with the Big Dye terminator kit and analyzed with a 377 ABI automated 96-lane sequencer (Perkin-Elmer, Branchburg, NJ). All electropherograms were reanalyzed by visual inspection to check for ambiguous bases and to correct misreads.
SAGE was performed on mRNA from MADCs and LPS-stimulated monocytes. Sequence files were analyzed with the SAGE software,28 the National Center for Biotechnology Information (NCBI) SAGE database (http://www.ncbi.nlm.nih.gov/SAGE/), and NCBI's sequence search tool (Advanced BLAST search,http://www.ncbi.nlm.nih.gov/BLAST/). After elimination of linker sequences and the repeated ditags, a total of 31 837 tag sequences from MADCs were analyzed.
Reverse transcriptase (RT)-PCR
Total RNAs (200 ng) were prepared by the use of RNAzol B. The RNA was reverse-transcribed in 50 μL of 10 mmol/L Tris-HCl (pH 8.3), 6.5 mmol/L MgCl2, 50 mmol/L KCl, 10 mmol/L dithiothreitol, 1 mmol/L of each dNTP, 2 μmol/L random hexamer, and 2.4 U/μL of Moloney murine leukemia virus reverse transcriptase for 1 hour at 42°C. The cDNA, corresponding to 40 ng of total RNA, was boiled for 3 minutes and quenched on ice before amplification by PCR. The conditions for PCR were as follows: in a 50-μL reaction, 0.15 μmol/L of each primer; 1.25 μmol/L of each dGTP, dATP, dCTP, and dTTP (Toyobo); 50 mmol/L KCl; 10 mmol/L Tris-HCl, pH 8.3; 0.15 mmol/L MgCl2; and AmpliTaq (Perkin-Elmer). The cycle number of PCR for each gene is shown in parentheses and primers used were as follows: CCR7 (28): sense 5′-TCCTTCTCATCAGCAAGCTGTC-3′, antisense 5′-GAGGCAGCCCAGGTCCTTGAAG-3′; actin-bundling protein (25): sense 5′-ATGGACCTGTCTGCCAATCAG-3′, antisense 5′-CTTTGATGTTGTAGGCGCCA-3′; interferon (IFN)-inducible protein 27 (30): sense 5′-TCTGCTCTCACCTCATCAGCA-3′, antisense 5′-CCTGGCATGGTTCTCTTCTCTG-3′; HEM45 (28): sense 5′-GGTGCTGTGCTGTACGACAAGT-3′, antisense 5′-CGGATTCTCTGGGAGATTTGAT-3′; EBI3 (30): sense 5′-TGTTCTCCATGGCTCCCTACGT-3′, antisense 5′-TACTTGCCCAGGCTCATTGTGG-3′; DC-LAMP (30): sense 5′-GGCCCTAGCTTAGCCCCTTATT-3′, antisense 5′-CTCCGAGGTGAAAAAACCGA-3′; cystatin F (28): sense 5′-TTCCCAGGACCTTAACTCACGT-3′, antisense 5′-GGTGTTTGTCATGGCTGTGGT-3′; CD23 (28): sense 5′-GGAGGAACTTCGAGCTGAACA-3′, antisense 5′-AGTTCCGAAGGCCAATCCA-3′; lysosomal acid lipase (28): sense 5′-TCTTCCCCAGAGTGCGTTTTt-3′, antisense 5′-CATGGAACACCAAGTTGGTGAT-3′; CD11b (28): sense 5′-TGTGAAAGGGCTCTGCTTCCT-3′, antisense 5′-TCTTAAAGGCATTCTTTCGGGC-3′; IgGFcR (28): sense 5′-CTTTTCCGTGCTTACCTGCAG-3′, antisense 5′-AAATCAGCATCCTGGGCCT-3′; factor XIII (28): sense 5′-ATTGGCCCTAGAAACTGCCCT-3′, antisense 5′-TGGACTTTTGCTTGGCCAGA-3′; MacMARCKS (40): sense 5′-CCACGTGAAAAGCAATGGAGA-3′, antisense 5′-TTCTGAGGCTGCACTAGCCTCT-3′; IL-12 p35 (40): sense 5′-CCTTCACCACTCCCAAAACCT-3′, antisense 5′-TGAAATTCAGGGCCTGCATC-3′; IL-12 p40 (30): sense 5′-GGATGCCCCTGGAGAAATGG-3′, antisense 5′-CTCCCAGCTGACCTCCACCT-3′; and adenylyl cyclase–associated protein (CAP) (28): sense 5′-GCACTGTTCGCGCAGATTAA-3′, antisense 5′-A CAATGCCCACCACGTCAT-3′.
Reaction mixtures were incubated in a Perkin-Elmer DNA Thermal Cycler (denaturation, 60 seconds at 94°C; annealing, 60 seconds at 58°C; extension, 120 seconds at 72°C).
Statistical analysis
Statistical significance between samples was calculated as described previously.29 To analyze the correlation coefficients between the different libraries, 58 540, 31 837, 31 837, 57 560, 55 856, and 57 463 tags from IMDCs, MADCs, LPS-stimulated monocytes (LPS-Mo), monocytes (Mo), macrophage colony-stimulating factor (M-CSF)-induced macrophages, and GM-CSF–induced macrophages,30 respectively, were normalized to 31 837, and then all pairwise Pearson correlation coefficients for each library-to-library comparison were calculated using all normalized gene expression measurements.
Results
Surface phenotype of LPS-stimulated MADCs
To identify genes specifically expressed in MADCs, we generated SAGE libraries from human MADCs. Peripheral blood CD14+monocytes were cultured with GM-CSF plus IL-4 for 5 days and then stimulated with LPS for 2 days. Under these culture conditions, the cells differentiated into nonadherent CD14−, CD1a−/+, CD80+, CD83+, CD86+, CD40high, and HLA-DRhighcells with the dendritic morphology of MADCs (Figure1), which validates the phenotype of MADCs.
Surface phenotype of human blood monocyte–derived DCs stimulated by LPS (MADCs).
Human monocytes were cultured in RPMI 1640 medium plus 7.5% FCS in the presence of GM-CSF (500 U/mL) and IL-4 (100 U/mL) for 5 days and then stimulated with LPS for 2 days. After culture, the cells were washed and then stained with various antibodies as described in “Materials and methods.” The data are shown as histograms depicting the number of cells exhibiting various fluorescence intensities. The dotted lines represent MADCs stained with specific antibodies, the solid histograms represent IMDCs stained with specific antibodies, and the solid lines represent isotype-matched control. Results are representative of 3 independent experiments.
Surface phenotype of human blood monocyte–derived DCs stimulated by LPS (MADCs).
Human monocytes were cultured in RPMI 1640 medium plus 7.5% FCS in the presence of GM-CSF (500 U/mL) and IL-4 (100 U/mL) for 5 days and then stimulated with LPS for 2 days. After culture, the cells were washed and then stained with various antibodies as described in “Materials and methods.” The data are shown as histograms depicting the number of cells exhibiting various fluorescence intensities. The dotted lines represent MADCs stained with specific antibodies, the solid histograms represent IMDCs stained with specific antibodies, and the solid lines represent isotype-matched control. Results are representative of 3 independent experiments.
SAGE tag abundance in MADCs
A total of 31 837 tag sequences from the MADC library allowed the identification of 10 962 different genes. Next, the expressed genes were searched through the GeneBank database to identify individual genes. Table 1 shows the top 50 transcripts in MADCs. The most frequently expressed gene in human MADCs was identified to be TARC (expression frequency, 2.62%), followed by ferritin H-chain gene (2.57%) and β2-microglobulin gene (2.06%). Overall, the genes expressed abundantly in the MADC library mostly consist of genes encoding major histocompatibility complex (MHC) class I and class II, chemokines, molecules related to protein synthesis, and cytoskeleton proteins. More information is available on the internet at http://www.prevent.m.u-tokyo.ac.jp/SAGE.html/.
Transcript profile in LPS-stimulated mature DCs (MADCs)
| Tag no. . | SAGE tag . | UniGene cluster . | Description . |
|---|---|---|---|
| 831 | GGCACAAAGG | 56742 | TARC |
| 617 | TTGGGGTTTC | 62954 | Ferritin, heavy polypeptide 1 |
| 655 | GTTGTGGTTA | 75415 | β-2-microglobulin |
| 338 | AACGGGGCCC | 97203 | MDC |
| 272 | GTTCACATTA | 84298 | CD74 antigen |
| 255 | GGGGAAATCG | 76293 | Thymosin beta 10 |
| 238 | CCTGTAATCC | Multiple match | |
| 211 | TGTGTTGAGA | 181185 | Eukaryotic translation elongation factor-1 α 1 |
| 210 | CAAATCCAAA | 255996 | Germinal center kinase–related protein kinase |
| 208 | TTGGTGAAGG | 75968 | Thymosin, beta 4, X chromosome |
| 183 | CTGACCTGTG | 77961 | Major histocompatibility complex, class I, B |
| 175 | CCCTGGGTTC | 111334 | Ferritin, light polypeptide |
| 172 | GTGCGCTGAG | 77961 | Major histocompatibility complex, class I. C |
| 188 | GGCTGGGGGC | 75721 | Profilin 1 |
| 168 | CCCATCGTCC | 249495 | Ribosomal protein S8 |
| 155 | GTGAAACCCC | Multiple match | |
| 154 | CAAGCATCCC | No match | |
| 148 | ATAGTAGCTT | 118400 | Actin-bundling protein |
| 147 | CCACTGCACT | Multiple match | |
| 128 | GCTTTATTTG | 180852 | Actin, gamma 1 |
| 111 | GTGCACTGAG | 181244 | Major histocompatibility complex, class I, A or C (77961) |
| 110 | TCCGTGGTTG | 78516 | Neuronal tissue–enriched acidic protein |
| 110 | TTGGTCCTCT | 108124 | Homosapiens mRNA for homolog to yeast ribosomal protein L41 |
| 108 | CGCCGACGAT | 21205 | Interferon-α–inducible protein (clone IFI-8-16) |
| 105 | GCAGCCATCC | 4437 | Ribosomal protein L28 |
| 103 | GCCTGCTGGG | 2706 | Glutathione peroxidase 4 (phospholipid hydroperoxidase) |
| 99 | GCCATAAAAT | 1908 | Proteoglycan 1, secretory granule |
| 94 | TGTCCTGGTT | 179685 | Cyclin-dependent kinase inhibitor 1A (p21, Cp1) |
| 83 | GTGAAACCCT | Multiple match | |
| 92 | CACCTAATTG | 181353 | Pre–B-cell colony-enhancing factor |
| 92 | AATCTGCGCC | 833 | Interferon-stimulated protein, 15 kd |
| 81 | TTTTCTGAAA | 78138 | Thioredoxin |
| 81 | ACTTTTTCAA | 149587 | Homo sapiens mRNA; cDNA DKFZpS64E1616 (from clone DKFZpS84E1616) |
| 76 | CTAAGACTTC | No match | |
| 74 | TGCAGCACGA | 77961 | Major histocompatibility complex, class I, C |
| 74 | TGCCTGCACC | 135084 | Cystatin C |
| 71 | GCCCCCAATA | 188261 | Lectin, galactoside-binding, soluble, 1 (galactin 1) |
| 71 | ACCCTTGGCC | No match | |
| 70 | ACGCAGGGAG | 180532 | Human heart mRNA for heat-shock protein 90, partial cos |
| 68 | GTGCTGAATG | 77385 | Myosin, light polypeptide 6, alkali, smooth muscle and nonmuscle |
| 67 | TTCTGCTTTC | 1852 | CCR7 |
| 66 | GTAGCGCCTC | 143212 | Cystatin F (leukocystatin) |
| 66 | GACGGCGCAG | 73946 | Endothelial cell growth factor 1 (platelet derived) |
| 64 | GGGCATCTCT | 76607 | Human HLA-DR α-chain mRNA |
| 61 | TTGGCCAGGC | Multiple match | |
| 59 | CCATTGCACT | 194382 | Ataxia telenglactasia mutated (includes complementation groups A, C, and D) |
| 59 | GAAGCAGGAC | 180370 | Collin 1 (nonmuscle) |
| 59 | ACTGTGGCGG | 112242 | ESTs |
| 59 | TTCATACACC | No match | |
| 59 | CACAAACGGT | 165453 | Ribosomal protein S27 (metallopanstimulin 1) |
| Tag no. . | SAGE tag . | UniGene cluster . | Description . |
|---|---|---|---|
| 831 | GGCACAAAGG | 56742 | TARC |
| 617 | TTGGGGTTTC | 62954 | Ferritin, heavy polypeptide 1 |
| 655 | GTTGTGGTTA | 75415 | β-2-microglobulin |
| 338 | AACGGGGCCC | 97203 | MDC |
| 272 | GTTCACATTA | 84298 | CD74 antigen |
| 255 | GGGGAAATCG | 76293 | Thymosin beta 10 |
| 238 | CCTGTAATCC | Multiple match | |
| 211 | TGTGTTGAGA | 181185 | Eukaryotic translation elongation factor-1 α 1 |
| 210 | CAAATCCAAA | 255996 | Germinal center kinase–related protein kinase |
| 208 | TTGGTGAAGG | 75968 | Thymosin, beta 4, X chromosome |
| 183 | CTGACCTGTG | 77961 | Major histocompatibility complex, class I, B |
| 175 | CCCTGGGTTC | 111334 | Ferritin, light polypeptide |
| 172 | GTGCGCTGAG | 77961 | Major histocompatibility complex, class I. C |
| 188 | GGCTGGGGGC | 75721 | Profilin 1 |
| 168 | CCCATCGTCC | 249495 | Ribosomal protein S8 |
| 155 | GTGAAACCCC | Multiple match | |
| 154 | CAAGCATCCC | No match | |
| 148 | ATAGTAGCTT | 118400 | Actin-bundling protein |
| 147 | CCACTGCACT | Multiple match | |
| 128 | GCTTTATTTG | 180852 | Actin, gamma 1 |
| 111 | GTGCACTGAG | 181244 | Major histocompatibility complex, class I, A or C (77961) |
| 110 | TCCGTGGTTG | 78516 | Neuronal tissue–enriched acidic protein |
| 110 | TTGGTCCTCT | 108124 | Homosapiens mRNA for homolog to yeast ribosomal protein L41 |
| 108 | CGCCGACGAT | 21205 | Interferon-α–inducible protein (clone IFI-8-16) |
| 105 | GCAGCCATCC | 4437 | Ribosomal protein L28 |
| 103 | GCCTGCTGGG | 2706 | Glutathione peroxidase 4 (phospholipid hydroperoxidase) |
| 99 | GCCATAAAAT | 1908 | Proteoglycan 1, secretory granule |
| 94 | TGTCCTGGTT | 179685 | Cyclin-dependent kinase inhibitor 1A (p21, Cp1) |
| 83 | GTGAAACCCT | Multiple match | |
| 92 | CACCTAATTG | 181353 | Pre–B-cell colony-enhancing factor |
| 92 | AATCTGCGCC | 833 | Interferon-stimulated protein, 15 kd |
| 81 | TTTTCTGAAA | 78138 | Thioredoxin |
| 81 | ACTTTTTCAA | 149587 | Homo sapiens mRNA; cDNA DKFZpS64E1616 (from clone DKFZpS84E1616) |
| 76 | CTAAGACTTC | No match | |
| 74 | TGCAGCACGA | 77961 | Major histocompatibility complex, class I, C |
| 74 | TGCCTGCACC | 135084 | Cystatin C |
| 71 | GCCCCCAATA | 188261 | Lectin, galactoside-binding, soluble, 1 (galactin 1) |
| 71 | ACCCTTGGCC | No match | |
| 70 | ACGCAGGGAG | 180532 | Human heart mRNA for heat-shock protein 90, partial cos |
| 68 | GTGCTGAATG | 77385 | Myosin, light polypeptide 6, alkali, smooth muscle and nonmuscle |
| 67 | TTCTGCTTTC | 1852 | CCR7 |
| 66 | GTAGCGCCTC | 143212 | Cystatin F (leukocystatin) |
| 66 | GACGGCGCAG | 73946 | Endothelial cell growth factor 1 (platelet derived) |
| 64 | GGGCATCTCT | 76607 | Human HLA-DR α-chain mRNA |
| 61 | TTGGCCAGGC | Multiple match | |
| 59 | CCATTGCACT | 194382 | Ataxia telenglactasia mutated (includes complementation groups A, C, and D) |
| 59 | GAAGCAGGAC | 180370 | Collin 1 (nonmuscle) |
| 59 | ACTGTGGCGG | 112242 | ESTs |
| 59 | TTCATACACC | No match | |
| 59 | CACAAACGGT | 165453 | Ribosomal protein S27 (metallopanstimulin 1) |
The top 50 transcripts expressed in MADCs are listed. The tag sequence represents the 10-bp SAGE lag. UniGene clusters are listed. More information is available on the Internet athttp://www.prevent.m.u-tokyo.ac.jp/SAGE.html/.
Comparison of expression patterns in MADCs
IMDC SAGE libraries were generated from human monocytes cultured in GM-CSF plus IL-4 plus TNF-α for 5 days.22 Under these culture conditions, the cells differentiated into nonadherent CD1a+, CD80low/−, CD86low/−, HLA-DR+, CD40+, and CD83− cells with the dendritic morphology of IMDCs (Figure 1). The 58 540 and 31 837 tag sequences from the IMDC and MADC libraries, respectively, were normalized to 31 837, and then comparison of the expressed genes between IMDCs and MADCs was performed. The expression levels of most of the transcripts in these cells were similar (Figure2). However, 225 transcripts were found to be statistically different (P < .01) between these types of cells. Expression levels of 95 of 225 genes were decreased in MADCs as compared with those in IMDCs. Conversely, 130 transcripts were expressed at higher levels in MADCs than in IMDCs. Table2 shows the top 50 increased transcripts in MADCs as compared with IMDCs. The most frequently increased transcript was identified to be germinal center kinase–related protein kinase (70-fold), followed by CCR7 (67-fold), cystatin F (66-fold), and so on. The transcripts increased in MADCs mainly consisted of genes encoding chemokines such as RANTES, ELC, PARC, MDC, and TARC; a chemokine receptor CCR7; enzymes such as germinal center kinase–related protein kinase, metallothionein 2A, and serine/threonine kinase 4; and IFN-inducible proteins such as IFN-stimulated protein 15 kd, IFN-α–inducible protein p27, IFN-α–inducible protein p78, HEM45 (IFN-stimulated gene 20 kd), IFN-α–inducible protein (clone IFI-6-16), and IFN-inducible protein 17. Table 2 compares the SAGE data for MADCs with those for monocytes, M-CSF–induced macrophages, and GM-CSF–induced macrophages. These data demonstrate that many of the genes, which are highly expressed in MADCs, are low in monocytes and macrophages.
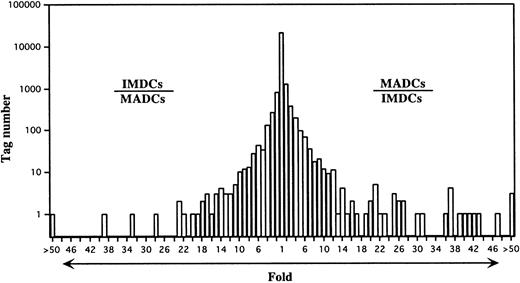
Comparison of gene expression frequency in IMDCs and MADCs.
A semilogarithmic plot reveals that 44 tags were decreased more than 10-fold in MADCs compared with IMDCs, whereas 71 tags were increased more than 10-fold in MADCs compared with IMDCs. Each number of tags was normalized to 31 837. The relative expression of each transcript was determined by dividing the number of tags observed in IMDCs or MADCs, as indicated. To avoid division by 0, a tag value of 1 for any tag that was not detectable in one sample was used. These ratios are plotted on the abscissa. The number of genes displaying each ratio is plotted on the ordinate.
Comparison of gene expression frequency in IMDCs and MADCs.
A semilogarithmic plot reveals that 44 tags were decreased more than 10-fold in MADCs compared with IMDCs, whereas 71 tags were increased more than 10-fold in MADCs compared with IMDCs. Each number of tags was normalized to 31 837. The relative expression of each transcript was determined by dividing the number of tags observed in IMDCs or MADCs, as indicated. To avoid division by 0, a tag value of 1 for any tag that was not detectable in one sample was used. These ratios are plotted on the abscissa. The number of genes displaying each ratio is plotted on the ordinate.
Increased transcripts in MADCs
| Fold . | n . | SAGE tag . | UniGene cluster . | Description . | |||||
|---|---|---|---|---|---|---|---|---|---|
| IMDC . | MADC . | LPS-Mo . | Mo . | M-CSF . | GM-CSF . | ||||
| 70 | 3 | 210 | 2 | 2 | 1 | 18 | CAAATCCAAA | 227400 | Germinal center kinase–related protein kinase |
| 67 | 1 | 67 | 6 | 0 | 0 | 0 | TTCTGCTTTC | 1652 | CCR7 |
| 66 | 1 | 86 | 0 | 0 | 0 | 0 | GTAGCGCCTC | 143212 | Cystatin F (leukocystatin) |
| 47 | 1 | 47 | 4 | 0 | 6 | 11 | GATCCCAACT | 118766 | Metallothionein 2A |
| 43 | 0 | 43 | 1 | 0 | 0 | 0 | GGCGTTTAGA | No match | |
| 42 | 0 | 42 | 11 | 0 | 0 | 0 | AAAAATCGGC | 226695 | RANTES |
| 41 | 2 | 62 | 48 | 4 | 0 | 1 | AATCTGCGCC | 833 | Interferon-stimulated protein, 15 kd |
| 40 | 0 | 40 | 0 | 0 | 0 | 0 | CCAGGGGAGA | 2867 | Interferon-α–inducible protein p27 |
| 38 | 1 | 39 | 3 | 1 | 1 | 0 | GGTGCCCAGT | 75607 | MARCKS, 8CK-L |
| 37 | 4 | 148 | 0 | 0 | 1 | 1 | ATAGTAGCTT | 118400 | Actin-bundling protein |
| 36 | 1 | 36 | 0 | 1 | 0 | 0 | CGGAGCCGGC | 34267 | ESTs |
| 31 | 0 | 31 | 0 | 0 | 0 | 0 | GCTCCCTACG | 185705 | EBI3 |
| 30 | 1 | 30 | 11 | 1 | 1 | 0 | AGTGCCGTGT | 76391 | Interferon-inducible protein p75 |
| 27 | 0 | 27 | 0 | 0 | 0 | 0 | CATCTCACTC | No match | |
| 27 | 0 | 27 | 0 | 0 | 0 | 0 | GCCCTGCTAC | 50002 | ELC |
| 26 | 0 | 26 | 54 | 4 | 1 | 1 | GCTTGCAAAA | 177781 | Superoxide dismutase 2 |
| 26 | 1 | 26 | 1 | 0 | 1 | 0 | CCTCCCTGCT | 90790 | ESTs |
| 25 | 1 | 25 | 0 | 0 | 0 | 0 | TGTTTCCTTA | 10887 | DC-LAMP |
| 25 | 1 | 25 | 20 | 4 | 2 | 0 | GGGTTTGTTT | 75889 | Proline-rich protein with nuclear targeting signal |
| 25 | 1 | 25 | 13 | 49 | 6 | 3 | TCACAGCTGT | 77054 | B-cell translocation gene 1, antiproliferative |
| 24 | 0 | 24 | 0 | 0 | 0 | 24 | TAAATCCAAA | 224982 | ESTs |
| 23 | 2 | 46 | 1 | 0 | 0 | 0 | GATCAATCAG | 16530 | PARC |
| 22 | 1 | 22 | 0 | 0 | 0 | 0 | GTGCCTTTTT | 204290 | cDNA DKFZp588N2119 |
| 21 | 0 | 21 | 0 | 0 | 0 | 0 | GCGGCTCCTG | No match | |
| 21 | 0 | 21 | 3 | 1 | 0 | 0 | AACCCCTGGA | 255647 | ESTs |
| 21 | 0 | 21 | 7 | 0 | 0 | 0 | AGCGGCTACA | 183487 | HEM46 (interferon-stimulated gene 20 kd) |
| 21 | 1 | 21 | 0 | 0 | 0 | 0 | GGCAGCCAGA | 75061 | MacMARCKS |
| 21 | 1 | 21 | 0 | 0 | 0 | 5 | AGCAAATCCA | No match | |
| 20 | 0 | 20 | 2 | 0 | 0 | 0 | CTTGCAAACC | 229838 | ESTs |
| 20 | 1 | 20 | 8 | 11 | 2 | 6 | ATCCCTCAGT | 181243 | Activating transcription factor 4 |
| 18 | 18 | 338 | 12 | 0 | 4 | 65 | AACGGGGCCC | 97203 | MDC |
| 17 | 0 | 17 | 11 | 2 | 1 | 0 | GTCTTAAAGT | 177781 | Superoxide dismutase 2, mitochondrial |
| 16 | 0 | 16 | 7 | 4 | 0 | 0 | TCCGCAAGGT | 37617 | ESTs |
| 16 | 1 | 16 | 0 | 0 | 0 | 0 | GCAGGCCAAG | Multiple match | |
| 15 | 7 | 108 | 24 | 14 | 70 | 30 | CGCCGACGAT | 21205 | Interferon-α–inducible protein (clone IFI-6-16) |
| 14 | 0 | 14 | 0 | 0 | 0 | 0 | TCTGTTGGAC | 218318 | ESTs |
| 14 | 0 | 14 | 6 | 0 | 0 | 0 | CAGGTGAAAC | Multiple match | |
| 14 | 1 | 14 | 4 | 6 | 1 | 3 | CCCCTGGCTG | 920 | Modulator recognition factor I |
| 14 | 8 | 110 | 21 | 6 | 11 | 7 | TCCGTGGTTG | 79518 | Neuronal tissue–enriched acidic protein |
| 13 | 0 | 13 | 0 | 8 | 0 | 0 | TTTGGGACCC | 270 | Sec7 and coiled/cell domains, binding protein |
| 12 | 68 | 831 | 15 | 0 | 0 | 1 | GGCACAAAGG | 86742 | TARC |
| 12 | 0 | 12 | 0 | 1 | 0 | 0 | CAGCCTGCGA | 35140 | Serine/threonine kinase 4 |
| 12 | 0 | 12 | 0 | 0 | 0 | 4 | GCAAATCCAA | No match | |
| 12 | 0 | 12 | 2 | 1 | 0 | 0 | ACCATTGGAT | 146360 | Interferon-induced protein 17 |
| 12 | 0 | 12 | 5 | 0 | 1 | 0 | GACAGATGGA | 83575 | ESTs |
| 12 | 1 | 12 | 0 | 0 | 0 | 0 | TTTCATCGTA | 106149 | ESTs |
| 12 | 1 | 12 | 0 | 1 | 0 | 0 | TGTGGAAACC | 80205 | plm-2 oncogene |
| 12 | 1 | 12 | 1 | 0 | 3 | 5 | ATGGTCTACG | 98593 | ESTs |
| 12 | 1 | 12 | 1 | 10 | 0 | 2 | GTGCCCGTGC | 83846 | Trisephosphate isomerase 1 |
| 12 | 5 | 59 | 59 | 1 | 15 | 11 | ACTGTGGCGG | 112242 | ESTs |
| Fold . | n . | SAGE tag . | UniGene cluster . | Description . | |||||
|---|---|---|---|---|---|---|---|---|---|
| IMDC . | MADC . | LPS-Mo . | Mo . | M-CSF . | GM-CSF . | ||||
| 70 | 3 | 210 | 2 | 2 | 1 | 18 | CAAATCCAAA | 227400 | Germinal center kinase–related protein kinase |
| 67 | 1 | 67 | 6 | 0 | 0 | 0 | TTCTGCTTTC | 1652 | CCR7 |
| 66 | 1 | 86 | 0 | 0 | 0 | 0 | GTAGCGCCTC | 143212 | Cystatin F (leukocystatin) |
| 47 | 1 | 47 | 4 | 0 | 6 | 11 | GATCCCAACT | 118766 | Metallothionein 2A |
| 43 | 0 | 43 | 1 | 0 | 0 | 0 | GGCGTTTAGA | No match | |
| 42 | 0 | 42 | 11 | 0 | 0 | 0 | AAAAATCGGC | 226695 | RANTES |
| 41 | 2 | 62 | 48 | 4 | 0 | 1 | AATCTGCGCC | 833 | Interferon-stimulated protein, 15 kd |
| 40 | 0 | 40 | 0 | 0 | 0 | 0 | CCAGGGGAGA | 2867 | Interferon-α–inducible protein p27 |
| 38 | 1 | 39 | 3 | 1 | 1 | 0 | GGTGCCCAGT | 75607 | MARCKS, 8CK-L |
| 37 | 4 | 148 | 0 | 0 | 1 | 1 | ATAGTAGCTT | 118400 | Actin-bundling protein |
| 36 | 1 | 36 | 0 | 1 | 0 | 0 | CGGAGCCGGC | 34267 | ESTs |
| 31 | 0 | 31 | 0 | 0 | 0 | 0 | GCTCCCTACG | 185705 | EBI3 |
| 30 | 1 | 30 | 11 | 1 | 1 | 0 | AGTGCCGTGT | 76391 | Interferon-inducible protein p75 |
| 27 | 0 | 27 | 0 | 0 | 0 | 0 | CATCTCACTC | No match | |
| 27 | 0 | 27 | 0 | 0 | 0 | 0 | GCCCTGCTAC | 50002 | ELC |
| 26 | 0 | 26 | 54 | 4 | 1 | 1 | GCTTGCAAAA | 177781 | Superoxide dismutase 2 |
| 26 | 1 | 26 | 1 | 0 | 1 | 0 | CCTCCCTGCT | 90790 | ESTs |
| 25 | 1 | 25 | 0 | 0 | 0 | 0 | TGTTTCCTTA | 10887 | DC-LAMP |
| 25 | 1 | 25 | 20 | 4 | 2 | 0 | GGGTTTGTTT | 75889 | Proline-rich protein with nuclear targeting signal |
| 25 | 1 | 25 | 13 | 49 | 6 | 3 | TCACAGCTGT | 77054 | B-cell translocation gene 1, antiproliferative |
| 24 | 0 | 24 | 0 | 0 | 0 | 24 | TAAATCCAAA | 224982 | ESTs |
| 23 | 2 | 46 | 1 | 0 | 0 | 0 | GATCAATCAG | 16530 | PARC |
| 22 | 1 | 22 | 0 | 0 | 0 | 0 | GTGCCTTTTT | 204290 | cDNA DKFZp588N2119 |
| 21 | 0 | 21 | 0 | 0 | 0 | 0 | GCGGCTCCTG | No match | |
| 21 | 0 | 21 | 3 | 1 | 0 | 0 | AACCCCTGGA | 255647 | ESTs |
| 21 | 0 | 21 | 7 | 0 | 0 | 0 | AGCGGCTACA | 183487 | HEM46 (interferon-stimulated gene 20 kd) |
| 21 | 1 | 21 | 0 | 0 | 0 | 0 | GGCAGCCAGA | 75061 | MacMARCKS |
| 21 | 1 | 21 | 0 | 0 | 0 | 5 | AGCAAATCCA | No match | |
| 20 | 0 | 20 | 2 | 0 | 0 | 0 | CTTGCAAACC | 229838 | ESTs |
| 20 | 1 | 20 | 8 | 11 | 2 | 6 | ATCCCTCAGT | 181243 | Activating transcription factor 4 |
| 18 | 18 | 338 | 12 | 0 | 4 | 65 | AACGGGGCCC | 97203 | MDC |
| 17 | 0 | 17 | 11 | 2 | 1 | 0 | GTCTTAAAGT | 177781 | Superoxide dismutase 2, mitochondrial |
| 16 | 0 | 16 | 7 | 4 | 0 | 0 | TCCGCAAGGT | 37617 | ESTs |
| 16 | 1 | 16 | 0 | 0 | 0 | 0 | GCAGGCCAAG | Multiple match | |
| 15 | 7 | 108 | 24 | 14 | 70 | 30 | CGCCGACGAT | 21205 | Interferon-α–inducible protein (clone IFI-6-16) |
| 14 | 0 | 14 | 0 | 0 | 0 | 0 | TCTGTTGGAC | 218318 | ESTs |
| 14 | 0 | 14 | 6 | 0 | 0 | 0 | CAGGTGAAAC | Multiple match | |
| 14 | 1 | 14 | 4 | 6 | 1 | 3 | CCCCTGGCTG | 920 | Modulator recognition factor I |
| 14 | 8 | 110 | 21 | 6 | 11 | 7 | TCCGTGGTTG | 79518 | Neuronal tissue–enriched acidic protein |
| 13 | 0 | 13 | 0 | 8 | 0 | 0 | TTTGGGACCC | 270 | Sec7 and coiled/cell domains, binding protein |
| 12 | 68 | 831 | 15 | 0 | 0 | 1 | GGCACAAAGG | 86742 | TARC |
| 12 | 0 | 12 | 0 | 1 | 0 | 0 | CAGCCTGCGA | 35140 | Serine/threonine kinase 4 |
| 12 | 0 | 12 | 0 | 0 | 0 | 4 | GCAAATCCAA | No match | |
| 12 | 0 | 12 | 2 | 1 | 0 | 0 | ACCATTGGAT | 146360 | Interferon-induced protein 17 |
| 12 | 0 | 12 | 5 | 0 | 1 | 0 | GACAGATGGA | 83575 | ESTs |
| 12 | 1 | 12 | 0 | 0 | 0 | 0 | TTTCATCGTA | 106149 | ESTs |
| 12 | 1 | 12 | 0 | 1 | 0 | 0 | TGTGGAAACC | 80205 | plm-2 oncogene |
| 12 | 1 | 12 | 1 | 0 | 3 | 5 | ATGGTCTACG | 98593 | ESTs |
| 12 | 1 | 12 | 1 | 10 | 0 | 2 | GTGCCCGTGC | 83846 | Trisephosphate isomerase 1 |
| 12 | 5 | 59 | 59 | 1 | 15 | 11 | ACTGTGGCGG | 112242 | ESTs |
The 50 transcripts displaying the largest increase in expression are listed. MADCs are listed by fold induction. n indicates the number of times the tag was identified. Conditions are described in Figure 2. More information is available on the Internet athttp://www.prevent.m.u-tokyo.ac.jp/SAGE.html/. In this table, each number of tags from IMDC (58 540), MADC (31 837), LPS-Mo (31 837, LPS-stimulated monocytes), Mo (57 580, monocytes), M-CSF (55 956, M-CSF–induced macrophages), and GM-CSF (57 483, GM-CSF–induced macrophages) was normalized to 31 837.
Table 3 shows the top 50 transcripts that are decreased in MADCs. The transcripts decreased in MADCs mainly consisted of genes encoding enzymes such as lipase A, α1-antichymotrypsin, glucose-6-phosphatase, and acid phosphatase type 5; and surface marker proteins such as CD52, CD11b, factor XIII, and CD23.
Decreased transcripts in MADCs
| Fold . | n . | SAGE tag . | UniGene cluster . | Description . | ||||
|---|---|---|---|---|---|---|---|---|
| IMDC . | MADC . | Mo . | M-CSF . | GM-CSF . | ||||
| 68 | 68 | 0 | 1 | 24 | 44 | AGAAGTGTCC | 85226 | Lipase A, lysosomal acid, cholesterol esterase |
| 39 | 38 | 0 | 4 | 10 | 6 | CACCACGGTG | 107325 | α-1-antichymotrypsin |
| 33 | 33 | 1 | 4 | 4 | 3 | AGCTGTCCCC | No match | |
| 28 | 58 | 2 | 38 | 71 | 196 | GGGGCAACAG | 214742 | CDW52 antigen (CAMPATH-1 antigen) |
| 23 | 23 | 0 | 7 | 7 | 5 | AGAAGCCGTG | 172831 | CD11b (p170) |
| 23 | 23 | 1 | 15 | 15 | 31 | CTGGCGCGAG | 83656 | Rho GDF dissociation inhibitor (GDI) β |
| 22 | 22 | 0 | 66 | 19 | 25 | ATGTGTAACG | 61256 | S100 calcium-binding protein A4 |
| 20 | 20 | 1 | 1 | 1 | 2 | TTTGCTCTCC | 75350 | Vinculin |
| 19 | 19 | 0 | 0 | 0 | 0 | CCTCTGGGCA | 167256 | P460 (cytochrome) oxidoreductase |
| 18 | 18 | 0 | 0 | 0 | 2 | CCAAGACTTC | No match | |
| 18 | 35 | 2 | 58 | 107 | 167 | GCGGTTGTGG | 79358 | Lysosomal-associated multiapanning membrane protein-5 |
| 17 | 52 | 3 | 1 | 1 | 1 | AGCCACCGCA | 242 | Glucose-6-phosphatase, catalytic |
| 17 | 17 | 0 | 3 | 24 | 3 | CCGACGGGCG | No match | |
| 17 | 17 | 1 | 8 | 6 | 5 | TGGCTGGCCA | 174142 | Colony-stimulating factor-1 receptor |
| 16 | 16 | 0 | 1 | 0 | 0 | TTGAGACCTC | 80424 | Coagulation factor XIII, A1 polypeptide |
| 15 | 15 | 0 | 2 | 1 | 3 | ACTGACTGCA | 23262 | Ribonuclease, RNase A family, k6 |
| 15 | 15 | 0 | 11 | 6 | 3 | CGTGTGCCTG | 108713 | ESTs |
| 15 | 15 | 1 | 3 | 15 | 12 | TGGCCATCTG | 184052 | ESTs |
| 14 | 14 | 0 | 0 | 8 | 17 | TCTTGATTTA | 74561 | α2-macroglobulin |
| 14 | 14 | 1 | 0 | 32 | 36 | TGTCCCAGCC | 1211 | Acid phosphatase-β, tartrate resistant |
| 14 | 14 | 1 | 6 | 11 | 7 | CTGCTAACCC | 170310 | ESTs |
| 14 | 14 | 1 | 14 | 14 | 23 | CCAGGCAGGG | 117339 | cDNA DKFZp586C1522 |
| 13 | 13 | 0 | 2 | 4 | 6 | GGGGGTGAAG | 76171 | CCAAT/enhancer-binding protein (C/EBP), α |
| 13 | 13 | 0 | 4 | 1 | 0 | AGTCTCTCTT | No match | |
| 13 | 25 | 2 | 12 | 13 | 8 | CCACACAAGC | 102737 | ESTs |
| 12 | 12 | 1 | 0 | 0 | 3 | ATGGAAGTCT | 155939 | Inosilol polyphosphate-5-phosphatase, 145 kd |
| 12 | 12 | 1 | 4 | 1 | 0 | AAAAGAAACT | 172182 | Poly(A)-binding protein-like 1 |
| 12 | 12 | 1 | 26 | 14 | 7 | TCACAAGCAA | 146763 | Nascent polypeptide-associated complex α polypeptide |
| 11 | 11 | 0 | 12 | 5 | 13 | CTGGCCCGAG | 83656 | Rho GDP dissociation inhibitor (GDI) β |
| 11 | 11 | 1 | 0 | 1 | 0 | GATACAGCCA | 1416 | CD33 |
| 11 | 11 | 1 | 0 | 0 | 1 | TGTGAACAAC | 129771 | ESTs |
| 11 | 11 | 1 | 3 | 1 | 0 | ACCCCCGGGC | Multiple match | |
| 11 | 11 | 1 | 4 | 10 | 13 | GTCTGAGCTC | 66915 | ESTs |
| 10 | 52 | 5 | 3 | 713 | 849 | TGGCCCCAGG | 132776 | Apolipoprotein C-1 |
| 10 | 10 | 0 | 0 | 0 | 8 | GCCTACCCGA | 23582 | GA733-1 |
| 10 | 10 | 0 | 0 | 3 | 11 | ACCCAGGGTA | 44234 | ESTs |
| 10 | 10 | 0 | 0 | 1 | 2 | CCCTCGGTCC | No match | |
| 10 | 10 | 0 | 2 | 14 | 8 | TCTCTGATGC | 8441 | cDNA DKF2pS86J021 |
| 10 | 10 | 0 | 2 | 9 | 3 | GCCACTACCC | 71475 | ESTs |
| 10 | 10 | 0 | 2 | 28 | 65 | ACTATTTCCA | 574 | Fructose-bisphosphatase 1 |
| 10 | 10 | 0 | 3 | 2 | 4 | AGGACACCGC | 77783 | c-src tyrosine kinase |
| 10 | 10 | 1 | 0 | 0 | 3 | CTGATCTCCA | Multiple match | |
| 10 | 10 | 1 | 9 | 6 | 0 | TGATGTTTGA | 75416 | KIAA0058 gene product |
| 9 | 28 | 3 | 47 | 17 | 11 | TCTGTACACC | 182740 | Aldolase β, fructose-bisphosphate |
| 9 | 27 | 3 | 2 | 15 | 4 | TGGCGTACGG | 159546 | ATP-binding cassette, subfamily D (ALD), member 1 |
| 9 | 9 | 0 | 0 | 23 | 18 | AAGATTGGTG | 1244 | CDS antigen (p24) |
| 9 | 9 | 0 | 0 | 0 | 0 | TGCAGAAGAA | 75182 | Mannose receptor, C type 1 |
| 9 | 9 | 0 | 0 | 1 | 0 | CCTCACTACC | 99307 | ESTs |
| 9 | 8 | 0 | 0 | 2 | 1 | GGTGTGCTTG | 155182 | Homo sapiens mRNA for KIAA1036 protein, complete cds |
| 9 | 9 | 0 | 0 | 0 | 2 | GGTCCCCTAC | 151761 | KIAA0100 gene product |
| Fold . | n . | SAGE tag . | UniGene cluster . | Description . | ||||
|---|---|---|---|---|---|---|---|---|
| IMDC . | MADC . | Mo . | M-CSF . | GM-CSF . | ||||
| 68 | 68 | 0 | 1 | 24 | 44 | AGAAGTGTCC | 85226 | Lipase A, lysosomal acid, cholesterol esterase |
| 39 | 38 | 0 | 4 | 10 | 6 | CACCACGGTG | 107325 | α-1-antichymotrypsin |
| 33 | 33 | 1 | 4 | 4 | 3 | AGCTGTCCCC | No match | |
| 28 | 58 | 2 | 38 | 71 | 196 | GGGGCAACAG | 214742 | CDW52 antigen (CAMPATH-1 antigen) |
| 23 | 23 | 0 | 7 | 7 | 5 | AGAAGCCGTG | 172831 | CD11b (p170) |
| 23 | 23 | 1 | 15 | 15 | 31 | CTGGCGCGAG | 83656 | Rho GDF dissociation inhibitor (GDI) β |
| 22 | 22 | 0 | 66 | 19 | 25 | ATGTGTAACG | 61256 | S100 calcium-binding protein A4 |
| 20 | 20 | 1 | 1 | 1 | 2 | TTTGCTCTCC | 75350 | Vinculin |
| 19 | 19 | 0 | 0 | 0 | 0 | CCTCTGGGCA | 167256 | P460 (cytochrome) oxidoreductase |
| 18 | 18 | 0 | 0 | 0 | 2 | CCAAGACTTC | No match | |
| 18 | 35 | 2 | 58 | 107 | 167 | GCGGTTGTGG | 79358 | Lysosomal-associated multiapanning membrane protein-5 |
| 17 | 52 | 3 | 1 | 1 | 1 | AGCCACCGCA | 242 | Glucose-6-phosphatase, catalytic |
| 17 | 17 | 0 | 3 | 24 | 3 | CCGACGGGCG | No match | |
| 17 | 17 | 1 | 8 | 6 | 5 | TGGCTGGCCA | 174142 | Colony-stimulating factor-1 receptor |
| 16 | 16 | 0 | 1 | 0 | 0 | TTGAGACCTC | 80424 | Coagulation factor XIII, A1 polypeptide |
| 15 | 15 | 0 | 2 | 1 | 3 | ACTGACTGCA | 23262 | Ribonuclease, RNase A family, k6 |
| 15 | 15 | 0 | 11 | 6 | 3 | CGTGTGCCTG | 108713 | ESTs |
| 15 | 15 | 1 | 3 | 15 | 12 | TGGCCATCTG | 184052 | ESTs |
| 14 | 14 | 0 | 0 | 8 | 17 | TCTTGATTTA | 74561 | α2-macroglobulin |
| 14 | 14 | 1 | 0 | 32 | 36 | TGTCCCAGCC | 1211 | Acid phosphatase-β, tartrate resistant |
| 14 | 14 | 1 | 6 | 11 | 7 | CTGCTAACCC | 170310 | ESTs |
| 14 | 14 | 1 | 14 | 14 | 23 | CCAGGCAGGG | 117339 | cDNA DKFZp586C1522 |
| 13 | 13 | 0 | 2 | 4 | 6 | GGGGGTGAAG | 76171 | CCAAT/enhancer-binding protein (C/EBP), α |
| 13 | 13 | 0 | 4 | 1 | 0 | AGTCTCTCTT | No match | |
| 13 | 25 | 2 | 12 | 13 | 8 | CCACACAAGC | 102737 | ESTs |
| 12 | 12 | 1 | 0 | 0 | 3 | ATGGAAGTCT | 155939 | Inosilol polyphosphate-5-phosphatase, 145 kd |
| 12 | 12 | 1 | 4 | 1 | 0 | AAAAGAAACT | 172182 | Poly(A)-binding protein-like 1 |
| 12 | 12 | 1 | 26 | 14 | 7 | TCACAAGCAA | 146763 | Nascent polypeptide-associated complex α polypeptide |
| 11 | 11 | 0 | 12 | 5 | 13 | CTGGCCCGAG | 83656 | Rho GDP dissociation inhibitor (GDI) β |
| 11 | 11 | 1 | 0 | 1 | 0 | GATACAGCCA | 1416 | CD33 |
| 11 | 11 | 1 | 0 | 0 | 1 | TGTGAACAAC | 129771 | ESTs |
| 11 | 11 | 1 | 3 | 1 | 0 | ACCCCCGGGC | Multiple match | |
| 11 | 11 | 1 | 4 | 10 | 13 | GTCTGAGCTC | 66915 | ESTs |
| 10 | 52 | 5 | 3 | 713 | 849 | TGGCCCCAGG | 132776 | Apolipoprotein C-1 |
| 10 | 10 | 0 | 0 | 0 | 8 | GCCTACCCGA | 23582 | GA733-1 |
| 10 | 10 | 0 | 0 | 3 | 11 | ACCCAGGGTA | 44234 | ESTs |
| 10 | 10 | 0 | 0 | 1 | 2 | CCCTCGGTCC | No match | |
| 10 | 10 | 0 | 2 | 14 | 8 | TCTCTGATGC | 8441 | cDNA DKF2pS86J021 |
| 10 | 10 | 0 | 2 | 9 | 3 | GCCACTACCC | 71475 | ESTs |
| 10 | 10 | 0 | 2 | 28 | 65 | ACTATTTCCA | 574 | Fructose-bisphosphatase 1 |
| 10 | 10 | 0 | 3 | 2 | 4 | AGGACACCGC | 77783 | c-src tyrosine kinase |
| 10 | 10 | 1 | 0 | 0 | 3 | CTGATCTCCA | Multiple match | |
| 10 | 10 | 1 | 9 | 6 | 0 | TGATGTTTGA | 75416 | KIAA0058 gene product |
| 9 | 28 | 3 | 47 | 17 | 11 | TCTGTACACC | 182740 | Aldolase β, fructose-bisphosphate |
| 9 | 27 | 3 | 2 | 15 | 4 | TGGCGTACGG | 159546 | ATP-binding cassette, subfamily D (ALD), member 1 |
| 9 | 9 | 0 | 0 | 23 | 18 | AAGATTGGTG | 1244 | CDS antigen (p24) |
| 9 | 9 | 0 | 0 | 0 | 0 | TGCAGAAGAA | 75182 | Mannose receptor, C type 1 |
| 9 | 9 | 0 | 0 | 1 | 0 | CCTCACTACC | 99307 | ESTs |
| 9 | 8 | 0 | 0 | 2 | 1 | GGTGTGCTTG | 155182 | Homo sapiens mRNA for KIAA1036 protein, complete cds |
| 9 | 9 | 0 | 0 | 0 | 2 | GGTCCCCTAC | 151761 | KIAA0100 gene product |
The 50 transcripts displaying the largest decrease in expression are listed. MADCs are listed by fold reduction. n indicates the number of times the tag was identified. Conditions are described in Figure 2. More information is available on the Internet athttp://www.prevent.m.u-tokyo.ac/jp/SAGE.html/. In this table, each number of tags from IMDC (58 540), MADC (31 837), Mo (57 560, monocytes), M-CSF (55 856, M-CSF–induced macrophages), and GM-CSF (57 463, GM-CSF–induced macrophages) was normalized to 31 837.
RT-PCR of genes selected in the SAGE analysis
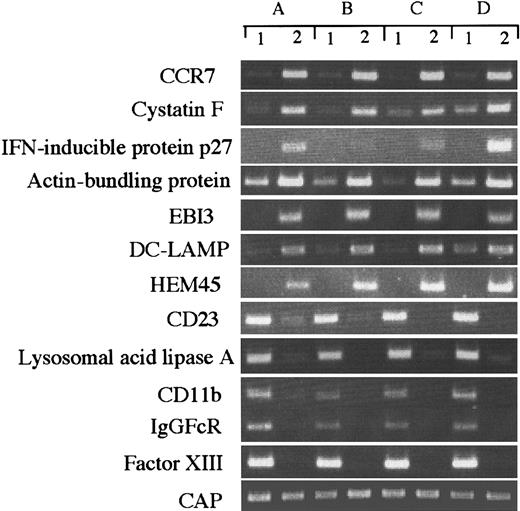
Although our data represent the average gene expression in cells obtained from 6 healthy donors, there could be differences in gene expression among individual donor-derived cells. To address this question, we arbitrarily selected 10 differently expressed genes and evaluated them in 4 donor-derived samples by RT-PCR (Figure3), and compared the expression of each transcript with the SAGE data. CAP was expressed almost equally in all cell types (IMDCs, tag number 11; MADCs, 9), but actin-bundling protein (IMDCs, 4; MADCs, 148), IFN-inducible protein (IMDCs, 0; MADCs, 46), HEM45 (IMDCs, 0; MADCs, 21), EBI3 (IMDCs, 0; MADCs, 31), and cystatin F (IMDCs, 1; MADCs, 66) were highly expressed in MADCs. On the other hand, CD23 (IMDCs, 11; MADCs, 1), lysosomal acid lipase (IMDCs, 68; MADCs, 0), CD11b (IMDCs, 23; MADCs, 0), IgGFcR (IMDCs, 9; MADCs, 1), and factor XIII (IMDCs, 16; MADCs, 0) were highly expressed in IMDCs. These results validate our SAGE data for IMDCs and MADCs and establish the general gene expression in these cells.
RT-PCR analysis of genes expressed differently in IMDCs and MADCs.
RT-PCR was performed on total RNA isolated from (1) IMDCs and (2) MADCs, as described in “Materials and methods.” A, B, C, and D indicate different donors.
RT-PCR analysis of genes expressed differently in IMDCs and MADCs.
RT-PCR was performed on total RNA isolated from (1) IMDCs and (2) MADCs, as described in “Materials and methods.” A, B, C, and D indicate different donors.
Comparison of gene expression profile of MADCs with that of LPS-stimulated monocytes
It is well known that LPS modulates the expression of numerous genes encoding proteins such as cytokines, chemokines, and transcriptional factors in monocytes. Thus, a comparison between MADCs and LPS-Mo (manuscript in preparation) was conducted to identify specific genes expressed only in MADCs. The 57 560 and 35 874 tag sequences from the Mo30 and LPS-Mo libraries, respectively, were normalized to 31 837 and were compared with the transcripts increased in MADCs (Table 2). The expression levels of RANTES (IMDCs, 0; MADCs, 42; LPS-Mo, 11; Mo, 0), IFN-stimulated protein 15 kd (IMDCs, 2; MADCs, 82; LPS-Mo, 48; Mo, 4), IFN-inducible protein P78 (IMDCs, 1; MADCs, 30; LPS-Mo, 11; Mo, 1), superoxide dismutase 2 (IMDCs, 0; MADCs, 26; LPS-Mo, 54; Mo, 4), activating transcription factor 4 (IMDCs, 1; MADCs, 20; LPS-Mo, 8; Mo, 11), HEM45 (IMDCs, 0; MADCs, 21; LPS-Mo, 7; Mo, 0), MDC (IMDCs, 18; MADCs, 336; LPS-Mo, 12; Mo, 0), IFN-inducible protein (IFI-6-16) (IMDCs, 7; MADCs, 106; LPS-Mo, 24; Mo, 14), and TARC (IMDCs, 68; MADCs, 831; LPS-Mo, 16; Mo, 0) were increased in LPS-stimulated DCs as well as in LPS-Mo, as compared with Mo (Table2). On the other hand, the expression levels of genes encoding proteins such as cystatin C (IMDCs, 1; MADCs, 66; LPS-Mo, 0; Mo, 0), IFN-α–inducible protein p27 (IMDCs, 0; MADCs, 40; LPS-Mo, 0; Mo, 0), actin-bundling protein (IMDCs, 4; MADCs, 148; LPS-Mo, 0; Mo, 0), EBI3 (IMDCs, 0; MADCs, 31; LPS-Mo, 0; Mo, 0), ELC (IMDCs, 0; MADCs, 27; LPS-Mo, 0; Mo, 0), DC-LAMP (IMDCs, 1; MADCs, 25; LPS-Mo, 0; Mo, 0), MacMARCKS (IMDCs, 1; MADCs, 21; LPS-Mo, 0; Mo, 0), serine/threonine kinase 4 (IMDCs, 0; MADCs, 12; LPS-Mo, 0; Mo, 1),pim-2 oncogene (IMDCs, 1; MADCs, 40; LPS-Mo, 0; Mo, 1), and several genes in ESTs, were highly specific for MADCs.
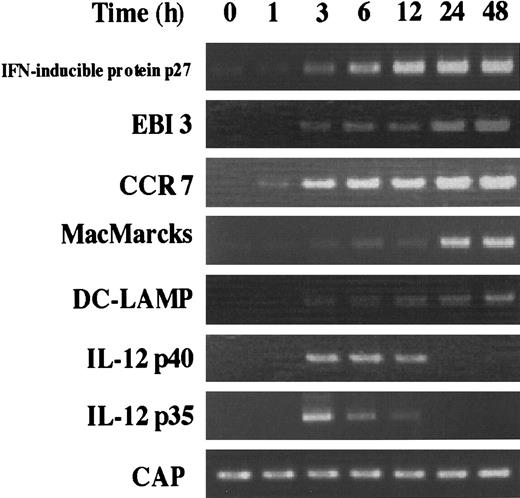
Time course of the induction of MADC-specific genes
During DC activation and maturation, the expression of several genes has been described to be increased. For example, it is already known that CCR7 is highly expressed in activated DCs,15,31 and DCs activated by several stimuli can produce IL-12.32-34 Here, EBI3, IFN-α–inducible protein p27, and MacMARCKS were newly identified as the inducible genes during DC activation and maturation. The time course of the induction of these genes was analyzed together with that of the CCR7, DC-LAMP, and IL-12 (p35 and p40) genes (Figure 4). Although IMDCs barely expressed the genes for IFN-α–inducible protein, EBI3, CCR7, and MacMARCKS, the level of mRNAs of most of these genes sharply increased within 3 hours after the stimulation with LPS and reached a maximum at 24 hours. IL-12 p35 and p40 mRNAs were transiently induced from 3 hours to 12 hours.
Kinetics of the induction of various mRNA expression by stimulating IMDCs with LPS.
RT-PCR was performed on total RNA isolated from IMDCs cultured in the presence of GM-CSF and IL-4 for 5 days, followed by 0, 1, 3, 6, 12, 24, and 48 hours of activation with LPS (100 μg/mL).
Kinetics of the induction of various mRNA expression by stimulating IMDCs with LPS.
RT-PCR was performed on total RNA isolated from IMDCs cultured in the presence of GM-CSF and IL-4 for 5 days, followed by 0, 1, 3, 6, 12, 24, and 48 hours of activation with LPS (100 μg/mL).
Correlation coefficients for all pairwise comparisons of libraries
To estimate the extent of similarity between any 2 libraries (monocytes, GM-CSF–induced macrophages, M-CSF–induced macrophages, IMDCs, and MADCs), we calculated each bivariate correlation coefficient. The correlation coefficients for all comparisons are shown in Table 4. Pearson correlation coefficients between GM-CSF– and M-CSF–induced macrophage libraries showed a high similarity, at 0.938. However, the extent of similarity between any other 2 libraries ranged from 0.460 to 0.659.
Correlation coefficients between the different libraries
| . | Mo . | GM . | M . | IMDC . |
|---|---|---|---|---|
| Mo | ||||
| GM | 0.518 | |||
| M | 0.561 | 0.938 | ||
| IMDC | 0.644 | 0.607 | 0.659 | |
| MADC | 0.511 | 0.460 | 0.511 | 0.586 |
| . | Mo . | GM . | M . | IMDC . |
|---|---|---|---|---|
| Mo | ||||
| GM | 0.518 | |||
| M | 0.561 | 0.938 | ||
| IMDC | 0.644 | 0.607 | 0.659 | |
| MADC | 0.511 | 0.460 | 0.511 | 0.586 |
Correlation coefficients for each library comparison were calculated as described in “Materials and methods.”
Mo indicates monocytes; GM, GM-CSF–induced macrophages; M, M-CSF–induced macrophages.
Discussion
In this study, we performed SAGE for LPS-stimulated mature DCs to obtain insights on the molecular level into the differentiation from IMDCs to MADCs.
Differential gene expression during DC maturation can be divided into 3 parts: a change of subcellular compartments to process antigens, a change of cell surface molecules to adhere to tissues and lymphocytes, and a change of molecules related to cell migration.
The differential gene expression of the multiple subcellular compartments that correlate with different stages of DC maturation may be involved in the loading of peptides on class I and class II molecules. The increased gene expression of several subcellular compartments, including enzymes such as cystatin F (IMDCs, 1; MADCs, 66), germinal center kinase–related protein kinase (IMDCs, 3; MADCs, 210), metallothionein 2A (IMDCs, 1; MADCs, 47), protein kinase C substrate, MARCKS 80K-L (IMDCs, 1; MADCs, 39) and DC-LAMP (IMDCs, 1; MADCs, 25), was also observed (P < .001). In contrast, many of the genes such as lipase A (IMDCs, 68; MADCs, 0), cystatin B (IMDCs, 58; MADCs, 15), cathepsin B (IMDCs, 104; MADCs, 45), and cystatin C (IMDCs, 243; MADCs, 74) were down-regulated significantly (P < .01). These regulations of gene expression may be related to the capability of antigen processing of IMDCs and MADCs. Moreover, many of the genes encoding IFN-inducible proteins were found to be increased. It is known that IFNs increase the expression of MHC antigens. In mice with a targeted disruption of the IFN-γ gene, reduced expression of macrophage MHC class II antigens has been observed.35 Furthermore, in IFN signal protein–deficient mice, IRF-1−/− mice, the expression of surface class I MHC is reduced.36 Therefore, the IFN-inducible proteins, which were found to be increased in MADCs, may be involved in the regulation of MHC class I and II.
The expression of the genes encoding cell surface molecules, such as class I molecules (IMDCs, 43; MADCs, 172), β2-microglobulin (IMDCs, 143; MADCs, 655), and Landinin (IMDCs, 0; MADCs, 10), was up-regulated (P < .01). In contrast, expression levels of the genes for class II molecules (IMDCs, 373; MADCs, 64), CD74 (IMDCs, 533; MADCs, 272), CD52 (IMDCs, 56; MADCs, 2), factor XIII (IMDCs, 16; MADCs, 0), CD9 (IMDCs, 9; MADCs, 0), CD23 (IMDCs, 11; MADCs, 1), CD11b (IMDCs, 23; MADCs, 0), and GA733-1 (IMDCs, 10; MADCs, 0) were down-regulated significantly (P < .05). The change in the expression of these genes encoding proteins related to cell surface molecules might be involved in adhesion to tissues and cells or capturing antigens. It is well known that bacterial products such as LPS increase the synthesis of MHC class I and II molecules in DCs. However, our data showed the down-regulation of genes of MHC class II–related molecules in MADCs as compared with IMDCs. Because it has been reported that the class II molecules are quickly synthesized and the expression is shut off in a short time after the synthesis of the class I molecules,37 this may be the reason for the apparent discrepancy.
Cell migration involves the coordinate activation of 2 classes of effector molecules, adhesion molecules and chemotactic factors.38 Many of the transcripts increased in MADCs consist of genes encoding chemokines such as TARC, MDC, RANTES, ELC, PARC, and a chemokine receptor CCR7 (Table 2). These results suggest that MADCs not only up-regulate the CCR7 to be recruited to secondary lymphoid organs to activate naive T lymphocytes as antigen-presenting cells, but also release chemotactic factors to attract memory-type Th1/Th2 polarized cells. In addition, these chemokines produced by MADCs at the inflammatory sites may also be involved in the recruitment of various types of leukocytes other than lymphocytes.
It is well known that LPS induces various types of genes encoding cytokines, chemokines, and transcriptional factors in monocytes. Thus, comparison of the gene expression profiles between MADCs and LPS-stimulated monocytes is important to negate the possibility that the identified genes are merely LPS-inducible genes. The expression of the genes of RANTES, IFN-inducible protein (IFI-6-16), IFN-inducible protein P78, superoxide dismutase 2, HEM45, activating transcription factor 4, MDC, IFN-α–inducible protein, and TARC was increased in MADCs as well as in LPS-stimulated monocytes as compared with monocytes. On the other hand, the genes for cystatin C, IFN-α–inducible protein p27, actin-bundling protein, EBI3, ELC, DC-LAMP, PARC, MacMARCKS, and several genes in ESTs were expressed in MADCs but not in monocytes stimulated by LPS. DC-LAMP is a lysosomal protein involved in remodeling of specialized antigen-processing compartments and in MHC class II–restricted antigen presentation, and its gene expression is known to be up-regulated in mature DCs induced by CD40L, TNF-α, and LPS.39 Therefore, DC-LAMP has been used as a DC maturation marker as CD83.40 Our results agree with these findings. Actin-bundling protein is known to be involved in the organization of the actin cytoskeleton in cytoplasmic extensions of nerve growth cones.41 Recently, it has been reported that actin-bundling protein plays a key role in forming dendritic processes in Langerhans cell maturation.42 Our data suggest that actin-bundling protein may also be involved in dendrite formation in monocyte-derived mature DCs, as in Langerhans cells. Interestingly, EBI3 has not been found to be expressed in any other types of cells or tissues available in the SAGE database so far, which suggests that EBI3 is a highly specific gene expressed in MADCs. Furthermore, we checked whether mature DCs induced by other stimuli express EBI3. TNF-α and anti-CD40 antibody also induced EBI3 mRNA expression in mature DCs, as LPS did (data not shown). These results indicate that mature DCs generally express EBI3. EBI3 is a soluble hematopoietic component related to the p40 subunit of IL-12 and has been reported to be expressed in Epstein-Barr virus–transformed B lymphocytes, tonsil, and spleen.43 Our results suggest that EBI3 may be expressed in mature DCs in tonsil and spleen. On the other hand, EBI3 forms a heterodimer with p35,44 and IL-12 production by DCs favors the differentiation of Th0 cells into Th1 cells.33,34,45 46 Therefore, the EBI3/p35 heterodimer is likely to be an important regulator for polarization into Th1 cells by mature DCs. On the other hand, kinetic analysis of IL-12 and EBI3 showed different expression patterns (Figure 4), suggesting that EBI3 gene expression is regulated independently from IL-12 gene expression. Establishment of the biologic role of EBI3 seems important.
In conclusion, SAGE allows both quantitative and simultaneous analysis of large numbers of transcripts. We investigated a total of 183 811 tags derived from 57 560, 58 540, 31 837, and 35 874 tags from monocytes, IMDCs, LPS-stimulated monocytes, and MADCs, respectively, and this allowed identification of 39 211 independent gene transcripts. The identification of the genes selectively expressed in human MADCs, including many novel genes, should provide molecular information to define different sets of resting and activated DCs and contribute to further understanding of the biologic function of DCs in the host defense system, and may also be useful for diagnosing or monitoring human diseases in which DCs play a role.
Acknowledgments
We are very grateful to Drs Victor E. Velculescu, Lin Zhang, Wei Zhou, Bert Vogelstein, and Kenneth W. Kinzler for their help in SAGE analysis.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Kouji Matsushima, Department of Molecular Preventive Medicine, School of Medicine, University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-0033, Japan; e-mail:koujim@m.u-tokyo.ac.jp.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal