Abstract
Congenital afibrinogenemia is a rare autosomal recessive disorder characterized by the complete absence of plasma fibrinogen and by a bleeding tendency ranging from mild to moderately severe. Beside a deletion of the almost entire Aα-chain gene, only 2 missense mutations in the C-terminal domain of the Bβ-chain have been very recently described as being associated with afibrinogenemia. We studied a Pakistani patient with unmeasurable plasma levels of functional and immunoreactive fibrinogen. Sequencing of the fibrinogen genes revealed a homozygous G→A transition at position +5 of intron 1 of the γ-chain gene. The predicted mutant fibrinogen γ-chain would contain the signal peptide, followed by a short stretch of aberrant amino acids, preceding a premature stop codon. To demonstrate the causal role of the identified mutation, we prepared expression vectors containing a region of the fibrinogen γ-chain gene spanning from exon 1 to intron 4 and carrying either a G or an A at position +5 of intron 1. Transient transfection of the mutated plasmid in HeLa cells, followed by RNA extraction and reverse transcriptase-polymerase chain reaction (RT-PCR) analysis, allowed us to demonstrate the production of an erroneously spliced messenger RNA (mRNA), retaining intron 1, as shown by direct sequencing. A normal splicing occurred in HeLa cells transfected with the wild-type plasmid. This is the first report of a mutation in the fibrinogen γ-chain gene causing afibrinogenemia and indicates that, in addition to the Aα and Bβ-chain genes, the γ-chain gene must also be considered in mutation screening for afibrinogenemia.
Introduction
Fibrinogen is a major plasma glycoprotein that is central to the blood clotting process1 and is also a primary participant in the acute phase response to injury and stress.2 It is synthesized in the liver and is secreted as a completely assembled hexamer of 340 kd, composed of 3 pairs of nonidentical but homologous polypeptide chains (Aα, Bβ, γ).3 The 3 fibrinogen chains are encoded by distinct species of messenger RNA (mRNA), resulting from the transcription of 3 single copy genes.4
Congenital afibrinogenemia (MIM 202400) is a rare autosomal recessive disorder described for the first time in 19205 and characterized by unmeasurable clottable fibrinogen and extremely low antigen levels in patient plasma.6 From the study of more than 150 cases reported thus far, a pattern of clinical symptoms can be compiled for this disorder.7,8 Diagnosis is often made at birth because of umbilical cord bleeding. Joint and mucosal bleeding, such as epistaxis, are also common symptoms, whereas gastrointestinal bleeding is less frequent and central nervous system bleeding is rare. Fresh frozen plasma or cryoprecipitate were widely used in the past to control bleeding symptoms, but fibrinogen concentrates that underwent viral inactivation are currently the best option for replacement therapy.7 8
Among fibrinogen congenital abnormalities, afibrinogenemia is the least characterized from a molecular point of view. So far, the molecular defects have been identified and the pathogenetic mechanism elucidated only for 3 afibrinogenemic kindreds. A homozygous 11- kilobase (kb) deletion of almost the entire Aα-chain gene has been reported in a Swiss family,9,10 and 2 different homozygous missense mutations in the Bβ-chain (L353R and G400D), leading to an impairment of fibrinogen secretion, were identified in 2 unrelated afibrinogenemic patients from Iran and Italy.6
In this paper, a Pakistani afibrinogenemic patient was studied. The proband had unmeasurable plasma levels of clottable and immunoreactive fibrinogen. Sequencing of fibrinogen genes, including exon-intron boundaries and promoter regions, allowed us to identify a homozygous G→A transition located at position +5 of intron 1 of the fibrinogen γ-chain gene. Production of the mutant transcript in transfected HeLa cells demonstrated the existence of an aberrant splicing of fibrinogen γ-mRNA, leading to a major γ-chain truncation associated with afibrinogenemia.
Materials and methods
Coagulation tests
Fibrinogen was measured in plasma by a functional assay based on fibrin polymerization time using a commercial kit (Laboratoire Stago, Asnieres, France) and enzyme immunoassay.11 The sensitivity of the functional assay was 5 mg/dL (normal range: 160-400 mg/dL) and that of the immunoassay was 0.02 mg/dL (normal range: 160-400 mg/dL).
DNA extraction
Genomic DNA was extracted from blood samples, using the Nucleon BACC1 kit (Amersham Pharmacia Biotech, Uppsala, Sweden). All examined subjects gave their informed consent before blood withdrawal.
Sequence analysis
DNA sequencing was performed on both strands either directly on purified polymerase chain reaction (PCR) products or on plasmids, using the Taq dye-deoxy terminator method and an automated 310 DNA sequencer (PE-Biosystems, Foster City, CA). All primers used for sequencing were designed on the basis of known sequences of the fibrinogen genes and intergenic regions (GenBank, accession numbers:M64982, M64983, M10014, U36478, and AF229198) and were purchased from Life Technologies (Inchinnan, Paisley, UK). Primer sequences can be provided on request. Factura and Sequence Navigator software packages (PE-Biosystems) were used for mutation detection.
Construction of expression vectors
Mammalian expression vector pTARGET (Promega, Milan, Italy) was used to transcribe either the wild-type or the mutant mRNA. A genomic DNA fragment spanning from exon 1 to intron 4 of the human fibrinogen γ-chain gene was amplified using the primer couple FGG-Ex1-F (5′-ATGAGTTGGTCCTTGCACAA-3′) and FGG-In4-R (5′-ACTAAATCAGTCTTGCAGAGC-3′). PCRs were carried out in a 50 μL reaction mixture containing 100 ng of genomic DNA (either from the proband or a healthy control individual), 2.5 units Taq DNA Polymerase (Sigma, St Louis, MO), 1× PCR buffer (10 mmol/L Tris-HCl pH 8.3, 50 mmol/L KCl, 1.5 mmol/L MgCl2 and 0.001% gelatin), 0.2 mmol/L dNTPs, and 0.4 μmol/L of each primer, in a PTC-100 thermal cycler (MJ-Research, Watertown, MA). Samples were subjected to 35 cycles of denaturation at 94°C for 30 seconds, annealing at 56°C for 30 seconds, and elongation at 72°C for 45 seconds, preceded by 3 minutes denaturation at 94°C and followed by 10 minutes elongation at 72°C. PCR products were inserted into pTARGET vector using the pTARGET Mammalian Expression T-Vector System kit (Promega). The 2 recombinant plasmids, hereafter referred to as pTarget-γ(Ex1-In4)-wt and pTarget-γ(Ex1-In4)-mut, were checked by sequencing.
Cell cultures, tranfections, and RNA extraction
Human cervix carcinoma HeLa cells were cultured in Dulbecco's modified Eagle's Medium containing 10% calf serum, antibiotics (100 IU/mL penicillin and 100 μg/mL streptomycin) and glutamine (1%). Cells were grown at 37°C in a humidified atmosphere of 5% CO2 and 95% air, according to standard procedures. Transfections were performed by the calcium phosphate technique, essentially as described by Wigler et al.12 HeLa cells were plated at a density of 2 × 106 per 10-cm diameter dish, and 24 hours later, tranfections were carried out, applying to the semiconfluent cells CaPO4-DNA precipitate containing 20 μg of either pTarget-γ(Ex1-In4)-wt or pTarget-γ(Ex1-In4)-mut plasmid. Cells were washed twice with phosphate-buffered saline (PBS) 16 hours after transfection, and the medium replaced with a fresh one. Forty-eight hours later, this medium was removed and total RNA was extracted from harvested HeLa cells, using the RNAWIZ kit (Ambion, Austin, TX) according to the manufacturer's instructions. All procedures were carried out at 0°C or 4°C using RNAse-free reagents and plasticware.
Analysis of splice-site mutation
First strand cDNA synthesis, starting from 1 μg of total RNA previously submitted to a DNAseI (Ambion) treatment, was carried out using random nonamers and the Enhanced Avian RT-PCR kit (Sigma). Of a total of 20 μL, 5 μL were used as template to amplify wild-type and mutant transcripts, using the primer couple FGG-Ex1-F and FGG-Ex3-R (5′-GTCTTCCAAAGACTGTAGATCC-3′). PCRs were carried out in 25 μL of a mixture, containing 1.5 units AccuTaq Polymerase (Sigma), 1× PCR buffer (50 mmol/L Tris-HCl, 15 mmol/L ammonium sulfate pH 9.3, 2.5 mmol/L MgCl2, and 0.1% Tween 20), 0.2 mmol/L dNTPs, and 0.4 μmol/L of each primer. The thermal profile consisted of 94°C for 3 minutes, followed by 35 cycles of 30 seconds at 94°C, 10 seconds at 58°C, and 30 seconds at 68°C. A final extension of 2 minutes at 68°C was performed at the end of PCR cycles.
Results
Patient data
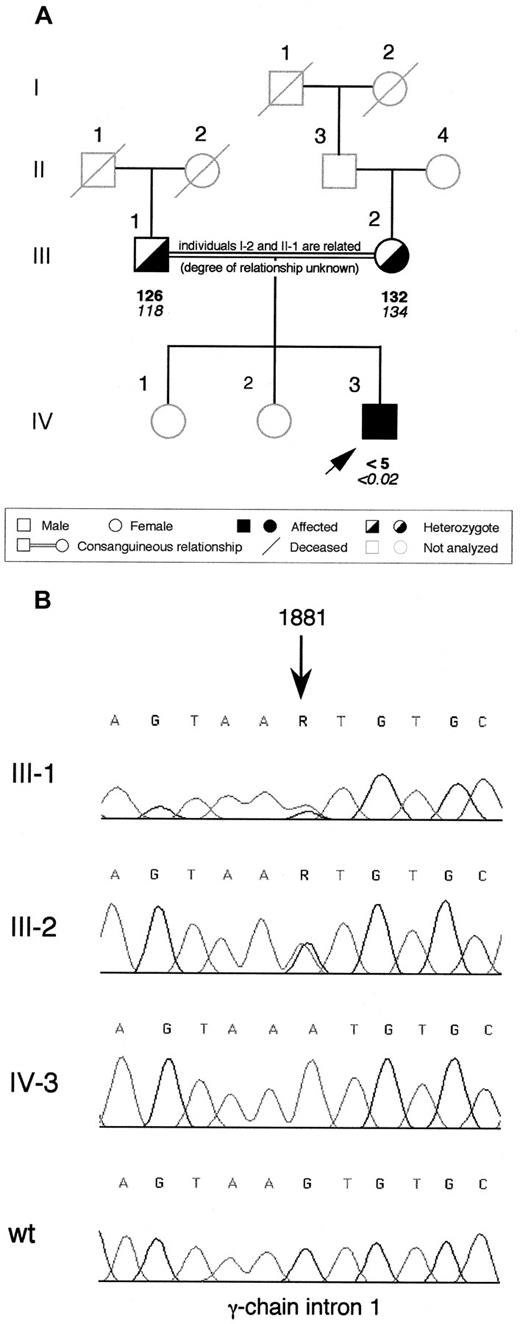
The proband is a 3-year-old Pakistani child born from a consanguineous marriage (Figure 1A; the maternal great-grandmother and the paternal grandfather were related but the degree of relationship is unknown). No bleeding complication occurred at birth, but after 3 weeks, the child presented with intracranial bleeding (subdural hematoma and ventricular hemorrhage) and afibrinogenemia was diagnosed. The child recovered from the hemorrhage after 3-month treatment with cryoprecipitate, but 6 months later, a new intracranial bleed (parenchimal hemorrhage with extension to the subdural space and ventricle) occurred and had to be surgically evacuated, administering fibrinogen concentrate. Since this episode, treatment with fibrinogen concentrate has been continued prophylactically and no new bleeding symptoms have been observed. The proband's parents and sisters are asymptomatic. Plasma fibrinogen levels were unmeasurable by clottable and immunologic assays in the proband at the time of the first diagnosis. The parents had reduced fibrinogen levels in plasma (Figure 1A).
Family pedigree of the afibrinogenemic proband and electropherograms showing the identified splice site mutation.
(A) Pedigree of the Pakistani family. Plasma functional fibrinogen levels (mg/dL) and immunoreactive fibrinogen levels (mg/dL) are indicated in this order below each symbol. The arrow indicates the proband. (B) Electropherograms showing the mutation identified in the afibrinogenemic Pakistani proband. The G→A transition, whose nucleotide position is indicated by an arrow (numbering according to GenBank accession number M10014), was present at the heterozygous state in both parents and absent in a healthy control individual (wt). Family members are labeled as in (A); R, A or G nucleotide.
Family pedigree of the afibrinogenemic proband and electropherograms showing the identified splice site mutation.
(A) Pedigree of the Pakistani family. Plasma functional fibrinogen levels (mg/dL) and immunoreactive fibrinogen levels (mg/dL) are indicated in this order below each symbol. The arrow indicates the proband. (B) Electropherograms showing the mutation identified in the afibrinogenemic Pakistani proband. The G→A transition, whose nucleotide position is indicated by an arrow (numbering according to GenBank accession number M10014), was present at the heterozygous state in both parents and absent in a healthy control individual (wt). Family members are labeled as in (A); R, A or G nucleotide.
Sequence analysis
The entire coding region, including exon-intron boundaries and approximately 500 base pairs (bp) of the promoter region of each fibrinogen gene of the proband was sequenced. Sequence analysis identified an homozygous G→A transition in intron 1 of the fibrinogen γ-chain at position 1881 (numbered according to GenBank accession number M10014) (Figure 1B). This nucleotide substitution (hereafter referred to as 1876 +5G→A) was located at the fifth position of intron 1 and might affect the correct splicing of γ-chain mRNA. The proband's parents were heterozygous for this mutation (Figure 1B). Two hundred aploid genomes from unrelated individuals belonging to 2 populations with different genetic background (100 from a Northern Italian and 100 from an Iranian control population) were also analyzed by dot blot hybridization with allele-specific oligonucleotide probes. The 1876 +5G→A mutation was absent in all of them (data not shown).
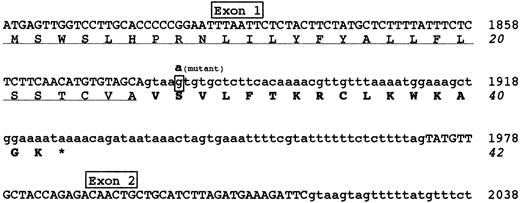
Figure 2 shows the predicted effect of the 1876 +5G→A mutation. The alteration of the splice donor site would determine the failure of splicing, causing a read-through of the downstream intronic sequence. The retention of intron 1 into mature mRNA would introduce 16 additional amino acids and a premature stop codon; accordingly, the predicted mutant fibrinogen γ-chain would consist of 26 normal amino acids (corresponding to the signal peptide), followed by a stretch of 16 aberrant amino acids.
Predicted effects of the 1876 +5G→A mutation.
Nucleotide and amino acid sequences surrounding the 1876 +5G→A mutation are shown. The G→A transition identified in intron 1 is expected to cause a read-through of the downstream intronic sequence, introducing an aberrant 16 amino acid polypeptide after the signal peptide. Exonic and intronic sequences are reported in upper or lower cases, respectively. The mutant nucleotide involved in 1876 +5G→A mutation is in bold-face type and shown above the wild-type boxed nucleotide. The signal peptide is underlined and the 16 aberrant amino acids are bolded. The premature stop codon, giving rise to a short protein of 42 amino acids, is indicated by an asterisk. Nucleotide sequence is numbered according to GenBank accession numberM10014.
Predicted effects of the 1876 +5G→A mutation.
Nucleotide and amino acid sequences surrounding the 1876 +5G→A mutation are shown. The G→A transition identified in intron 1 is expected to cause a read-through of the downstream intronic sequence, introducing an aberrant 16 amino acid polypeptide after the signal peptide. Exonic and intronic sequences are reported in upper or lower cases, respectively. The mutant nucleotide involved in 1876 +5G→A mutation is in bold-face type and shown above the wild-type boxed nucleotide. The signal peptide is underlined and the 16 aberrant amino acids are bolded. The premature stop codon, giving rise to a short protein of 42 amino acids, is indicated by an asterisk. Nucleotide sequence is numbered according to GenBank accession numberM10014.
As shown in Table 1, by sequencing the 3 fibrinogen genes, we also detected a few trivial differences from the reported sequences (GenBank, accession numbers: M64982, M64983, andM10014). Four variations had already been reported to be common in a control population.13 Two new nucleotide differences were found within introns of the Aα-chain gene. In particular, a T→G transversion was observed at position 1943, whereas the other variation was represented by 3 consecutive transitions (G→A, A→G, and G→A) at positions 3223-3225 (Table 1). These base substitutions were also found, in the homozygous state, in PCR-amplified DNA fragments from 13 normal healthy controls. All the above variations are likely to represent sequence conflicts, probably because of sequencing errors.
Trivial nucleotide substitutions detected in fibrinogen chain genes of the patient.
| Fibrinogen chain . | Position . | Substitution . | Reference . |
|---|---|---|---|
| Aα-chain | Exon 1 | A57 → G | 13 |
| Exon 1 | T60 → C | 13 | |
| Intron 3 | T1943 → G | This study | |
| Intron 4 | 3223GAG3225 → AGA | This study | |
| Bβ-chain | Intron 6 | 6939GAGAGA6944 → GAGA | 13 |
| γ-chain | Intron 1 | 1953TC1954 → CT | 13 |
| Fibrinogen chain . | Position . | Substitution . | Reference . |
|---|---|---|---|
| Aα-chain | Exon 1 | A57 → G | 13 |
| Exon 1 | T60 → C | 13 | |
| Intron 3 | T1943 → G | This study | |
| Intron 4 | 3223GAG3225 → AGA | This study | |
| Bβ-chain | Intron 6 | 6939GAGAGA6944 → GAGA | 13 |
| γ-chain | Intron 1 | 1953TC1954 → CT | 13 |
Production of γ-chain messenger RNA in HeLa cells
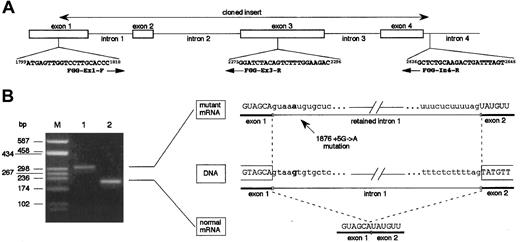
To verify whether the 1876 +5G→A mutation determines the retention of intron 1 into the mature mRNA, mutant γ-chain mRNA was transiently produced in HeLa cells. For this purpose, we prepared 2 expression vectors, pTarget-γ(Ex1-In4)-wt and pTarget-γ(Ex1-In4)-mut, by cloning a PCR-amplified genomic DNA fragment of the fibrinogen γ-chain gene, as described in “Materials and methods.” As shown in Figure 3A, this fragment spanned from nucleotide position 1799, corresponding to the first position of the ATG translation start codon, to nucleotide position 2646, corresponding to the 5′ part of intron 4 (numbering according to GenBank accession number M10014). Orientation of the inserts and absence of newly unanticipated changes due to PCR errors in the cloned regions were checked by sequencing. Both pTarget-γ(Ex1-In4)-wt and pTarget-γ(Ex1-In4)-mut plasmids were independently transfected in HeLa cells and total RNA was extracted after 48 hours. Reverse transcriptase-polymerase chain reaction (RT-PCR) assays on purified DNAseI-treated RNA were performed using the exonic primers FGG-Ex1-F (nucleotide position 1799-1818) and FGG-Ex3-R (nucleotide position 2296-2275) (Figure 3A). RT-PCR performed on RNA extracted from pTarget-γ(Ex1-In4)-wt–transfected HeLa cells allowed us to detect the expected 213-bp long fragment (constituted by 78-bp of exon 1, 45-bp corresponding to the entire exon 2, and 90-bp of exon 3) (Figure 3B, lane 2). The same amplification, carried out using RNA extracted from pTarget-γ(Ex1-In4)-mut–transfected HeLa cells as template, resulted in a longer product of about 300 bp, as evaluated by gel electrophoresis (Figure 3B, lane 1). The greater length of this mutant transcript was compatible with the retention into the mature mRNA of the 96-bp long intron 1. This hypothesis was confirmed by direct sequencing of the PCR product, which spanned along 309 bp and contained the complete intron 1 sequence. These data demonstrate that 1876 +5G→A mutation alters the intron 1 splice-donor site and results in an abnormal γ-mRNA.
Functional analysis of the effect of splice-donor site mutation 1876 + 5G→A on γ-chain mRNA.
(A) Schematic representation of the 5′ portion of the fibrinogen γ-chain gene. Primers used in cloning experiments (FGG-Ex1-F and FGG-In4-R) and in RT-PCR assays (FGG-Ex1-F and FGG-Ex3-R) are reported. Numbers preceding and following each oligonucleotide indicate primer position (numbering according to GenBank accession number M10014); arrows indicate primer orientation. (B) Left panel: RT-PCR products obtained with primers FGG-Ex1-F and FGG-Ex3-R, separated on a 2% agarose gel. Lane M: molecular weight marker (pUC8-HaeIII); lane 1: RT-PCR product amplified from transfected HeLa cells expressing mutant mRNA; lane 2: RT-PCR product amplified from transfected HeLa cells expressing wild-type mRNA. Right panel: schematic representation of the normal and aberrant splicing events. Nucleotide at position +5 of intron 1 is in bold-face type in both wild-type and mutant sequences.
Functional analysis of the effect of splice-donor site mutation 1876 + 5G→A on γ-chain mRNA.
(A) Schematic representation of the 5′ portion of the fibrinogen γ-chain gene. Primers used in cloning experiments (FGG-Ex1-F and FGG-In4-R) and in RT-PCR assays (FGG-Ex1-F and FGG-Ex3-R) are reported. Numbers preceding and following each oligonucleotide indicate primer position (numbering according to GenBank accession number M10014); arrows indicate primer orientation. (B) Left panel: RT-PCR products obtained with primers FGG-Ex1-F and FGG-Ex3-R, separated on a 2% agarose gel. Lane M: molecular weight marker (pUC8-HaeIII); lane 1: RT-PCR product amplified from transfected HeLa cells expressing mutant mRNA; lane 2: RT-PCR product amplified from transfected HeLa cells expressing wild-type mRNA. Right panel: schematic representation of the normal and aberrant splicing events. Nucleotide at position +5 of intron 1 is in bold-face type in both wild-type and mutant sequences.
Discussion
Congenital quantitative deficiencies have been identified for different proteins of the coagulation and anticoagulation systems, including factor V,14 factor VII,15 factor VIII,16 protein C,17 protein S,18 and antithrombin III.19 In these cases, probands' DNA and/or RNA analysis allowed the identification of several genetic abnormalities responsible for reduced plasma levels of the corresponding protein. Several mutations, such as deletions, missense mutations, nonsense mutations, and splice-site abnormalities have been described to be associated with protein deficiency. So far, inherited afibrinogenemia has been associated only with a gross homozygous deletion of the almost the entire Aα-chain gene9,10 and with 2 homozygous missense mutations in the Bβ-chain gene.6 In the case of the Aα-chain gene deletion, fibrinogen was unmeasurable in plasma because no Aα-chains were synthesized, whereas both Bβ-chain missense mutations caused afibrinogenemia by impairing fibrinogen secretion.
In this study, by direct sequence analysis of the 3 fibrinogen genes in a Pakistani afibrinogenemic patient, we identified a novel point mutation, the first localized in the γ-chain gene, responsible for afibrinogenemia: a homozygous G→A transition at the fifth nucleotide of intron 1 (1876 +5G→A). The mutation was present in the heterozygous state in both the consanguineous proband's parents, who had reduced plasma levels of clottable and immunoreactive fibrinogen compatible with an heterozygous phenotype. Gene frequency analysis in 100 healthy subjects confirmed the absence of this mutation in the normal population. Because a Pakistani population sample was not available for this study, 2 different control populations, one from Europe (Italy) and one from the Middle East (Iran), were analyzed to exclude possible differences in allelic frequencies due to a particular genetic background.
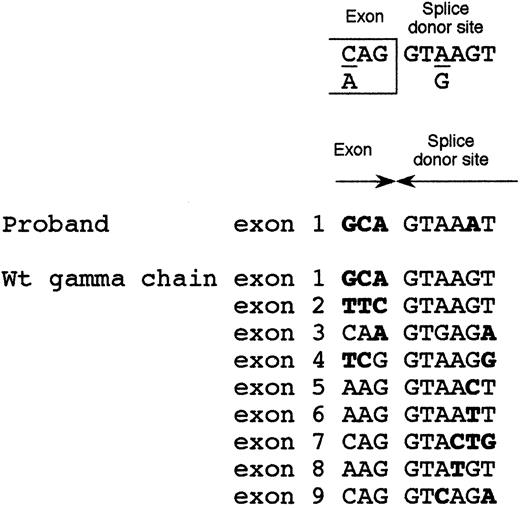
Because the 1876 +5G→A intronic mutation was localized in proximity of the exon 1-intron 1 boundary, we postulated its involvement in inactivating the intron 1 splice-donor site. The consensus sequence of the splice-donor site20 consists of 6 conserved nucleotides (Figure 4). The splice-donor site of intron 1 of γ-fibrinogen gene perfectly shared this consensus. Moreover, even if some degree of degeneration at position +5 between the 9 splice-donor sites of the γ-chain gene can be observed, in no instance can a substitution of the conserved guanine with adenine at position +5, that was detected in our afibrinogenemic patient, be observed (Figure 4). Only 2 other splicing abnormalities have been described in fibrinogen genes21,22 and both were in the fibrinogen γ-chain gene. One was an A→G transition in intron 8, leading to an insertion of 15 amino acids after residue Q350 as a consequence of an alternative splicing event. The resulting dysfunctional fibrinogen molecule, called Paris I, was not associated with bleeding or thrombosis.21 The second mutation, associated with hypofibrinogenemia, was an A82G amino acid substitution that could activate a cryptic 5′ splice site in exon 4. The aberrant splicing would produce an abnormal γ-chain truncated after residue 97 (Fibrinogen Dunedin).22 However, the effect of these 2 mutations was analyzed at the protein level, whereas the underlying molecular mechanism was only postulated.
Alignment of the 9 splice-donor sites of fibrinogen γ-chain gene.
Top: splice-donor site consensus sequence.20 Middle: intron 1 splice-donor site of the afibrinogenemic Pakistani patient. Bottom: alignment of the 9 wild-type splice-donor sites of the fibrinogen γ-chain gene. Bases not matching the consensus sequence are in bold-face type.
Alignment of the 9 splice-donor sites of fibrinogen γ-chain gene.
Top: splice-donor site consensus sequence.20 Middle: intron 1 splice-donor site of the afibrinogenemic Pakistani patient. Bottom: alignment of the 9 wild-type splice-donor sites of the fibrinogen γ-chain gene. Bases not matching the consensus sequence are in bold-face type.
Our Pakistani afibrinogenemic patient had unmeasurable levels of plasma fibrinogen, thus preventing a protein investigation and suggesting a study at mRNA level. Because fibrinogen expression is confined mainly to the liver23 and we had no access to liver biopsy specimens of the Pakistani patient, we adopted an ex vivo approach to demonstrate the aberrant splicing of fibrinogen mutant γ-mRNA. Fibrinogen nonexpressing HeLa cells24 were chosen as an experimental system to transiently produce wild-type and mutant γ-mRNAs. RT-PCR assays demonstrated that the 1876 +5G→A mutation determined the introduction of the intron 1 sequence into mature mRNA and, consequently, the very severe truncation of the mutant protein (Figure 2). This truncation involves the entire normal γ-chain protein, except the signal peptide. From this point of view, it can be considered as the counterpart of the 11-kb deletion of the Aα-chain gene,9,10 even though the underlying genetic mechanism is completely different. In both cases, the virtually complete absence of one of the 3 chains would prevent the secretion of fibrinogen into the circulation, because fully assembled fibrinogen is the only form of the molecule that is secreted.25
In summary, we identified the first truncation of the fibrinogen γ-chain associated with afibrinogenemia. This new mutation elucidates a possible correlation between genotype and clinical phenotype. In fact, all homozygous fibrinogen-chain truncations identified so far, with the exception of fibrinogen Milano III,26 cause quantitative deficiencies: the more extended the truncation, the more severe the fibrinogen deficiency, ranging from mild to severe hypodysfibrinogenemia27-29 up to afibrinogenemia.9,10 The recently described cases of hypofibrinogenemia associated with heterozygous missense mutations,13,22,30 may in some cases represent the phenotypic effect of mutations that, at the homozygous state, could cause afibrinogenemia, like the βL353R and βG400D mutations.6
Acknowledgments
We thank family members for their participation in this study.
Supported by the Ministero dell'Università e della Ricerca Scientifica e Tecnologica (MURST 60%), by IRCCS Maggiore Hospital, Milan, Italy, and by Progetto Giovani 1998.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Note added in proof
Author notes
Maria Luisa Tenchini, Dipartimento di Biologia e Genetica per le Scienze mediche, via Viotti, 3/5-20133 Milano, Italy; e-mail:marialuisa.tenchini@unimi.it.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal