Abstract
To understand the molecular basis of exocytosis in human neutrophils, the role of syntaxin 6 and SNAP-23 in neutrophil degranulation was examined. Human syntaxin 6 was cloned and identified as a 255-amino acid protein with a carboxy-terminal transmembrane region and two coiled-coil domains. Syntaxin 6 was localized mainly in the plasma membrane of human resting neutrophils, whereas SNAP-23 was located primarily in the mobilizable tertiary and specific granules. SNAP-23 was translocated to the cell surface, colocalizing with syntaxin 6, on neutrophil activation. In vitro binding studies established that SNAP-23 binds to syntaxin 6. Coimmunoprecipitation assays indicated that SNAP-23 interacts with syntaxin 6 in vivo, and this interaction was dramatically increased on neutrophil activation. Antibodies against SNAP-23 inhibited Ca++ and GTP-γ-S–induced exocytosis of CD67-enriched specific granules, but they hardly affected exocytosis of the CD63-enriched azurophilic granules, when introduced into electropermeabilized neutrophils. Anti–syntaxin 6 antibodies prevented exocytosis of both CD67- and CD63-enriched granules in electropermeabilized neutrophils. These data show that syntaxin 6 and SNAP-23 are involved in human neutrophil exocytosis, demonstrating that vesicle SNAP receptor-target SNAP receptor (v-SNARE– t-SNARE) interactions modulate neutrophil secretion. Syntaxin 6 acts as a target for secretion of specific and azurophilic granules, whereas SNAP-23 mediates specific granule secretion.
Introduction
Polymorphonuclear neutrophils play a major role in the surveillance system of the host organism against foreign invaders and constitute one of the primary mediators of the acute inflammatory response. Exocytosis of distinct cytoplasmic granules present in human neutrophils plays a critical role in neutrophil biology and seems to regulate important neutrophil functions in both inflammation and infection, such as adhesion, diapedesis, generation of reactive oxygen metabolites, and release of lytic enzymes. A remarkable feature of human neutrophils is the presence of 4 distinct types of cytoplasmic granules: azurophilic or primary granules, specific or secondary granules, gelatinase-rich tertiary granules, and alkaline phosphatase-rich granules or phosphasomes, also named as secretory vesicles.1-7 Exocytosis of the distinct granule populations may occur independently.8-11 Unlike azurophilic granules, which are hardly mobilized, specific and tertiary granules are readily exocytosed on cell activation. These mobilizable granules contain many components involved in the adhesion and extravasation of human neutrophils, including adhesion molecules, extracellular matrix proteases, and enzymes implicated in the generation of soluble mediators of inflammation.3,7,9,10,12-16 Furthermore, these cytoplasmic mobilizable granules, together with the alkaline phosphatase-rich granules, constitute a reservoir of plasma membrane proteins that are translocated to the cell surface following neutrophil activation. However, little is known regarding the mechanisms that lead to membrane docking and fusion between cytoplasmic granules and cell surface in human neutrophils. Microtubules have been reported to be required for neutrophil degranulation in response to different stimuli,17,18 but they do not discriminate among exocytosis of the distinct granules.17
SNARE (soluble N-ethylmaleimide-sensitive fusion [NSF] factor attachment protein receptor) proteins have been found to mediate vesicle secretion in essentially all organisms investigated, from yeast to human, leading to the postulation of the SNARE hypothesis.19-22 This constitutes an attractive model for vesicle-membrane fusion in eukaryotic cells. According to the SNARE hypothesis, docking and fusion of vesicles with the plasma membrane is modulated by the specific interaction of vesicle proteins (v-SNAREs) with target plasma membrane-located proteins (t-SNAREs). v-SNAREs are type II integral membrane proteins located on vesicles and oriented in such a way that most of the protein is in the cytosol. t-SNAREs are membrane proteins that are associated with target membranes and are also oriented toward the cytoplasm. Genetic and immunological evidences indicate that human neutrophils contain a wide number of SNARE proteins23-26 that could be related to their high secretory capacity and to the presence of distinct neutrophil cytoplasmic granules with different exocytic capabilities. It has been suggested that the selective presence of certain SNARE proteins in distinct granule membranes could be an explanation for the independent mobilization of the different granule populations present in human neutrophils during cell activation.24 26
We have previously found the expression of SNAP-23 present in 2 isoforms, A and B,24 and syntaxin 6 in human neutrophils as well as in the human leukemic cell line HL-6024,26considered a good cell culture model for human neutrophils.27 The sequence of the SNAP-23A isoform corresponded to the previously described SNAP-23 complementary DNA (cDNA).24,28 SNAP-23A is the major isoform expressed in human neutrophils,24 and thereby it will be referred to as SNAP-23 in this report. Our previous studies indicated that expression of SNAP-23 and syntaxin 6 was increased during neutrophil differentiation of HL-60 cells,24,26 suggesting a putative role for these SNARE proteins in some specialized functions of mature neutrophils. SNAP-23 has been shown to bind efficiently to syntaxins 1, 2, 3, and 4 in vitro.28 In this report, we have cloned and sequenced the human syntaxin 6 from peripheral blood neutrophils and investigated the subcellular localization and functional role of syntaxin 6 and SNAP-23 in these cells. The present study provides evidence for the interaction of syntaxin 6 and SNAP-23 in vitro and in vivo and for their involvement in neutrophil exocytosis.
Materials and methods
Antibodies
Polyclonal antibodies against human SNAP-23 were produced by subcutaneous injections of glutathione-S-transferase (GST)–SNAP-23 fusion protein into rabbits. To avoid possible cross-reactivity with SNAP-25, the antiserum was incubated overnight at 4°C with GST–SNAP-25 coupled to glutathione-agarose beads (Pharmacia, Uppsala, Sweden) and centrifuged. Supernatant was then immunoabsorbed to GST–SNAP-23 coupled to glutathione-agarose beads, eluted with 4 mol/L MgCl2, and dialyzed with phosphate-buffered saline (PBS) to obtain affinity-purified anti–SNAP-23 antiserum. Anti–syntaxin 6 3D10 monoclonal antibody (MoAb)29 was kindly provided by Dr J. Bock (Stanford University Medical Center, Stanford, CA). Specific MoAbs against human CD63 and CD67 were from Central Laboratory of the Blood Transfusion Service (Amsterdam, The Netherlands). Biotinylated antimouse immunoglobulin G (IgG) and biotinylated antirabbit IgG, used for immunoblotting, were from Amersham (Buckinghamshire, UK). Fluorescein isothiocyanate (FITC)-conjugated antimouse immunoglobulin was from Dakopatts (Glostrup, Denmark), phycoerythrin-conjugated antirabbit immunoglobulin was from Sigma (St Louis, MO), and CY2-conjugated antirabbit immunoglobulin was from Pharmacia. P3X63 myeloma culture was used as a negative control.
Neutrophil isolation and activation
Neutrophils were obtained from fresh heparinized human peripheral blood by dextran sedimentation and centrifugation on Ficoll-Hypaque, followed by hypotonic lysis of residual erythrocytes as previously described.17,26 Freshly isolated human neutrophils were resuspended at 3-5 × 106 cells/mL of Hepes/glucose buffer (150 mmol/L NaCl, 10 mmol/L Hepes, 5 mmol/L KOH, 1.2 mmol/L MgCl2, 1.3 mmol/L CaCl2, 5.5 mmol/L glucose, pH 7.5) and incubated at 37°C for 10 minutes with 100 ng/mL phorbol-12-myristate-13-acetate (PMA). Release of gelatinase, lactoferrin, β-glucuronidase, and peroxidase following neutrophil activation with PMA was determined as previously described.5,9 10
Subcellular fractionation
Resting and PMA-activated neutrophils were resuspended in 50 mmol/L Tris-HCl, pH 7.5 containing 2 mmol/L phenylmethylsulfonyl fluoride (PMSF), and then disrupted by repeated freeze–thaw. Homogenates were centrifuged at 1200 rpm in a Sorvall T 6000D centrifuge for 10 minutes, and the supernatant, representing the postnuclear extract, was saved. After centrifugation of the postnuclear extract at 45 000 rpm in a TLA rotor for 90 minutes at 4°C using a Optima TL ultracentrifuge (Beckman Instruments, Palo Alto, CA), supernatant (cytosol fraction) and pellet (membrane fraction) were saved. Pellets were resuspended in 50 mmol/L Tris-HCl, pH 7.5, containing 2 mmol/L PMSF.
To prepare the distinct subcellular fractions, freshly prepared neutrophils (about 3-5 × 108) were gently disrupted as described previously.3,30 The postnuclear fraction was fractionated in 15%-40% (w/w) continuous sucrose gradients, by centrifugation at 4°C for 10 minutes at 25 000 rpm in a Beckman L8-70B ultracentrifuge, using a SW27 rotor, as described previously,3,30 and 2 mmol/L PMSF was added at each fraction. Subcellular fractions were assayed for marker enzymes for each organelle, namely 5′-nucleotidase (plasma membrane), latent alkaline phosphatase (phosphasomes), gelatinase (tertiary granules), lactoferrin (specific granules), and peroxidase (azurophilic granules) as described previously.30 31 Membranes from each fraction were obtained by diluting the fractions with 50 mmol/L Tris-HCl, pH 8.0, 100 mmol/L NaCl and centrifugation at 29 000 rpm for 90 minutes at 4°C using a 30-type rotor (Beckman Instruments). The pellets, representing the membranous fractions, were resuspended in 50 mmol/L Tris-HCl, pH 7.5 containing 2 mmol/L PMSF, and stored at −20°C until use.
Reverse transcriptase-polymerase chain reaction (RT-PCR)
Total RNA was extracted from human neutrophils according to the protocol of Chomczynski and Sacchi.32 Total RNA (15 μg), primed with oligo-dT, was reverse-transcribed into cDNA with 30 units of AMV reverse transcriptase from Promega (Madison, WI) in a final volume of 20 μL. The mixture was incubated at 37°C for 2 hours and stored at −20°C until use. The PCR mixture (50 μL) contained the template cDNA (1-2 μL), 20 pmol of the corresponding primers, 0.2 mmol/L dNTP, 2.5 mmol/L MgCl2, 5 units of EcoTaq DNA polymerase derived from Thermus aquaticus (ECOGEN, Barcelona, Spain). PCR reactions were performed in GeneAmp PCR System model 9600 (PerkinElmer, Norwalk, CT). The primers used are shown as follows, where the nucleotide numbers indicate the primer location in the corresponding sequence of rat syntaxin 6 obtained from the GenBank/EMBL database (accession number, U56815): set 1 forward—nt 206-230 5′-TGCCCAGGGATTGTTCCAGAGATGG-3′; reverse—nt 856-880 5′-AGGACCGCGAAGAGGATGGCTATGG-3′; set 2 forward—nt 147-169 5′-ATGTCCATGGAGGACCCCTTCTT-3′; and reverse—nt 900-920 5′-CCACCATCACAGCACTAGGAA-3′.
Primers were designed by using the PCgene program for DNA analysis from Intelligenetics (Mountain View, CA). The conditions for PCR amplification using a thermal cycler were as follows: 1 cycle at 95°C for 5 minutes as an initial denaturation step, then denaturation at 95°C for 30 seconds, annealing at 62°C (set 1) or 67°C (set 2) for 30 seconds, and extension at 72°C for 90 seconds (30 cycles), followed by further incubation for 15 minutes at 72°C (1 cycle). An aliquot of the PCR reaction was analyzed on a 2% agarose gel in 1 × TAE (40 mmol/L Tris-acetate, 1 mmol/L EDTA, pH 8.0) and checked for the expected PCR products.
cDNA cloning and sequencing
The PCR products were directly cloned into the pCR 2.1 vector, using the TA cloning kit (Invitrogen, San Diego, CA) following the manufacturer's indications. DNA sequencing was performed by thermal cycle sequencing using the Cy5 AutoCycle sequencing kit (Pharmacia Biotech) and a PE Applied Biosystems 377 DNA Sequencer (PerkinElmer). DNA sequencing was performed on both strands from 10 independent cDNA clones.
Production of SNAP-23 fusion protein
Full-length coding sequence for SNAP-23, corresponding to the SNAP-23A isoform,24 was amplified by the PCR by using oligonucleotides flanked by EcoRI and XhoI cleavage sites and was subsequently subcloned into the bacterial expression vector pGEX-4T-1 (Pharmacia Biotech), obtaining the in-frame recombinant proteins composed of GST fused to the N terminus of recombinant SNAP-23. Escherichia coli cells expressing GST or GST–SNAP-23 fusion protein were grown in 400 mL of 2 × YT-G medium to A600 = 0.9, induced by the addition of 0.1 mmol/L isopropyl-β-D-thiogalacto-pyranoside, and harvested after 4 hours. Cells were pelleted, resuspended in 20 mL PBS, and sonicated on ice 4 × 30 seconds. Triton X-100 (1%, v/v) was added to the lysate and mixed for 30 minutes at 4°C. Suspension was centrifuged at 12 000 rpm for 10 minutes in an SS34 rotor at 4°C. The supernatant was mixed with 0.4 mL of a 50% slurry of glutathione-Sepharose 4B (Pharmacia Biotech) for 30 minutes at room temperature with gentle agitation. Beads were sedimented and washed 3 times with PBS. Fusion protein or GST was eluated from the beads with 200 μL elution buffer (20 mmol/L glutathione, 100 mmol/L Tris-HCl, pH 8.0, 120 mmol/L NaCl), analyzed by SDS-polyacrylamide gel electrophoresis and visualized by Coomassie Blue staining. Eluates were used to immunize rabbits and for in vitro binding assays after extensive dialysis against PBS, containing 0.5% Triton X-100. Full-length SNAP-23 recombinant protein was obtained by treating the respective glutathione-Sepharose-immobilized fusion proteins, derived from pGEX-4T-1, with 3 units thrombin/100 μL beads for 1 hour at room temperature in 20 mmol/L Tris, pH 7.2, 150 mmol/L NaCl, 0.5% Triton X-100. Supernatant containing the purified SNAP-23 protein was collected, and thrombin was inhibited by 1 mmol/L PMSF.
In vitro binding assays
Freshly isolated human neutrophils (2 × 108cells) were homogenized in 3 mL of 150 mmol/L NaCl, 10 mmol/L Hepes-KOH, pH 7.4, containing 1 mmol/L PMSF, 4 μg/mL leupeptin, and 4 μg/mL aprotinin using a Potter homogenizer. Triton X-100 (0.5%, v/v) was added to the homogenate, and the mixture was incubated at 4°C for 1 hour with constant agitation. Mixture was clarified by centrifugation at 45 000 rpm for 2 hours at 4°C in a TLA rotor, and supernatant was used for binding to GST and GST–SNAP-23 fusion protein. Glutathione-Sepharose 4B beads preincubated with equal amounts (100 μg) of GST-SNAP-23 or GST were added to 1 mL of the supernatant and incubated with gentle mixing for 3 hours at 4°C. Beads were then sedimented, washed 5 times in ice-cold PBS, with gentle mixing, and proteins bound to matrix were eluated with 50 μL elution buffer (20 mmol/L glutathione, 100 mmol/L Tris-HCl, pH 8.0, 120 mmol/L NaCl) for 30 minutes with gentle mixing. Eluates were separated from the beads by centrifugation at 12 000 rpm for 10 minutes in a microfuge and processed for SDS-12% polyacrylamide gel electrophoresis and Western blotting.
SDS-polyacrylamide gel electrophoresis and Western blotting
Proteins were separated by SDS-12% polyacrylamide gels according to standard procedures as described previously.31 Immunoblotting was performed as described previously,25 with slight modifications. After blocking for 5 hours at room temperature with 5% powdered defatted milk in TBS buffer (50 mmol/L Tris-HCl, pH 8.0, 150 mmol/L NaCl) containing 0.05% Tween 20, blots were incubated for 2 hours with anti–SNAP-23 polyclonal antibody at a dilution 1:400 in TBS buffer containing 0.05% Tween 20 or with anti–syntaxin 6 3D10 MoAb at a dilution of 1:1000 in TBS buffer containing 0.05% Tween 20. Antibody reactivity was monitored with biotinylated antimouse IgG or antirabbit IgG (both diluted at 1:1000 in TBS buffer), using an enhanced chemiluminescence detection system (Amersham).
Coimmunoprecipitation
Resting and activated cells (5 × 106 cells) were lysed with 60 μL lysis buffer (20 mmol/L Tris-HCl, 100 mmol/L KCl, 0.9% Triton X-100, 10% glycerol, 2 mmol/L orthovanadate, 2 mmol/L PMSF). Whole cell lysates were centrifuged in a microfuge at 12 000 rpm for 20 minutes at 4°C, and the supernatant was precleared by incubation for 2 hours at 4°C with 100 μL protein A-Sepharose (20% in lysis buffer). Antibodies to SNAP-23 were precoupled to protein A-Sepharose by incubation for 2 hours at 4°C in lysis buffer. The precoupled beads were pelleted, washed twice with lysis buffer, and added to the supernatant from the preclearing step. Lysate and antibody were incubated for 1 hour at 4°C with constant rotation, then Sepharose beads were pelleted and washed 5 times with lysis buffer. Then, 30 μL SDS sample buffer (50 mmol/L Tris-HCl pH 6.8, 100 mmol/L dithiothreitol, 2% SDS, 0.1% bromophenol blue, 10% glycerol) was added, samples were subjected to SDS-12% polyacrylamide gel electrophoresis, and proteins were transferred to nitrocellulose and immunoblotted as described above.
Electropermeabilization
Neutrophils were permeabilized immediately before use, following a method described previously33 34 with slight modifications. An aliquot of 6 × 106 cells was washed once and resuspended in 0.6 mL of ice-cold electropermeabilization buffer (120 mmol/L KCl, 10 mmol/L NaCl, 1 mmol/L KH2PO4, 10 mmol/L glucose, 20 mmol/L Hepes, pH 7.0). The suspension was transferred to a BTX cuvette and subjected to 2 discharges of 5 kV/cm, 25 μF, and 72 Ω using a BTX electroporator (Biotechnologies & Experimental Research, San Diego, CA). The cells were stirred gently between pulses, using a plastic pipette. Permeabilized cells were immediately transferred to a plastic tube in which the corresponding antibodies were added and incubated for 3 minutes at room temperature. Then, the cells were transferred to 37°C. After a 5-minute incubation with 5 μg/mL cytochalasin B at 37°C, the cells were stimulated with 1 μmol/L Ca++ (0.1 mmol/L CaCl2, 5.37 mmol/L MgCl2, 5 mmol/L HEDTA) and 50 μmol/L GTP-γ-S for 10 minutes at 37°C. The free Ca++ concentration in the incubation was checked by Fura-2 measurement. Then, cells were fixed with 1% paraformaldehyde and processed for immunofluorescence flow cytometry.
Immunofluorescence flow cytometry
Propidium iodide and FITC fluorescence were analyzed by using a FACScan flow cytometer (Becton Dickinson, San Jose, CA). Linear and logarithmic immunofluorescence intensities were determined as described previously,13 and the P3X63 myeloma culture supernatant was used as a negative control. Cell surface expression of CD63 and CD67 was measured in paraformaldehyde-fixed electropermeabilized neutrophils by incubation for 30 minutes at 4°C with 2 μg/mL of anti-CD63 or anti-CD67 MoAbs. After washing with PBS, FITC-conjugated antimouse immunoglobulin, previously diluted 1:50 in PBS, was added, and the incubation proceeded for an additional period of 30 minutes at 4°C. Finally, cells were washed with PBS, and their fluorescence was estimated with a FACScan flow cytometer at a linear scale.
After paraformaldehyde fixation of electropermeabilized neutrophils, addition of the secondary antibody FITC-conjugated antimouse immunoglobulin only detected the anti-CD3 and anti-CD67 mouse antibodies bound to the cell surface, but not in the interior of the cell. Thus, in the absence of anti-CD63 and anti-CD67 MoAbs, no staining was obtained when the secondary antibody FITC-conjugated antimouse immunoglobulin was added to paraformaldehyde-fixed electropermeabilized neutrophils incubated before fixation with the distinct antibodies used in the functional assays.
Confocal laser scanning microscopy
Resting and PMA-activated cells were resuspended in Hepes/glucose buffer (3 × 106 cells/mL), and an aliquot (100 μL) of the cell suspension was cytospun at 1000 rpm for 3 minutes (cytospin 3, Shandon Scientific, Cheshire, UK). Confocal microscopy was carried out as described previously.10 In brief, cells were washed with PHEM buffer (60 mmol/L Pipes, 25 mmol/L Hepes, 10 mmol/L EGTA, 3 mmol/L MgCl2, pH 7), and fixed in 4% (w/v) paraformaldehyde in PHEM buffer for 10 minutes. After extensive washing with PBS, cells were permeabilized for 5 minutes with −20°C acetone and then washed again with PBS. Fixed cells were blocked with 5% goat serum in PBS for 2 hours at room temperature, washed with PBS, and incubated with primary antibodies for 1 hour in a humidified chamber. The primary antibodies used in this study were anti–SNAP-23 rabbit polyclonal antibody (diluted 1:500 in PBS) and 3D10 anti–syntaxin 6 mouse MoAb (diluted 1:200 in PBS). The samples were then washed with PBS and incubated for 45 minutes with secondary antibodies, using CY2-conjugated antirabbit antibody (diluted 1:200 in PBS) or FITC-conjugated antimouse antibody (diluted 1:200 in PBS). For colocalization studies, the primary antibodies were added consecutively. Then, cells were washed with PBS and incubated subsequently with phycoerythrin-conjugated antirabbit antibody (diluted 1:200 in PBS) and FITC-conjugated antimouse antibody (diluted 1:200 in PBS) for 45 minutes in a humidified chamber at room temperature in the dark. Slides were then washed extensively with PBS and mounted in the aqueous-medium Crystal/Mount (Biomeda, Foster City, CA). Negative controls were routinely prepared by omitting the primary antibodies or by using an irrelevant antibody, showing no fluorescence staining of the samples. Colocalization of both antigens was analyzed by excitation of both fluorochromes in the same section. Fluorescence was visualized with a Zeiss LSM 310 laser scan confocal microscope (Zeiss, Switzerland).
Results
Cloning and sequencing of syntaxin 6 from human neutrophils
We have previously demonstrated by RT-PCR that syntaxin 6 is expressed in both human neutrophils and HL-60 cells differentiated toward neutrophils.26 By using 2 sets of overlapping PCR oligonucleotides designed from rat syntaxin 6, we generated DNA fragments by RT-PCR from peripheral blood neutrophil messenger RNA (mRNA), which were subsequently cloned and sequenced. Following this strategy, we sequenced the human homologue of rat syntaxin 6 from human neutrophils. The amino acid sequence of human syntaxin 6 showed a 95.3% identity to the corresponding rat syntaxin 6 coding region. The human syntaxin 6 cDNA codes for a protein sequence of 255 amino acids with a deduced molecular mass of about 29.2 kd and an isoelectric point (pI) of 4.6. Syntaxin 6 shows a transmembrane region at the carboxy terminal (residues 235-255), and it has been previously reported to be an integral membrane protein.35 The primary structure of syntaxin 6 shows 2 regions predicted to form coiled coils. The first region spans residues 41-74, including a leucine-zipper–like region with 5 heptad repeats of leucine, isoleucine, and valine; the second region, spanning residues 174-227, includes a leucine-zipper with 5 heptad repeats of leucine (Figure1). These leucine-zipper–like regions could be putatively involved in the formation of dimers with itself or with other proteins. The syntaxin 6 protein (Figure 1) also has 2 potential sites for N-glycosylation (N-131 and N-185), 5 potential protein kinase C phosphorylation sites (S-86, S-94, T-137, S-187, and S-230), 5 potential casein kinase II phosphorylation sites (S-2, T-39, T-46, S-69, and S-213), and one N-myristoylation site (G-181). Human syntaxin 6 shows a strong homology with human syntaxin 10 (61.2% identity), which also shows potential coiled-coil domains and a carboxy-terminal hydrophobic tail as a transmembrane region.36 Interestingly, the amino-terminal leucine-zipper–like region of the syntaxin 6, comprising amino acids 41-74, is 88.2% identical to the corresponding region in syntaxin 10 with only 4 conservative changes. This amino-terminal heptad repeat region has been reported to be required for α-SNAP binding in rat syntaxin 6.35
Primary structure of human syntaxin 6.
The nucleotide and predicted amino acid sequences of human syntaxin 6 are available from GenBank/EMBL database under accession numberAJ002078. The putative transmembrane region is boxed, and the 2 potential coiled-coil domains are underlined with the heptad repeats filled in black.
Primary structure of human syntaxin 6.
The nucleotide and predicted amino acid sequences of human syntaxin 6 are available from GenBank/EMBL database under accession numberAJ002078. The putative transmembrane region is boxed, and the 2 potential coiled-coil domains are underlined with the heptad repeats filled in black.
Subcellular localization of SNAP-23 and syntaxin 6 in human neutrophils
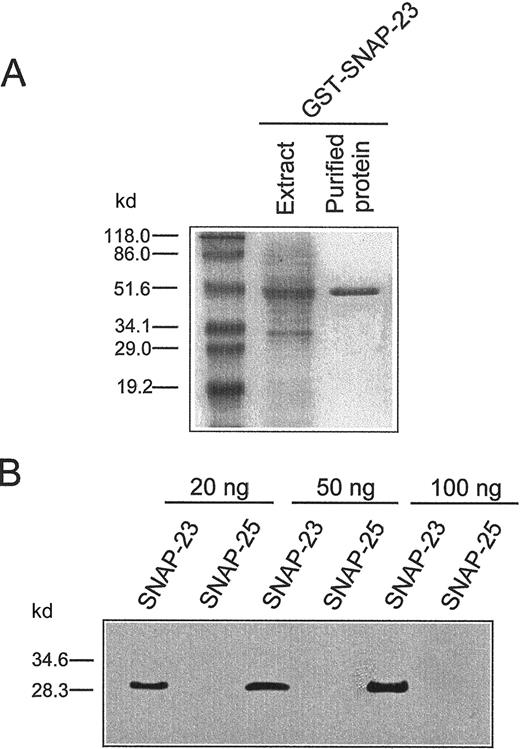
We investigated the subcellular localization of SNAP-23 and syntaxin 6 in both resting and activated human neutrophils. To this aim, we generated a specific affinity-purified antiserum against human SNAP-23 protein by immunizing rabbits with a fusion protein containing the entire open reading frame of SNAP-23 constructed in pGEX-4T-1 vector (Figure 2A). This anti–SNAP-23 polyclonal antibody detected SNAP-23 protein (Figure 2B) but failed to recognize SNAP-25 (Figure 2B), a related SNARE protein.24 28
Generation of GST–SNAP-23 fusion protein and characterization of anti–SNAP-23 antibody.
(A) Bacterial extract and purified GST–SNAP-23 fusion protein were run in SDS-polyacrylamide gel electrophoresis and stained with Coomassie Blue. (B) Characterization of the specificity of the polyclonal rabbit antibody against SNAP-23. Different amounts of purified recombinant human SNAP-23 and rat SNAP-25 proteins were electrophoresed and blotted with anti-SNAP-23 antibody. Bands were visualized by using an enhanced chemiluminescence system. The molecular masses (kd) of protein markers are indicated on the left.
Generation of GST–SNAP-23 fusion protein and characterization of anti–SNAP-23 antibody.
(A) Bacterial extract and purified GST–SNAP-23 fusion protein were run in SDS-polyacrylamide gel electrophoresis and stained with Coomassie Blue. (B) Characterization of the specificity of the polyclonal rabbit antibody against SNAP-23. Different amounts of purified recombinant human SNAP-23 and rat SNAP-25 proteins were electrophoresed and blotted with anti-SNAP-23 antibody. Bands were visualized by using an enhanced chemiluminescence system. The molecular masses (kd) of protein markers are indicated on the left.
We next determined the SNAP-23 location in both resting and activated human neutrophils. Anti–SNAP-23 antiserum recognized a single band of about 29 kd, corresponding to SNAP-23 in the postnuclear extract and in the membrane fraction, but not in the soluble fraction containing the cytosol (Figure 3A). This band was eliminated by preincubating the antiserum with soluble recombinant SNAP-23 protein (data not shown). These data indicated that SNAP-23 was membrane bound in human neutrophils. The anti-SNAP-23 antiserum was able to recognize both human recombinant SNAP-23A (29 kd) and SNAP-23B (26 kd) isoforms (data not shown). Thus, these results indicate that SNAP-23A constitutes the major SNAP-23 isoform expressed in human neutrophils, as previously suggested from the mRNA expression data.24 The molecular mass detected for human SNAP-23 (29 kD) is in good agreement with previous estimates for mouse and rat SNAP-23, 29 and 30 kD, respectively.37 38
Subcellular localization of SNAP-23 and syntaxin 6 in resting and activated human neutrophils.
(A,C) Localization of SNAP-23 and syntaxin 6 in the membrane fraction. Equal amounts of postnuclear extract (E), soluble (S), and membrane (M) proteins (60 μg), prepared from resting human neutrophils as described in “Materials and methods,” were run on SDS-polyacrylamide gels and analyzed by immunoblotting, using anti–SNAP-23 polyclonal antibody (A) or 3D10 anti–syntaxin 6 MoAb (C). (B,D) Subcellular distribution of SNAP-23 and syntaxin 6 in resting and PMA-activated human neutrophils (PMNs). Equal amounts of total protein (30 μg), prepared from the subcellular fractions 2-8 from resting and activated human neutrophils prepared as described in “Materials and methods,” were run on SDS-polyacrilamide gels and analyzed by immunoblotting, using anti–SNAP-23 polyclonal antibody (B) or 3D10 anti–syntaxin 6 MoAb (D). Fractions enriched in plasma membrane (PM), tertiary granules (TG), specific granules (SG), and azurophilic granules (AG) were analyzed. The molecular masses (kd) of protein markers are indicated on the left.
Subcellular localization of SNAP-23 and syntaxin 6 in resting and activated human neutrophils.
(A,C) Localization of SNAP-23 and syntaxin 6 in the membrane fraction. Equal amounts of postnuclear extract (E), soluble (S), and membrane (M) proteins (60 μg), prepared from resting human neutrophils as described in “Materials and methods,” were run on SDS-polyacrylamide gels and analyzed by immunoblotting, using anti–SNAP-23 polyclonal antibody (A) or 3D10 anti–syntaxin 6 MoAb (C). (B,D) Subcellular distribution of SNAP-23 and syntaxin 6 in resting and PMA-activated human neutrophils (PMNs). Equal amounts of total protein (30 μg), prepared from the subcellular fractions 2-8 from resting and activated human neutrophils prepared as described in “Materials and methods,” were run on SDS-polyacrilamide gels and analyzed by immunoblotting, using anti–SNAP-23 polyclonal antibody (B) or 3D10 anti–syntaxin 6 MoAb (D). Fractions enriched in plasma membrane (PM), tertiary granules (TG), specific granules (SG), and azurophilic granules (AG) were analyzed. The molecular masses (kd) of protein markers are indicated on the left.
To examine which membrane compartment of human neutrophils SNAP-23 is associated with, we performed immunoblotting analysis of membranes prepared from subcellular fractions enriched in individual organelles. To this aim, cells were gently disrupted, and postnuclear fractions obtained from resting human neutrophils were separated by rate zonal centrifugation under conditions that resolved cytosol, plasma membrane, gelatinase-rich tertiary granules, specific granules, and azurophilic granules as previously described.3,30 Secretory vesicles, also named phosphasomes, assayed by latent alkaline phosphatase, were not resolved from the plasma membrane fraction under the fractionation conditions used.30 Membranes prepared from the distinct subcellular fractions were used for immunoblotting with the anti–SNAP-23 polyclonal antibody. As shown in Figure 3B, SNAP-23 was localized mainly in the membranes prepared from subcellular fractions 4-6, enriched in the tertiary and specific granules. A secondary localization for SNAP-23 was found at the plasma membrane (Figure 3B). When similar subcellular fractionation experiments were carried out from human neutrophils activated with 100 ng/mL PMA for 10 minutes, we found that SNAP-23 was translocated from the above cytoplasmic granules toward the plasma membrane (Figure 3B). This subcellular translocation was correlated with a high release of gelatinase (88% secretion) and lactoferrin (76% secretion), markers for gelatinase-rich tertiary and specific granules, respectively. However, peroxidase and β-glucuronidase were hardly released (< 8% secretion) under this PMA treatment, indicating that azurophilic granules were not mobilized. Thus, we found a high correlation between the extent of the translocation of SNAP-23 from the fractions enriched in tertiary and specific granules (fractions 4-6 in Figure 3B) to the plasma membrane and the extent of the secretion of these cytoplasmic granules on neutrophil activation.
The subcellular localization of syntaxin 6 in human neutrophils was studied by using the 3D10 MoAb against rat syntaxin 6, which binds to the amino-terminal 25 amino acids of the protein.29This amino acid region is identical to the corresponding region in human syntaxin 6. This antibody recognized a single band of about 32 kd in human neutrophil extract (Figure 3C). Syntaxin 6 was located exclusively in the membrane fraction with no detectable amount in the cytosolic fraction (Figure 3C). To analyze the subcellular localization of syntaxin 6, we used the membranes prepared from the subcellular fractions enriched in individual organelles described above for SNAP-23 localization studies and performed immunoblotting studies. Figure 3D shows that syntaxin 6 was localized mainly in subcellular fractions 2 and 3, enriched in plasma membrane in both resting and activated neutrophils.
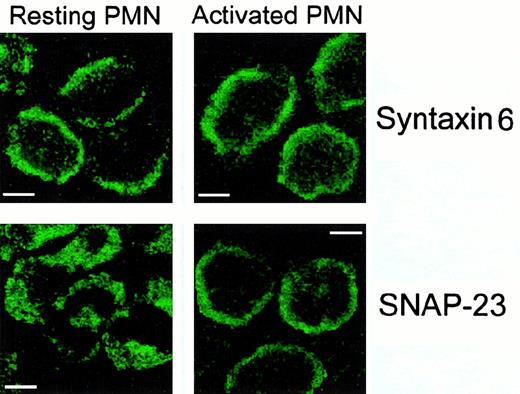
The above subcellular localization patterns of SNAP-23 and syntaxin 6 in human neutrophils were confirmed by immunofluorescence confocal laser scanning microscopy. Immunofluorescence images showed a major localization for SNAP-23 in the cytoplasmic granules of resting neutrophils with a secondary location at the cell surface (Figure4A). However, SNAP-23 was detected mainly at the cell surface of activated neutrophils by confocal microscopy (Figure 4A). By using the 3D10 MoAb for syntaxin 6 localization, we found the most prominent fluorescent labeling throughout the cell surface in both resting and activated human neutrophils (Figure 4B). Thus, SNAP-23 was located mainly in the cytoplasmic granule membranes and translocated to the plasma membrane on cell activation, whereas syntaxin 6 was associated with the plasma membrane in both resting and activated neutrophils.
Localization of SNAP-23 and syntaxin 6 in resting and activated human neutrophils (PMNs) by confocal laser scanning microscopy.
Resting and PMA-activated cells were permeabilized and incubated with anti-SNAP-23 or anti–syntaxin 6 antibodies as described in “Materials and methods.” Bar, 4 μm.
Localization of SNAP-23 and syntaxin 6 in resting and activated human neutrophils (PMNs) by confocal laser scanning microscopy.
Resting and PMA-activated cells were permeabilized and incubated with anti-SNAP-23 or anti–syntaxin 6 antibodies as described in “Materials and methods.” Bar, 4 μm.
Colocalization of syntaxin 6 and SNAP-23 at the cell surface of activated human neutrophils
We next examined whether these 2 SNARE proteins colocalized on human neutrophil activation by double staining and confocal microscopy. Resting cells showed a major localization of syntaxin 6 at the plasma membrane, whereas SNAP-23 was located mainly in cytoplasmic granules (Figure 5). On neutrophil activation with PMA, SNAP-23 was translocated to the cell surface, and we found a significant colocalization of both syntaxin 6 and SNAP-23 at the cell surface (Figure 5).
Colocalization of SNAP-23 and syntaxin 6 in activated human neutrophils (PMNs) by confocal laser scanning microscopy.
Resting and PMA-activated human neutrophils were permeabilized and processed for double-labeled immunofluorescence with anti–SNAP-23 and anti–syntaxin 6 antibodies. The immuno-decorated areas were detected with the appropriate secondary antibodies coupled to phycoerythrin (red fluorescence for anti-SNAP-23) or FITC (green fluorescence for anti–syntaxin 6) by using confocal laser scanning microscopy, as described in “Materials and methods.” Overlay of the adjacent panels (right panels) shows colocalization of SNAP-23 and syntaxin 6 at the cell surface of activated human neutrophils. Areas of colocalization are yellow. Bar, 3 μm.
Colocalization of SNAP-23 and syntaxin 6 in activated human neutrophils (PMNs) by confocal laser scanning microscopy.
Resting and PMA-activated human neutrophils were permeabilized and processed for double-labeled immunofluorescence with anti–SNAP-23 and anti–syntaxin 6 antibodies. The immuno-decorated areas were detected with the appropriate secondary antibodies coupled to phycoerythrin (red fluorescence for anti-SNAP-23) or FITC (green fluorescence for anti–syntaxin 6) by using confocal laser scanning microscopy, as described in “Materials and methods.” Overlay of the adjacent panels (right panels) shows colocalization of SNAP-23 and syntaxin 6 at the cell surface of activated human neutrophils. Areas of colocalization are yellow. Bar, 3 μm.
In vitro interaction between syntaxin 6 and SNAP-23
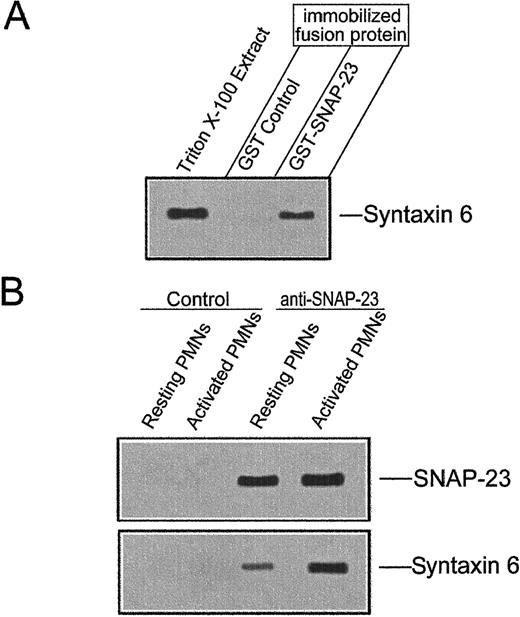
Previous reports have shown that SNAP-23 is able to bind to syntaxins 1, 2, 3, and 428 in vitro and to interact with syntaxin 4 in vivo.39 40 Because syntaxin 6 colocalized with SNAP-23 in activated neutrophils, we examined whether recombinant SNAP-23 could interact with syntaxin 6. Recombinant GST-SNAP-23 and GST immobilized on glutathione-Sepharose 4B beads were incubated with Triton X-100–solubilized neutrophil extract. Proteins specifically bound to the matrix were then eluted with glutathione and analyzed by SDS-12% polyacrylamide gel electrophoresis and Western blotting, using the 3D10 anti–syntaxin 6 MoAb. Syntaxin 6 bound to GST–SNAP-23 beads but not to beads coupled to the control protein GST (Figure6A).
Interaction between SNAP-23 and syntaxin 6.
(A) In vitro binding of syntaxin 6 to GST-SNAP-23. Unfused GST and full-length GST–SNAP-23 fusion protein were immobilized on glutathione-Sepharose beads and incubated with Triton X-100–extracts of human neutrophils. Then, bound proteins were eluted, analyzed by SDS-polyacrylamide gel electrophoresis, and blotted with 3D10 anti–syntaxin 6 MoAb. Triton X-100 extract of human neutrophils was used as a positive control. The position of syntaxin 6 is indicated. The experiment shown is representative of 3 performed. (B) Coimmunoprecipitation of SNAP-23 and syntaxin 6 from resting and activated human neutrophils. SNAP-23 was immunoprecipitated from detergent-solubilized whole cell extracts of resting and PMA-activated human neutrophils. Immunoprecipitates were electrophoresed in SDS-polyacrylamide gels and immunoblotted with the anti–SNAP-23 or anti–syntaxin 6 antibodies. Lysates were also immunoprecipitated with P3X63 myeloma culture supernatant as a negative control (Control). The positions of SNAP-23 and syntaxin 6 are indicated. Experiments shown are representative of 3 performed.
Interaction between SNAP-23 and syntaxin 6.
(A) In vitro binding of syntaxin 6 to GST-SNAP-23. Unfused GST and full-length GST–SNAP-23 fusion protein were immobilized on glutathione-Sepharose beads and incubated with Triton X-100–extracts of human neutrophils. Then, bound proteins were eluted, analyzed by SDS-polyacrylamide gel electrophoresis, and blotted with 3D10 anti–syntaxin 6 MoAb. Triton X-100 extract of human neutrophils was used as a positive control. The position of syntaxin 6 is indicated. The experiment shown is representative of 3 performed. (B) Coimmunoprecipitation of SNAP-23 and syntaxin 6 from resting and activated human neutrophils. SNAP-23 was immunoprecipitated from detergent-solubilized whole cell extracts of resting and PMA-activated human neutrophils. Immunoprecipitates were electrophoresed in SDS-polyacrylamide gels and immunoblotted with the anti–SNAP-23 or anti–syntaxin 6 antibodies. Lysates were also immunoprecipitated with P3X63 myeloma culture supernatant as a negative control (Control). The positions of SNAP-23 and syntaxin 6 are indicated. Experiments shown are representative of 3 performed.
In vivo interaction between syntaxin 6 and SNAP-23
We next investigated whether the interaction between syntaxin 6 and SNAP-23 occurred also in vivo by coimmunoprecipitation experiments. SNAP-23 was immunoprecipitated from resting and activated neutrophils, and after SDS-12% polyacrylamide gel electrophoresis and transfer to nitrocellulose filters, the SNAP-23 immunoprecipitates were probed for SNAP-23 and syntaxin 6 (Figure 6B). SNAP-23 and syntaxin 6 coimmunoprecipitated in both resting and activated human neutrophils, but this in vivo interaction between both SNARE proteins was highly increased on neutrophil activation (Figure 6B).
Incorporation of antibodies into electropermeabilized neutrophils
The translocation of SNAP-23 from mobilizable cytoplasmic granules to the cell surface during neutrophil activation, together with the colocalization and in vivo interaction of both syntaxin 6 and SNAP-23 in the cell surface of activated neutrophils, prompted us to investigate whether these SNARE proteins could play a role in neutrophil exocytosis. Electropermeabilized neutrophils were used to permit access of specific antibodies into the neutrophil cytoplasm in whole and functional neutrophils to study the putative functional role of SNAP-23 and syntaxin 6 in neutrophil exocytosis. The efficiency of the permeabilization procedure was established by measuring the incorporation of propidium iodide and the FITC-conjugated anti-CD3 MoAb into the human neutrophils. Propidium iodide binds to DNA and RNA but is unable to stain intact cells. Anti-CD3 MoAb recognizes the CD3 lymphocyte marker but does not react with any antigen in human neutrophils. Thus, the analysis of the incorporation of anti-CD3 MoAb was used as a marker to examine the efficacy and capacity of the electropermeabilization procedure to permit molecules of great size, including antibodies, into the cytoplasm. Control and permeabilized cells were exposed to propidium iodide and FITC-conjugated anti-CD3 MoAb and analyzed by using flow cytometry. Two discharges of 5 kV/cm were found to render the membrane of more than 95% of the cells permeable to the probes, as illustrated in Figure7. Less than 1% of the intact cells were stained by propidium iodide (molecular mass, 668) or by FITC-conjugated anti-CD3 MoAb (molecular mass greater than 150 000), whereas most of the cells were stained by these fluorescent probes following exposure to the electric field (Figure 7), indicating that electroporated neutrophils were rendered permeable to these molecules. Electropermeabilized cells preserved the fluorescent agents into the cytoplasm after 15 minutes incubation (Figure 7). Activation of electropermeabilized neutrophils with Ca++ and GTP-γ-S did not affect the incorporation of both propidium iodide or anti-CD3 MoAb (Figure 7).
Incorporation of propidium iodide (PI) and FITC-conjugated anti-CD3 MoAb into electropermeabilized resting and activated human neutrophils.
Intact (Control) and electropermeabilized (Elect.) neutrophils were incubated with 10 μg/mL PI or 24 μg/mL FITC-conjugated anti-CD3 MoAb for 3 minutes at room temperature. The electropermeabilized cells were preincubated with 5 μg/mL cytochalasin B for 5 minutes at 37°C and subsequently incubated for 10 minutes at 37°C in the absence (Elect. [15 minutes]) or in the presence (Elect. [Act.]) of 1 μmol/L Ca++ and 50 μmol/L GTP-γ-S. Then, intact and electropermeabilized neutrophils were analyzed for anti-CD3 MoAb and PI incorporation by flow cytometry as described in “Materials and methods.” The results shown are representative of 3 independent determinations.
Incorporation of propidium iodide (PI) and FITC-conjugated anti-CD3 MoAb into electropermeabilized resting and activated human neutrophils.
Intact (Control) and electropermeabilized (Elect.) neutrophils were incubated with 10 μg/mL PI or 24 μg/mL FITC-conjugated anti-CD3 MoAb for 3 minutes at room temperature. The electropermeabilized cells were preincubated with 5 μg/mL cytochalasin B for 5 minutes at 37°C and subsequently incubated for 10 minutes at 37°C in the absence (Elect. [15 minutes]) or in the presence (Elect. [Act.]) of 1 μmol/L Ca++ and 50 μmol/L GTP-γ-S. Then, intact and electropermeabilized neutrophils were analyzed for anti-CD3 MoAb and PI incorporation by flow cytometry as described in “Materials and methods.” The results shown are representative of 3 independent determinations.
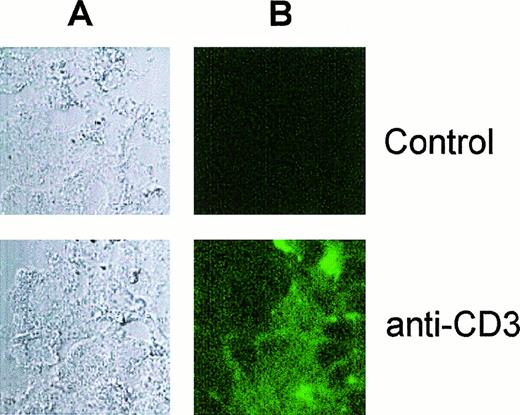
To further demonstrate the incorporation of antibodies into electroporated neutrophils, we carried out confocal microscopy analysis and found that neutrophils were loaded with FITC-conjugated anti-CD3 MoAb following the electroporation procedure described above (Figure8).
Visualization of electropermeabilized neutrophils loaded with antibody by confocal laser scanning microscopy.
Electropermeabilized neutrophils were incubated in the absence (Control) or in the presence of FITC-conjugated anti-CD3 MoAb (anti-CD3) as described in Figure 7 and analyzed by confocal microscopy. The corresponding light transmission (A) and fluorescence (B) images are shown.
Visualization of electropermeabilized neutrophils loaded with antibody by confocal laser scanning microscopy.
Electropermeabilized neutrophils were incubated in the absence (Control) or in the presence of FITC-conjugated anti-CD3 MoAb (anti-CD3) as described in Figure 7 and analyzed by confocal microscopy. The corresponding light transmission (A) and fluorescence (B) images are shown.
Effect of antibodies directed against SNAP-23 and syntaxin 6 on granule exocytosis in electropermeabilized human neutrophils
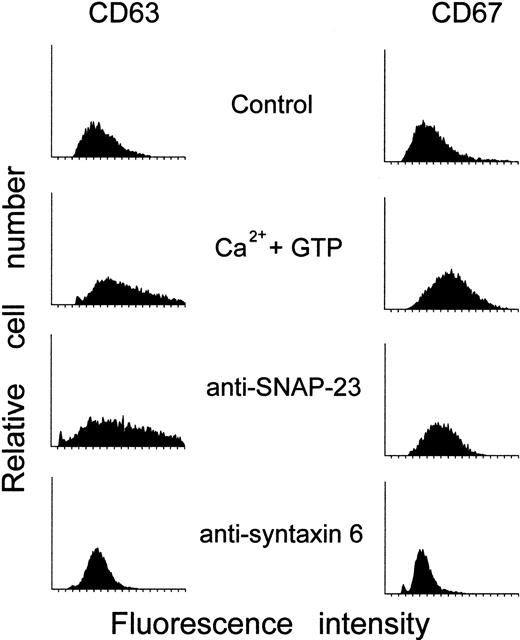
We next analyzed whether SNAP-23 and syntaxin 6 serve a functional role in neutrophil exocytosis. We found that electropermeabilized neutrophils were able to undergo exocytosis of specific and azurophilic granules on cell activation with Ca++ and GTP-γ-S (Figure9), corroborating previous reports.34 Degranulation was studied by measuring up-regulation of the granule membrane markers CD63 and CD67 in electropermeabilized neutrophils (Figure 9). This method has been shown to measure efficiently neutrophil degranulation in electropermeabilized neutrophils avoiding centrifugation,34 which could potentially destabilize the cells. Following paraformaldehyde fixation of electropermeabilized neutrophils, we monitored the expression of both CD63 and CD67 only at the cell surface, allowing us to determine neutrophil degranulation. Immunoelectron microscopy studies have shown that CD63 is present exclusively in the azurophilic granules,41 whereas CD67 is located in the specific granules42 of resting neutrophils. Thus, up-regulation of CD63 and CD67 parallels the secretion of azurophilic and specific granules, respectively, as has been previously reported in both intact and electropermeabilized neutrophils.34 Incubation of electropermeabilized neutrophils with anti–SNAP-23 polyclonal antibodies inhibited CD67 up-regulation in a dose-dependent manner (Figures 9 and 10) but had no effect on CD63 up-regulation, after cell activation with Ca++ and GTP-γ-S. Incubation of electropermeabilized neutrophils with anti–syntaxin 6 MoAb blocked both CD63 and CD67 up-regulation following cell activation with Ca++ and GTP-γ-S. (Figures9 and 10). In contrast, rabbit nonimmune immunoglobulins or an irrelevant mouse MoAb, such as anti-CD20 MoAb, had no effect on neutrophil degranulation (Figure 10). The present findings indicate that anti–SNAP-23 antibodies inhibit secretion of specific granules without affecting secretion of azurophilic granules, whereas anti–syntaxin 6 MoAb prevents secretion of both types of cytoplasmic granules in activated human neutrophils.
Effect of anti–SNAP-23 and anti–syntaxin 6 antibodies on the cell surface expression of CD63 and CD67 in activated electropermeabilized human neutrophils.
Electropermeabilized cells were incubated with 8 μg/mL anti–SNAP-23 or anti–syntaxin 6 for 3 minutes at room temperature, then were preincubated with 5 μg/mL cytochalasin B for 5 minutes at 37°C, and activated with 1 μmol/L Ca++ and 50 μmol/L GTP-γ-S for 10 minutes at 37°C. The cells were then fixed with 1% paraformaldehyde and assayed for CD63 and CD67 antigen expression as described in “Materials and methods.” Untreated electropermeabilized cells (Control) and electropermeabilized cells stimulated with 1 μmol/L Ca++ and 50 μmol/L GTP-γ-S for 10 minutes at 37°C (Ca++ and GTP) in the absence of anti–SNAP-23 or anti–syntaxin 6 antibodies were run in parallel. The results shown are representative of 3 independent determinations.
Effect of anti–SNAP-23 and anti–syntaxin 6 antibodies on the cell surface expression of CD63 and CD67 in activated electropermeabilized human neutrophils.
Electropermeabilized cells were incubated with 8 μg/mL anti–SNAP-23 or anti–syntaxin 6 for 3 minutes at room temperature, then were preincubated with 5 μg/mL cytochalasin B for 5 minutes at 37°C, and activated with 1 μmol/L Ca++ and 50 μmol/L GTP-γ-S for 10 minutes at 37°C. The cells were then fixed with 1% paraformaldehyde and assayed for CD63 and CD67 antigen expression as described in “Materials and methods.” Untreated electropermeabilized cells (Control) and electropermeabilized cells stimulated with 1 μmol/L Ca++ and 50 μmol/L GTP-γ-S for 10 minutes at 37°C (Ca++ and GTP) in the absence of anti–SNAP-23 or anti–syntaxin 6 antibodies were run in parallel. The results shown are representative of 3 independent determinations.
Effect of anti–SNAP-23 and anti–syntaxin 6 antibodies on the up-regulation of CD63 and CD67 cell surface expression in activated electropermeabilized human neutrophils.
Electropermeabilized human neutrophils (PMNs) were incubated for 3 minutes at room temperature in the absence (Control) or in the presence of CD20 MoAb (2 μg/ml) or in the presence of increasing concentrations (8, 16, and 20 μg/mL) of anti–SNAP-23 and anti–syntaxin 6 antibodies. Cells were then incubated with 5 μg/mL cytochalasin B for 5 minutes at 37°C and activated with 1 μmol/L Ca++ and 50 μmol/L GTP-γ-S for 10 minutes at 37°C. Subsequently, cells were fixed with 1% paraformaldehyde for 2 minutes at room temperature and treated as described in “Materials and methods” for CD63 and CD67 cell surface expression. Data are expressed as percentage of the increase of CD63 and CD67 cell surface expression on electropermeabilized PMN activation compared with regard to the up-regulation of CD63 and CD67 detected in control Ca++ and GTP-γ-S–stimulated electropermeabilized neutrophils in the absence of any antibody (Control), considered as 100% of increase in cell surface antigen expression. Mean values ± SD of 3 independent determinations are shown.
Effect of anti–SNAP-23 and anti–syntaxin 6 antibodies on the up-regulation of CD63 and CD67 cell surface expression in activated electropermeabilized human neutrophils.
Electropermeabilized human neutrophils (PMNs) were incubated for 3 minutes at room temperature in the absence (Control) or in the presence of CD20 MoAb (2 μg/ml) or in the presence of increasing concentrations (8, 16, and 20 μg/mL) of anti–SNAP-23 and anti–syntaxin 6 antibodies. Cells were then incubated with 5 μg/mL cytochalasin B for 5 minutes at 37°C and activated with 1 μmol/L Ca++ and 50 μmol/L GTP-γ-S for 10 minutes at 37°C. Subsequently, cells were fixed with 1% paraformaldehyde for 2 minutes at room temperature and treated as described in “Materials and methods” for CD63 and CD67 cell surface expression. Data are expressed as percentage of the increase of CD63 and CD67 cell surface expression on electropermeabilized PMN activation compared with regard to the up-regulation of CD63 and CD67 detected in control Ca++ and GTP-γ-S–stimulated electropermeabilized neutrophils in the absence of any antibody (Control), considered as 100% of increase in cell surface antigen expression. Mean values ± SD of 3 independent determinations are shown.
Discussion
The data reported here show the involvement of syntaxin 6 in granule secretion in human neutrophils through its interaction with SNAP-23, demonstrating for the first time the functional involvement of SNARE proteins in neutrophil exocytosis. The SNARE hypothesis predicts that vesicle docking with target membranes requires the interaction of v- and t-SNAREs, which provide specificity to the fusion process.19 We report here the cloning and characterization of human syntaxin 6 that acts as a t-SNARE in the docking and fusion of neutrophil granules. Our data indicate that the primary structure of human syntaxin 6 has a carboxyl-terminal transmembrane region and two regions with a high probability to form coiled coils. One coiled-coil region, comprising residues 174-227, shows a perfect leucine-zipper containing 5 heptad repeats of leucine positioned just before the transmembrane anchor (Figure 1). The presence of a leucine-zipper and an additional leucine-zipper–like layer in syntaxin 6 suggests that this SNARE is able to form strong interactions with other leucine-zipper–like containing proteins. Because of the high homology between syntaxins 6 and 10, it could be envisaged that both proteins play a similar functional role. Syntaxins 6 and 10 have been previously reported to be located mainly in the trans-Golgi network of several cell types, leading to the suggestion that these proteins mediate a trans-Golgi trafficking event, perhaps targeting to endosomes in mammalian cells.29,36 However, we report here a major location of syntaxin 6 in the plasma membrane of human neutrophils as assessed by subcellular fractionation and immunofluorescence confocal microscopy assays. This disagreement may be partially explained by the fact that neutrophils are unique, terminal-differentiated cells, with a low capacity for macromolecule biosynthesis, and where regulated degranulation of individual cytoplasmic granules plays a crucial role. In addition, human neutrophils reduce considerably the Golgi region during their development1 and show a rather low constitutive secretion. In this regard, we have previously reported that human neutrophils do not express syntaxin 10.26
The antibodies against syntaxin 6 and SNAP-23 recognized bands at apparent molecular masses of about 32 kd and 29 kd, respectively, in human neutrophils estimated by SDS-polyacrylamide gel electrophoresis and immunoblotting. These molecular masses are higher than the predicted molecular masses based on the open-reading frames of the respective cloned cDNAs (29.2 kd for human syntaxin 6 and 23.3 kd for human SNAP-23) and could be in part due to posttranslational modifications of the proteins that would alter both the size and shape of the proteins. In this context, SNAP-23 has been shown to be palmitoylated in vivo.43
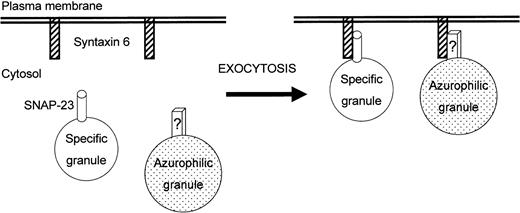
Figure 11 depicts a model for the involvement of SNARE proteins in human neutrophil exocytosis, based on the present results. The fact that anti–syntaxin 6 MoAb blocks almost completely neutrophil degranulation suggests that this protein plays a key role in docking and subsequent fusion of the distinct intracellular granules present in human neutrophils. The predominant plasma membrane localization of syntaxin 6 suggests that this protein serves as a t-SNARE in neutrophil secretion. The translocation of SNAP-23 from mobilizable cytoplasmic granules to the cell surface and its interaction with syntaxin 6, together with the functional assays on electropermeabilized neutrophils, lead to the suggestion that SNAP-23 acts mainly as a v-SNARE in human neutrophils, modulating degranulation of specific granules. However, SNAP-23 does not participate in the exocytosis of azurophilic granules. This finding supports the notion that different neutrophil cytoplasmic granules with distinct exocytic capabilities may contain different subsets of SNARE proteins that could direct their secretion independently.
Model for the involvement of syntaxin 6 and SNAP-23 in neutrophil exocytosis.
This schematic diagram is designed to portray a currently plausible mechanism for the involvement of syntaxin 6 and SNAP-23 in the human neutrophil exocytosis. Interaction of both proteins, acting as t- and v-SNAREs, provides specificity to the fusion process. Exocytosis of specific granules is mediated by interaction of syntaxin 6 with SNAP-23, whereas exocytosis of azurophilic granules would involve interaction of syntaxin 6 with a still unknown structure (denoted as “?”). See text for further details.
Model for the involvement of syntaxin 6 and SNAP-23 in neutrophil exocytosis.
This schematic diagram is designed to portray a currently plausible mechanism for the involvement of syntaxin 6 and SNAP-23 in the human neutrophil exocytosis. Interaction of both proteins, acting as t- and v-SNAREs, provides specificity to the fusion process. Exocytosis of specific granules is mediated by interaction of syntaxin 6 with SNAP-23, whereas exocytosis of azurophilic granules would involve interaction of syntaxin 6 with a still unknown structure (denoted as “?”). See text for further details.
Here we report the in vitro and in vivo binding of syntaxin 6 with SNAP-23. This interaction is dramatically increased during neutrophil activation. SNAP-23 shows a 59% amino acid identity with SNAP-25, with the highest identity at the amino- and carboxyl-terminal parts of the molecules,24,28 but, unlike SNAP-25, it is not cleaved by botulinum neurotoxins A or E.44-46 SNAP-25 and SNAP-23 are not integral membrane proteins and associate with the membrane by attachment of palmitate groups to cysteine residues present in a “palmitoylation domain.” SNAP-25 and SNAP-23 contain 2 similar palmitoylation domains with 4 and 5 cysteine residues, respectively, and these cysteine clusters have been shown to be required for SNAP-25 and SNAP-23 palmitoylation in vivo.43 The binding domain of SNAP-25 to syntaxin 1 encompasses most of the amino-terminal half of SNAP-25, including its palmitoylation sites.47 This syntaxin 1–binding domain is preserved in SNAP-23 and includes a leucine-zipper–like domain with a very high probability to form a coiled coil (residues 45-79 in SNAP-23 amino acid sequence) that could be putatively involved in their interaction with syntaxin 6.
SNAP-23 as well as its homologue SNAP-25 were initially reported to be located exclusively at the plasma membrane of different cells leading to the notion that these proteins act as t-SNAREs. However, recent reports also indicate that SNAP-23 can be detected in intracellular vesicles in rat kidney cells37 and in endosomal compartments in HepG2 and HT4 cells.44 Furthermore, the SNAP-23 homologue, SNAP-25, has also been detected in neutrophil cytoplasmic granules,25 synaptic vesicles,48-50 and chromaffin granules.51,52Here, we report evidence for the major localization of SNAP-23 in intracellular granules (specific and tertiary granules) of resting human neutrophils that are readily mobilized to the plasma membrane on cell activation. Because tertiary granules fuse readily with the cell surface under very mild experimental conditions,8,31 we can suggest that part of the SNAP-23 localized in the plasma membrane of resting cells may be due to fusion of tertiary granules with the cell surface during neutrophil isolation. The experiments described here suggest that SNAP-23 may function as a v-SNARE in human neutrophils, mediating fusion of the specific granules with the cell surface. The involvement of SNAP-23 in neutrophil exocytosis is in agreement with reports showing that SNAP-23 can function in regulated exocytosis in insulin secretion45 and in platelet α-granule release.53 In this regard, SNAP-25 has been also involved in exocytosis in neurons and endocrine cells.45,54 Our data indicate that anti–SNAP-23 antibodies inhibit exocytosis of CD67-containing granules. CD67 has been previously detected in specific granules.42 However, because specific and tertiary granules share a number of constituents, and because SNAP-23 seems to be translocated to the cell surface from both specific and tertiary granules on cell activation, a putative role for SNAP-23 in the secretion of both granules in human neutrophils is suggested. In this regard, the data herein reported indicate a high correlation between SNAP-23 translocation from the cytoplasmic granules to the plasma membrane and secretion of gelatinase-rich tertiary granules.
The data herein reported suggest that different combinations of SNARE proteins regulate exocytosis of the distinct cytoplasmic granules in human neutrophils. Because exocytosis plays a key role in neutrophil physiology, the understanding of the molecular mechanisms of neutrophil secretion is of major pharmacological interest and raises the possibility that proteins mediating neutrophil exocytosis may serve as appropriate targets for anti-inflammatory agents.
Acknowledgments
We are grateful to Drs E. Barbosa and S. Callejo for their help in confocal microscopy experiments.
Supported by grant PB95-0713 from the Dirección General de Investigación Cientı́fica y Técnica (DGICYT), grant FIS98/0313 from the Fondo de Investigación Sanitaria, and grant 08.1/0004/98 from the Comunidad de Madrid.
B.M.-M. and S.M.N. are recipients of fellowships from the Ministerio de Educación y Cultura of Spain.
The nucleotide sequence data for human syntaxin 6 reported in this work has the GenBank/EMBL accession number AJ002078.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Faustino Mollinedo, Centro de Investigación del Cáncer, Instituto de Biologı́a Molecular y Celular del Cáncer, CSIC-Universidad de Salamanca, Campus Miguel de Unamuno, E-37007 Salamanca, Spain; e-mail:fmollin@usal.es.




![Fig. 7. Incorporation of propidium iodide (PI) and FITC-conjugated anti-CD3 MoAb into electropermeabilized resting and activated human neutrophils. / Intact (Control) and electropermeabilized (Elect.) neutrophils were incubated with 10 μg/mL PI or 24 μg/mL FITC-conjugated anti-CD3 MoAb for 3 minutes at room temperature. The electropermeabilized cells were preincubated with 5 μg/mL cytochalasin B for 5 minutes at 37°C and subsequently incubated for 10 minutes at 37°C in the absence (Elect. [15 minutes]) or in the presence (Elect. [Act.]) of 1 μmol/L Ca++ and 50 μmol/L GTP-γ-S. Then, intact and electropermeabilized neutrophils were analyzed for anti-CD3 MoAb and PI incorporation by flow cytometry as described in “Materials and methods.” The results shown are representative of 3 independent determinations.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/96/7/10.1182_blood.v96.7.2574/5/m_h81900189007.jpeg?Expires=1769114528&Signature=QJ-oNNmEmU0QlydzR0ujK~hP9PEJjS9hPG0I65HnW-Xb6hyMtqNaO8uFlXhHtBMPOfIciJXiWNXwrCDmukHgOo7vP8Z2FSSgbC3UVoxJB7-hCZ5VjjrQA-mIvHSMmLzbyYZ3H6JECm~zNYfGaeceSbdBfx9NewqL-xLEw952SBoZavREQuzjicdLRW9UWrxTTRrKfkFwZ7rWGr6HhTSa3bPUsxK3rxvyS71jt2uwGUcGxpVShnm4xZiF8-QMFxamjwEzjG5212bVoTvrUiNJtaFqaEcOLhSZU7KpfrUYgJK8ZQNbX6f9aYUSqdVjW3fQ0OYf8NXv5kHhPbuJAVeffg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal